ANALYSIS ON GENETIC DIVERSITY OF AA-GENOME ORYZA SPECIES BY ISSR MARKERS
DUAN Shi-hua, ZHENG Zhuo, LUO Qiang, LONG Wei-xiong,LIAO Fo-cai
ANALYSIS ON GENETIC DIVERSITY OF AA-GENOME ORYZA SPECIES BY ISSR MARKERS
*DUAN Shi-hua1,2, ZHENG Zhuo1, LUO Qiang1, LONG Wei-xiong1,LIAO Fo-cai1
(1. School of Life Science, Jinggangshan University, Ji’an, Jiangxi 343009, China;2. Institute of Eco-Environment and Resources, Jinggangshan University, Ji’an, Jiangxi 343009, China)
In order to determine genetic diversity of the AA-genomespecies (Poaceae), inter-simple sequence repeat (ISSR) markers from a total of 62 rice accessions collected worldwide were analyzed. These accessions encompassed six wild (.,.,..,., and) and two cultivated (.and.) species. 21 selected ISSR primers that produced consistent and repeatable banding patterns revealed significant polymorphisms among the 62 rice accessions with an overall gene diversity () of 0.527, indicating the power of ISSR markers in studying genetic diversity ingermplasm. The consensus tree constructed on the basis of the pairwise Jaccard similarity coefficients of the ISSR banding pattern revealed an evident genetic variation relationships of the AA-genomespecies with high bootstrap value supports. It is concluded from this study that thespecies from different continents possessed close linkages and current classification of the AA-genomespecies suggested by Vaughan (1989) remains valid, particularly in relation to that of the Asian wild rice. The knowledge will be useful forthe effective utilization of AA-genome wildspecies in rice breeding programs.
genetic diversity; ISSR;;AA-genome
The genusL. (Poaceae)comprises approximately 24 taxa, including two cultivated and 22 wild species[1]. Species in the genus are of a great value in human’s daily life and for rice breeding. Asian cultivated rice (L.) had its origin in Asia and now cultivated worldwide. As a world important cereal crop, Asian cultivated rice provides staple foods for nearly one half of the global population. African cultivated rice (.Steud.) was domesticated in West Africa and only grown in a limited farming system in Africa[2]. The wildspecies, containing the AA, BB, CC, EE, FF, and GG genomes for diploids or combination of the BBCC, CCDD, HHJJ and HHKK genomes for tetraploids, are widely distributed in pan-tropics of the world[1, 3].Among these, wild specieswith the AAgenome are the most importantand accessible genetic resources for rice breeding, because the cultivated rice,.and., also shares the same AAgenome. As a consequence, the transferring of alien genes from these close wild relatives to cultivated ricecan easily be achieved through sexual hybridization in breeding programs. A few successful examples of using the AA-genome wild relatives in rice breeding have been reported, such as the introduction into cultivated rice of a male sterility (MS) gene from.for hybrid rice production[4]and a gene resistant to grassy stunt virus from.for virus resistant[5]. In addition, a large number of useful traits that can substantially increase the yield of rice, or resist to insect and disease pests have been identified in the AA-genome wild rice[6]. Undoubtedly, the more efficient utilization of the close wild relatives of rice will facilitate the effective rice breeding, in addition to strategic rice germplasm conservation. All these are essentially rely on the better understanding of genetic relationships of theAA-genomespecies, including the cultivated rice species[2].
Despite the importance in rice breeding and genetic diversity studies, the genetic relationships of the AA-genome species are particularly problematic due to considerable morphological overlapping among species and variation within species[7]. In addition, genetic relationships of these species have not been fully resolved, which results in a considerable inconsistency and even contradiction in interpreting the species relationships. A great effort has been made to investigate relationships between the cultivated riceand its closely relatedspecies through analyses of morphological characteristics[8], isozyme polymorphisms[9], and variation in different types of molecular markers[10-14]. Certainly, data generated from these studies have provided solid data for illustrating the morphological variation pattern and genetic relationships of the AA-genome species. However, some points regarding the AA-genomespecies still remain incongruous.
