Small auxin upregulated RNA(SAUR)gene family in maize:Identification,evolution,and its phylogenetic comparison with Arabidopsis,rice,and sorghum
Yuzhu Chen,Xi Haoand Jun Cao
1Institute of Life Science,Jiangsu University,Zhenjiang 212013,China,2Industrial Crop Institute,Henan Academy of Agricultural Sciences,Zhengzhou 450002,China. †These authors contributed equally to this work.*Correspondence:cjinfor@163.com
INTRODUCTION
Auxin plays a critical role in plant growth and development by modulating the expression of genes involved in several physiological processes(Woodward and Bartel 2005;Teale et al.2006;Hoffmann et al.2011).These regulated genes mainly include three different early auxin-responsive gene families.They are the auxin/indoleacetic acid(Aux/IAA)gene family,Gretchenhagen-3(GH3)gene family,and small auxin-up RNA(SAUR)gene family(Hagen and Guilfoyle 2002).Aux/IAA genes encode Aux/IAA proteins that function as negative regulators of auxin response proteins(Ulmasov et al.1997).GH3 genes encode IAA-amido synthetases that conjugate free IAA with amino acids,that is,they convert the active IAA to an inactive form(Staswick et al.2005).SAURs are primary auxin response genes involved in the auxin-signaling pathway.Their expression could be induced within 2–5 min by active auxin,indicating that auxin plays an important role in transcriptional regulation of SAUR genes(Franco et al.1990).In addition,many SAUR genes are also regulated post-transcriptionally due to a highly conserved downstream(DST)element in their 3′-untranslated region(UTR)that contributes to mRNA instability in an auxinindependent manner(Newman et al.1993;Park et al.2012).Therefore,regulation of SAURs may occur at the transcriptional,posttranscriptional,and protein levels(Esmon et al.2006;Spartz et al.2012).
The first SAUR gene was originally identified in soybean hypocotyls(McClure and Guilfoyle 1987).Subsequently,some homologous genes have been identified in a wide range of plants,and genomic sequencing projects have revealed that SAURs are present as a large gene family in Physcomitrella(18 members)(Rensing et al.2008),Arabidopsis(over 70 members)(Hagen and Guilfoyle 2002),rice(58 members)(Jain et al.2006),and sorghum(71 members)(Wang et al.2010).Recently,by means of a bioinformatics approach,99 and 134 SAUR genes were also identified in two Solanaceae species,tomato and potato,respectively(Wu et al.2012).Despite many SAUR genes being identified in different plant species,only a small number of them have been functionally characterized recently.Some Arabidopsis SAUR proteins can be bound to calmodulin,suggesting that they may mediate responses to changes in intracellular calcium level(Yang and Poovaiah 2000;Reddy et al.2002;Knauss et al.2003).SAUR genes are observed mainly expressed in growing hypocotyls or other elongating tissues,implying that they play a role in the regulation of cell elongation(Gee et al.1991;Gil and Green 1997;Knauss et al.2003;Chae et al.2012).In contrast,the Arabidopsis SAUR32 gene was not auxin induced and caused apical hook opening and a short hypocotyl when overexpressed(Park et al.2007).Similarly,transgenic lines overexpressing OsSAUR39 decreased shoot growth and auxin transport,suggesting that OsSAUR39 may negatively regulate auxin biosynthesis and transport(Kant et al.2009).The addition of an N-terminal GFP or epitope tag can dramatically increase the stability of Arabidopsis SAUR19–24 protein,resulting in increased hypocotyl and leaf size,defective apical hook maintenance and altered tropic responses.These findings demonstrate that SAUR19–24 function as positive effectors of cell expansion through the modulation of auxin transport(Spartz et al.2012).Recently,research by Hou et al.(2013)suggests that Arabidopsis SAUR36 positively regulates leaf senescence by mediating auxin.
Maize,an important cereal crop,has become a model plant for genetic and other basic biological research.The availability of the maize genome sequence has provided an excellent opportunity for whole-genome annotation,classification,and comparative genomics research(Schnable et al.2009).Although SAUR genes have been extensively characterized in Arabidopsis,rice,and other species(Hagen and Guilfoyle 2002;Jain et al.2006;Rensing et al.2008;Wang et al.2010;Wu et al.2012),and two maize SAUR genes have been identified(Yang and Poovaiah 2000;Knauss et al.2003),a systemic analysis of the SAUR gene family in maize has not been reported.In this study,79 putative SAUR genes in the maize genome were identified and characterized.Analyses of sequence phylogeny,gene organization,adaptive evolution,auxin-responsive cis-elements,and expression profiling were performed to provide insights into the evolutionary mechanisms of the maize SAUR protein family.The results maybe provide a biological reference for future studies on the functions of the SAUR genes.
RESULTS
Identification and annotation of the SAUR genes in maize
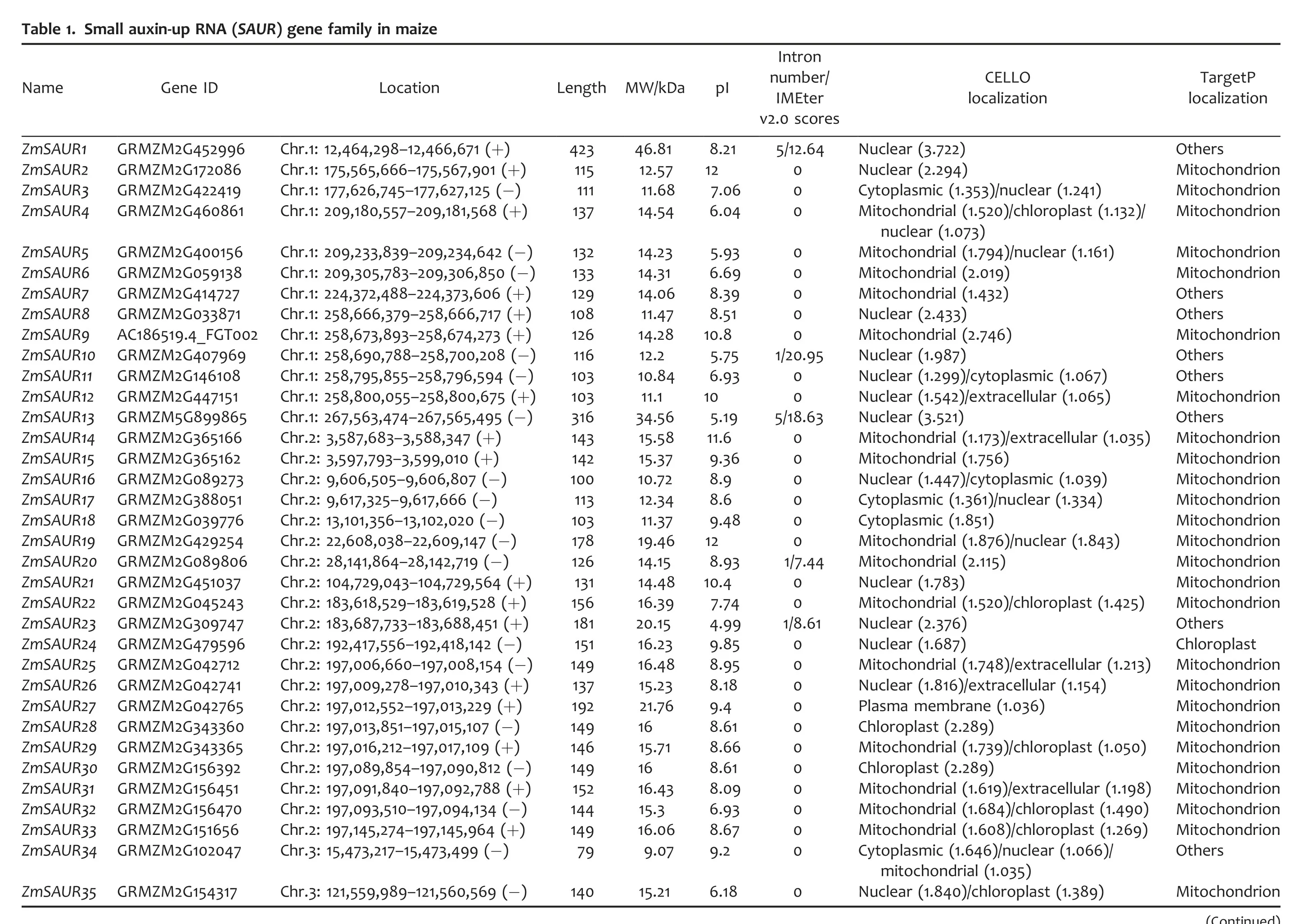
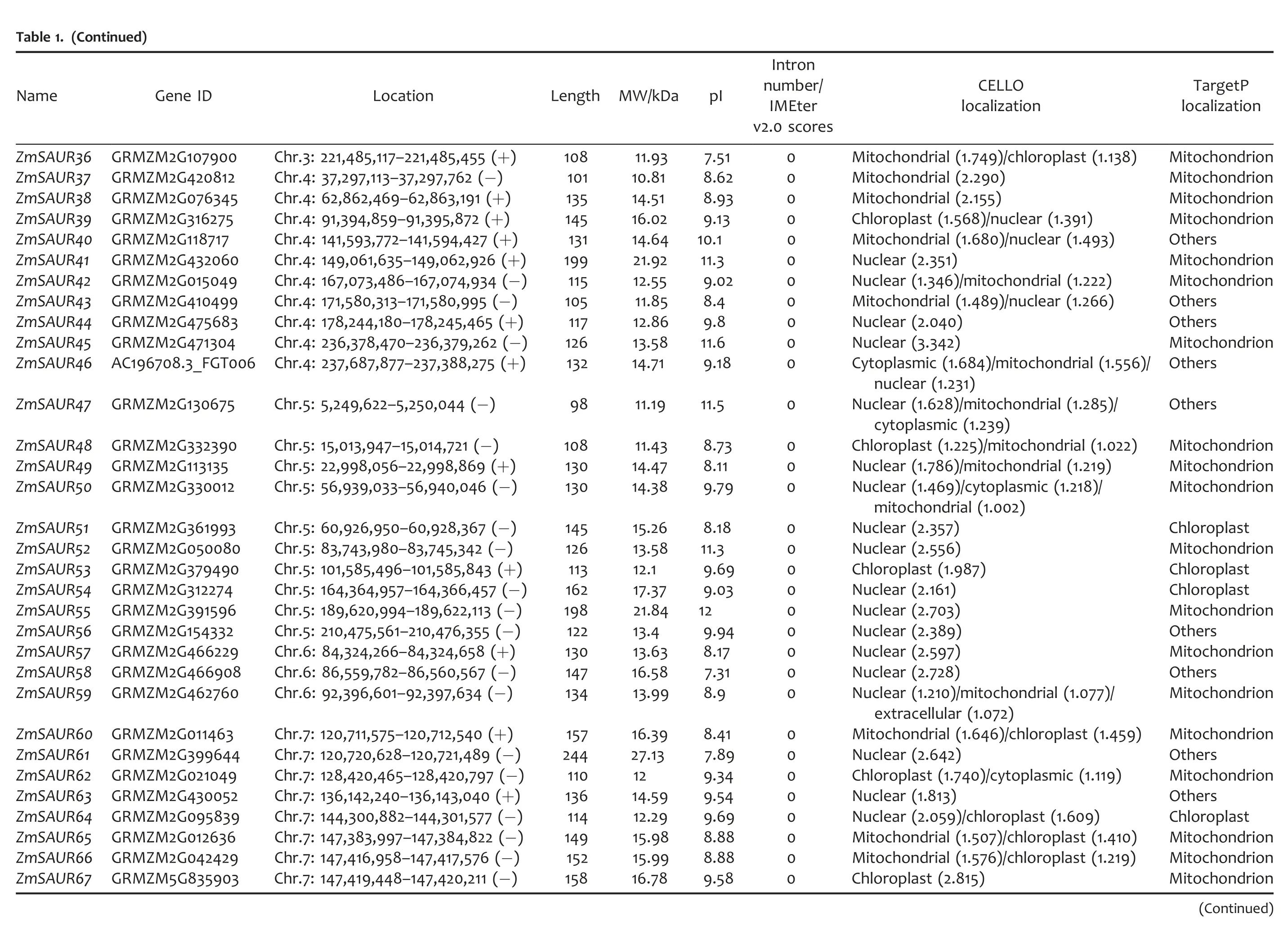
To identify members of the SAUR gene family in maize,we first searched relevant databases using the rice and sorghum SAUR protein sequences as queries.The maize sequences returned from such searches were confirmed to encode SAUR by the Conserved Domain Databases (CDD) (Marchler-Bauer et al.2009)and Pfam(Punta et al.2012)searches for the presence of an auxin-inducible signature