Inter-simple sequence repeats (ISSR) are typical dominant molecular markers that have been extensively used to detect genetic variation among plants at different scales—from the major lineages of terrestrial plants to closely related taxa[15]. The ISSR technique is widely applied because it is rapid, inexpensive, and simple to perform, in addition to the ability to perform analyses without the need for prior DNA sequence data and requirement of very little starting DNA template[16]. Of the ISSR method, it has been reported to produce a more complex marker pattern, which is advantageous when differentiating closely related taxa[17]. Furthermore, ISSR markers possess excellent reproducibility, because they are designed to anneal to a microsatellite sequence with longer primers and allowed higher annealing temperatures to be used.
Variation patterns of ISSRs from a large set of samples of the AA-genomespecies were analyzed in this study. The objectives of the analysis were to evaluate genetic diversity within and among the of the AA-genomespecies. Result from such an analysiswill provideuseful information forthe genetic relationships of AA-genome wildspecies and effective utilization of the beneficial genes harbored by the close wildrelatives for the genetic improvement of cultivated rice.
1 Materials and methods
1.1 Plant materials
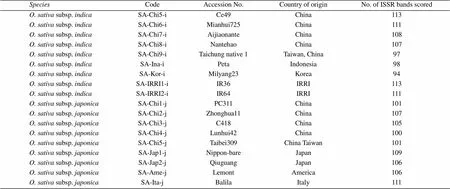
Sixty-twoaccessions representing 22 Asian cultivated rice (13.subsp.and nine.subsp.) and its close relative species containing the AA genomes were included in this study for ISSR analysis(Table 1). All the accessions of Asian cultivated rice used were provided by the Laboratory of Plant Developmental Genetic, College of Life Sciences, Wuhan University, China. Other wild accessions were donated by the International Rice Genebank (IRGB) of the International Rice Research Institute (IRRI) in Los Baños, the Philippines.
DNA samples were isolated from 3-5 seedlings of germinated seeds from each accession following the method of Dellaporta et al. (1983)[18]. Quality and quantity of isolated DNA were estimated by a spectrophotometer using the standard amount of lambda DNA (MBI, Fermentas, Hanover, MD, USA) after the DNA samples were separated on agrose gels, and visually by ethidium bromide staining on the agrose gels.
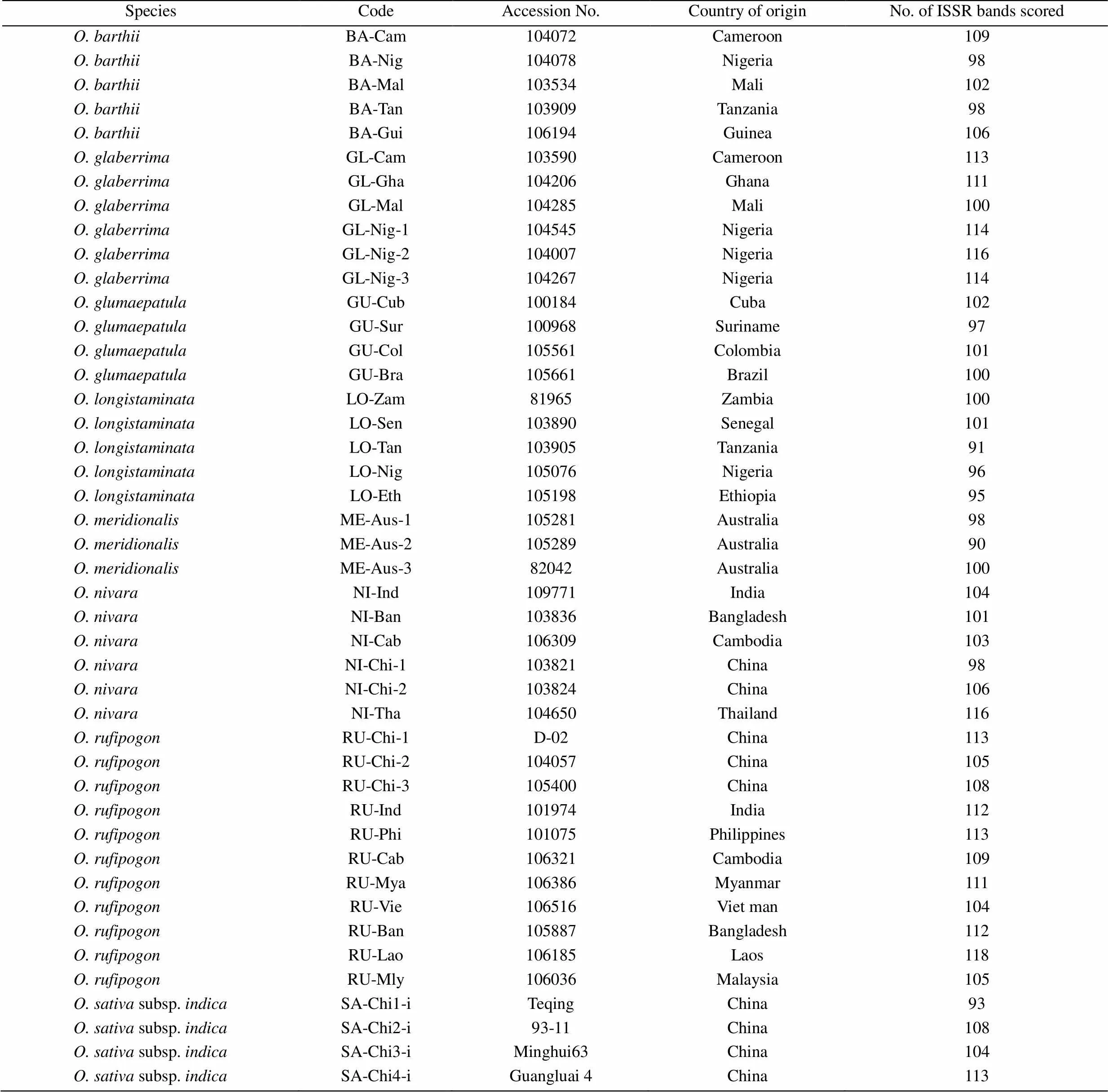
Table 1 The AA-genome Oryza species used for ISSR analysis with their code, accession number, country of origins, and number of amplified bands by 21 selected ISSR primers
1.2 PCR amplification and electrophoresis
A total of 42 ISSR primers were screened for PCR amplification. Of these, 21 primers[19]were selected for this study because of their ideal repeatability and polymorphisms among the representative rice varieties, IR36, IR64 and Nipponbare, used for mark screening. All primers were synthesized by SBS Biotech. Co. Ltd., Shanghai. Amplification of DNA was performed in 10 mM of Tris-HCl pH 9.0, 50 mM of KCl, 1.5 mM of MgCl2, 0.25 mM of dNTPs, 2% formamide, 0.2 µM of primer, 0.5 mM of spermidine, 0.8 U of Taq DNA polymerase (MBI, Fermentas, Hanover, MD, USA) and 20 ng of DNA per 20-µL reaction using a Double Engine thermocycler (MJ Research, USA) for 40 cycles. After initial denaturation for 5-min at 94 °C, each cycle comprised 1-min denaturation at 94 °C, 1-min annealing at 49 °C, 1.5-min extension at 72 °C with a final extension for 10-min at 72 °C at the end of 40 cycles. The annealing temperature was usually adjusted according to themof the primer being used in the reaction. A negative control which contained all PCR components except rice genomic DNA (replaced by water) was included in every experiment to test for DNA contamination of the reagents.
Amplified products were mixed with bromophenol blue gel-loading dye and analyzed by electrophoresis on a 2% agarose gel using 1×Tris Acetate EDTA (TAE) buffer pH 8.0 at room temperature. Gels were stained with ethidium bromide and bands were visualized and photographed under UV light by Gel DOC 2000 (BIO-RAD Laboratories-Segrate, Milan, Italy). Molecular weights were estimated using DNA markers DL2000 (TaKaRa, Biotech. Co., Ltd., Dalian, China). All the patterns generated were repeated at least three times in order to obtain reproducible data.