conserved in other SAUR proteins(McClure et al.1989).As a result,79 SAUR genes were identified in maize(Table 1).All 79 ZmSAURs were named according to their position from the top to the bottom of the maize chromosomes 1–10.Maize SAUR genes encode polypeptide of 79–423 amino acids in length,with a predicted molecular mass range of 9.07–46.81 kDa and theoretical pIs ranging from 4.39 to 12.3(Table 1).Further analyses using protein subcellular localization prediction software,CELLO v2.5 Server(http://cello.life.nctu.edu.tw/)(Yu et al.2004),predicted the probable protein localization for each of the different candidate SAURs in maize.It was found that over 60%of maize SAUR proteins possess signal sequences for targeting the nucleus.Some other members were predicted to localize from cytoplasm,chloroplast to mitochondria(Table 1).We also used the TargetP(Emanuelsson et al.2000)for predicting the localization of ZmSAUR proteins,and found that most of the members were predicted to localize in the mitochondrial and chloroplast(Table 1).
Phylogenetic analyses of the SAUR genes in maize,Arabidopsis,rice,and sorghum
In order to analyze the phylogenetic organization of the SAUR family,we performed a phylogenetic analysis of 79 maize SAURs,77 Arabidopsis SAURs,55 rice SAURs,and 71 sorghum SAURs by generating a phylogenetic tree based on neighbor joining(NJ)method using MEGA 5(Tamura et al.2011).Based on their phylogenetic relationships,we divided these SAURs into 16 groups,designated from Groups I to XVI(Figures 1 and S1).While the nodes at the base of the larger clades were