1.3 Analysis of ISSR data and construction of a dendrogram
DNA fragments (bands) from the AA-genomesamples generated by the elected ISSR primers was coded as a unit character and scored as a binary form, i.e. 1 for presence and 0 for absence, respectively. Only those fragments that were reproducible and about 0.5mm apart on the gels were considered for scoring. The number of ISSR fragments, the percentage of polymorphic loci, and Nei’s unbiased genetic diversity [= (/-1)(1-∑P2)] for each species (subspecies) were calculated manually[20]. Cluster analysis was performed to infer genetic relationships among the AA-genomeindividuals on the basis of the ISSR data. The Jaccard (1908) coefficient was calculated from the binary data matrix for all pairwise combinations of accessions using NTSYS-PC 2.02[21-22]. The unweighted pair-group method for the arithmetic average (UPGMA) was used to construct a dendrogram on the basis of the similarity matrix data. By using this program, the dendrogram was reconstructed 1,000 times by repeated sampling with replacement, and the frequency with which a particular grouping was identified was taken to reflect the strength of the clustering.
2 Results
2.1 Genetic diversity between and within the Oryza species
Based on the data matrix generated from the 281 fragments amplified by the 21 selected primers among the 62 accessions of the AA-genomespecies, genetic diversity parameters were calculated. The percentage of polymorphic fragments () varied from 30.48%~66.92% among the different species or subspecies, with an overall value of 99.90% for all the species (Table 2). The overall gene diversity () for the AA-genomespecies was 0.527. Thevalues showed a considerable variation among different species, with the highest values in.(0.278) and.(0.235), and the lowest in.(0.058) and.(0.108) (Table 2). Since the unbiasedvalues may still considerably affected by the sample size (number of accessions) of different species, an equal number (three) of accessions was randomly selected from each species and re-analyzed forvalues. Results showed that the variation pattern ofvalues obtained from each species with three accessions remained basically the same, although the values were generally slightly lower than those of the original sample sets (data not shown).
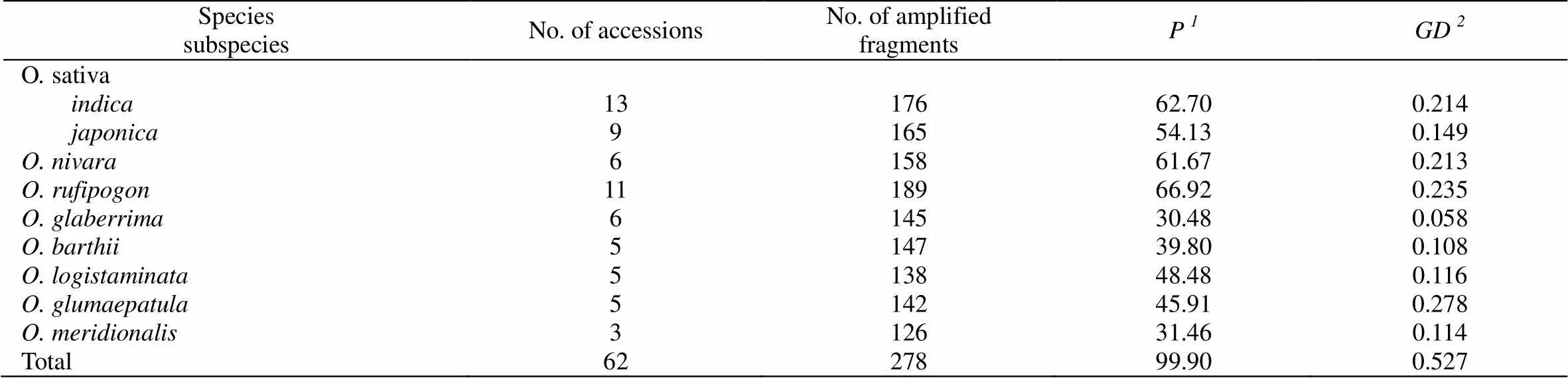
Table 2 Sampling ISSR survey and genetic diversity of AA-genome Oryza genus species
= percentage of polymorphic fragments;
= Gene Diversity (Nei 1978),= (/-1)(1-∑P2) where is the frequency of theth allele at each ISSR locus in each species (subspecies) andis the number of individuals for species (subspecies).
2.2 Genetic variation relationships between the Oryza species
The pairwise Jaccard similarity coefficients calculated based on the ISSR polymorphic data matrix were varying from 0.183 to 0.958 among the AA-genomeaccessions. A dendrogram based on the genetic similarities of all the accessions was constructed (Fig. 1). Nine major clusters involving the 62 AA-genomeaccessions were identified at the similarity coefficient value around 0.55 on the dendrogram with the high bootstrap support (72%~100%). The clustering of the 62 accessions was essentially corresponded to the traditional classification of the AA-genomespecies, with an unexpected pattern for the two subspecies (and) of Asian cultivated rice.(very high bootstrap value support, 100%). The cluster analysis evidently showed the preferential association of the AA-genomespecies with each other, particularly with their geographical origins. The Asian wild and cultivatedspecies (uppermost clusters on the dendrogram) were grouped together (bootstrap value 79%) with the close association among the.,., and.subsp.accessions, and somewhat separation of the.subsp.accessions. Considerable variation was observed within each cluster of this group. The Australian wild.and South American wild.accessions were grouped together (bootstrap value 81%) with a medium degree of variation, although a small sample size was included. The African cultivated rice.and its wild ancestral species.were included in the same group (bootstrap value 90%), but showed their well separation, having a strong bootstrap support (100%). Relatively low genetic variation was found among the.and.accessions, compared with accessions from other groups. The African perennial wild species.was grouped separately from all other AA-genome species, with a relatively high value of bootstrap support (79%) and a certain variation among the accessions.

Nice clusters were resolved at the similarity coefficient level of 0.6, which is closely associated with the current classification of the eight AA-genomespecies (includingandsubspecies). Bootstrap values (P) are indicated at the corresponding node of each cluster. The identity codes for each accession refer to those in Table 1
Fig.1. A UPGMA dendrogram of the AA-genomeaccessions based on the pairwise Jaccard (1908) similarity coefficient of the ISSR banding patterns.
3 Discussion
The genusincludes two cultivated species (the Asian.and African.) and six wild species from Asia(..), Africa(.,.), South America (.), and Australia (.), containing the AA genome[1]. These species are the most important and accessible genetic resources in the genusfor rice breeding because of their close genomic relationships. As a result, the agronomically beneficial traits (genes) can be easily transferred to cultivated rice by simple breeding methods as such sexual hybridization. Since the past several decades, tens of thousands of AA-genomeaccessions (most of them are cultivated species) have been collected by breeders, conservationists, and geneticists, and stored in germplasm banks over the world[5]However, only a very small portion of these collections has been successfully utilized in rice breeding. This is partially due to the limited understanding of their genetic diversity and complex relationships. The application of molecular markers (i.e., RFLPs, AFLPs, ISSRs, and SSRs) to characterize the stored genetic resources have facilitated the better understanding of these resources by determining and quantifying the genetic uniqueness of each germplasm accession[23], and therefore will certainly enhance our relevant knowledge on the rice germplasm.
In this study, a total of 278 polymorphic and repeatable ISSR fragments were amplified from 62 AA-genomeaccessions (representing eight species) collected from a wide range of geographical distribution, although the intra-specific variation patterns were remarkably different. This suggests the power of ISSR fingerprints as a tool for genetic diversity analysis. Even though only a small number of accessions were included in this study, the analysis demonstrated abundant genetic diversity within and among the AA-genomespecies. The result is generally in agreement with the estimation of genetic studies variation in these species based on previous[11, 24-26]. Therefore, the AA-genomegermplasm is of great importance and value in the breeding programs for broadening genetic background of cultivated rice varieties. As suggested by previous studies, the abundant genetic diversity of the AA-genomespecies is possibly caused by their adaptation to diverged ecological habitats of a wide geographical distribution, in addition to frequent introgressions among the sympatric species[27-28]. The abundant genetic diversity accumulated through the adaptive evolutionary process will continue to offer a great opportunity for selecting elite germplasm for rice improvement. The accessions of Asian cultivated rice and its putative ancestral species (..) demonstrated relatively high genetic diversity in this study. This is likely attributed to the wide cultivation of the cultivated species in different agricultural ecosystems, and the broad range of distribution and habitats of the wild ancestral species, where rich genetic diversity was accumulated. On the other hand, the accessions of African cultivated rice and its ancestor.showed relatively low genetic diversity, which may be largely imputed to the narrow cultivation area of the cultivar, and the low level of outcrossing rate of the ancestor. Other species showed a considerably variable level of genetic diversity among their accessions, although limited sample size may influence such diversity pattern. A large sample size involving more collections of each species may provide a more precise genetic diversity pattern of the AA-genomespecies.
[1] Vaughan, D A. The wild relatives of rice[M]. (Manila, Philippines :IRRI,), 1994.
[2] Bellon M R, Brar D S, Lu B R,et al. Rice genetic resources[M]. In Dwoling, N G., Greenfield S M., and Fischer K S. eds (Sustainability of Rice in the Global Food System. Califolia: Pacific Basin St udy Center and IRRI, Manila), 1998:251-283.
[3] Ge S, Sang T, Lu B R., et al. Phylogeny of rice genomes with emphasis on origins of allotetraploid species[J]. Proc. Natl. Acad. Sci. USA, 1999,96: 14400-14405.
[4] Yuan L P. Advantages and constraints to use of hybrid rice varieties[M]. In Wilson, ed (International Workshop on Apomixis in rice. Hunan Hybrid Rice Research Center, Changsha, China), 1993.
[5] Khush G S. Disease and insect resistance in rice[J]. Adv. Agronom, 1977, 29: 265-361.
[6] Brar D S, Dalmacio R, Elloran R, et al. Gene transfer and characterization of introgression from wildspecies into rice[M]. In Khush, G.A. ed (Rice genetics Ⅱ. IRRI, Manila), 1996: 477-486.
[7] Lu B R, Naredo M B E, Juliano A B, et al. Preliminary studies on taxonomy and biosystematics of the AA-genome,species (Poaceae)[M]. In Jacobs, S.W.L., and Everett, J. eds (Grasses, Systematics and Evolution. Melbourne, Australia: CSIRO),2000:51-58.
[8] Morishima H, Oka H I. The pattern of interspecific variation in the genus: its quantitative representation by statistical methods[J]. Evolution, 1960, 14: 153-165.
[9] Second G. Origin of the genetic diversity of cultivated rice: study of the polymorphism scored at 40 isozyme loci[J]. Jpn. J.Genet, 1982, 57: 25-57
[10] Ishii T, Nakano T, Maeda H, et al Phylogenetic relationships in A-genome species of rice as revealed by RAPD analysis[J]. Genes Genet. Syst, 1996, 71: 195-201.
[11] Lu B R, Zheng K L, Qian H R, et al. Genetic differentiation of wild relatives of rice as referred by the RFLP analysis[J]. Theor. Appl. Genet., 2002, 106: 101-106.
[12] Ren FG, Lu B R, Li S Q, et al. comparative study of genetic relationships among the AA-genomespecies using RAPD and SSR markers[J]. Theor. Appl. Genet., 2003, 108: 113-120.
[13] Park K C, Kim N H, Kim N S. Genetic variations of AA genomespecies measured by MITE-AFLP[J]. Theor. Appl. Genet., 2003,107: 203-209.
[14] Duan S H, Zheng Z, Hu N F, et al. Genetic diversity of common wild-rice and Asian cultivated rice by ISSR fingerprints[J].Journal of Jinggangshan University : Natural Science, 2010, 31:118-124(in Chinese).
[15] Zeitkiewicz E, Rafalski A, Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification[J]. Genomic, 1994, 20: 176-183.
[16] Joshi S P, Gupta V S, Aggarwal R K, et al. Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus[J]. Theor. Appl. Genet., 2000, 100: 1311-1320.
[17] Parsons B J, Newbury H J, Jackson M T. et al. Contrasting genetic diversity relationships are revealed in rice (L.) using different marker types[J]. Mol. Breeding, 1997, 3: 115-125.
[18] Dellaporta S L, Wood J, Hick J B. A plant DNA minipreparation: version II[J]. Plant Mol. Biol. Rep. ,1983, 1: 19-21.
[19] Qian W, Ge S, Hong D Y. Genetic variation within and among populations of a wild-ricefrom China detected by RAPD and ISSR markers[J]. Theor. Appl. Genet., 2001, 102: 440-449.
[20] Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals[J]. Genetics,1978, 89: 583-590.
[21] Jaccard P. Nouvelles rescherches sur la distribution florale[J]. Bulletin de la Societe Vaudoise des Sciences Naterelles, 1908, 44: 223-270.
[22] Rohlf F J. NTSYS-pc, numerical taxonomy and multivariate analysis system, version 2.02[M]. (New York: Exeter Software, Setauket), 1998.
[23] Brown S M., Kresovich S. Genome mapping in plants[M]. In Paterson, A.H. ed (Clandes, New York), 1996: 85-93.
[24] Sun CQ, WangXK, Li ZC, et al. Comparison of the genetic diversity of common wild-rice (Griff.) and cultivated rice (.L.) using RFLP markers[J]. Theor. Appl. Genet.,2001, 102: 157-162.
[25] Wang M X., Zhang H L, Zhang D L, et al. Geographical genetic diversity anddivergence of common wild rice (Griff. ) in China[J]. Chinese Science Bulletin, 2008, 53: 1-8(in Chinese).
[26] Gao L Z, Zhang C H, Li D Y, et al. Genetic diversity withingermplasms reserved in Chinese field gene banks of wild rice as revealed by microsatellite markers[J]. Biodiversity and Conservation, 2006, 15: 4059-4077(in Chinese).
[27] Wang X.K, Sun C Q. Origin and differentiation of Chinese cultivation rice[M]. Beijing: China Agricultural University Press, 1996.
[28] Song Z P, Xu X, Wang B, et al. Genetic diversity in the northernmostpopulations estimated by SSR markers[J]. Theor Appl Genet, 2003, 107: 1492- 1499.
基于ISSR标记的稻属AA基因组遗传多样性分析
*段世华1,2,郑 卓1,罗 强1,龙伟雄1,廖佛才1
(1. 井冈山大学生命科学学院,江西,吉安 343009;2. 井冈山大学生态环境与资源研究所,江西,吉安 343009)
为了确定稻属AA基因组物种间的遗传差异和系统进化关系,62份来自广泛地理分布的水稻品系被用于ISSR标记分析。这些品系包含有6个野生稻种(.,.、.,.,., 和.)和2个栽培稻种(.和.)。21条能产生良好重复性条带模式的ISSR引物被筛选出,并在62个水稻品系中揭示出非常好的多态性。全部样品的基因多样性为0.527,同时显示出ISSR标记在稻属物种遗传多样性研究中具有强大的作用。根据ISSR条带模式,利用Jaccard配对相似系数构建的一致性树状图,显示出具有良好自展支持率的稻属AA基因组遗传多样性关系。结果表明,来自不同大陆的稻属物种具有较近的亲缘关系,尤其是亚洲野生稻物种与Vaughan1989年建立的稻属分类系统具有良好的一致性。研究结果将对稻属AA基因组野生稻在水稻育种实践中的有效利用具有非常重要的意义。
遗传多样性;ISSR;稻属;AA基因组
S511 Document code:A
10.3969/j.issn.1674-8085.2013.03.024
2013-03-27
Modified date:2013-04-08
This work was supported by the Natural Science Foundation of Jiangxi Province (2009GZN0071) and the Science & Technology(S&T) Plan Projects of Jiangxi Provincial Education Department (GJJ12464).
Biographies: * DUAN Shi-hua (1968-), male, from Yongxin, Ji’an, Jiangxi Province, Ph. D, Professor, Mainly engaged in molecular biology research(E-mail:shihua_duan@yahoo.com.cn);
ZHENG Zhuo (1973-) , male, from Zhuxi Shiyan, Hubei Province, Ph. D, Deputy Professor, Mainly engaged in rice genetic and breeding research.(E-mail:zhengzhuodai@yahoo.com.cn);
LUO Qiang (1990-), male, from Nankang Ganzhoun, Jiangxi Province, undergraduate(E-mail:luoqian90@163.com);
LONG Wei-xiong (1991-), male, from Yongxin, Ji’an, Jiangxi Province, undergraduate(E-mail:lwx1991@yahoo.com.cn);
LIAO Fo-cai (1989-), male, from Xunwug Ganzhoun, Jiangxi Province, undergraduate(E-mail:liaofocai1989@163.com).
1674-8085(2013)03-0100-07