not well supported(low scores in the bootstrap analyses),nodes at the base of many smaller clades were robust(bootstrap values>70%)(Figure S1).This may be due to reasons of considerable divergence between them.Group XV contained 29 members and constituted the largest clade in the SAUR phylogeny(Table 2).Interestingly,using phylogenetic analyses,some SAUR genes were suggested to form species-specific clades or sub-clades after the divergence of these species in this study.As seen in Figure 1,4 and 21 Arabidopsis SAUR genes formed the Groups VI and X clades,respectively.In this study,we also found that some monocot-specific SAUR clades formed Groups I,II,III,IV,and XV(Table 2).Within these monocot-specific SAUR clades,species-specific SAUR subclades were also found.
Chromosomal location,duplication events,and synteny analyses
Phylogenetic analyses also showed several pairs of SAUR proteins.These paralogous SAUR members were closely related and they evolved from relatively recent gene duplications(Cao and Shi 2012).To further investigate the relationship between genetic divergence within the SAUR family and gene duplication and loss in maize and sorghum genomes,chromosomal location of each SAUR gene was determined.The results showed that the SAUR genes were dispersed throughout the maize and sorghum genomes(Figures 2 and S2).Within identified duplication events,12 of 28 pairs(ZmSAUR23/ZmSAUR61,ZmSAUR15/ZmSAUR79,ZmSAUR56/ZmSAUR44,ZmSAUR24/ZmSAUR63,ZmSAUR41/ZmSAUR55, ZmSAUR19/ZmSAUR76, ZmSAUR57/ZmSAUR51,ZmSAUR49/ZmSAUR7, ZmSAUR52/ZmSAUR45, ZmSAUR74/ZmSAUR59,ZmSAUR9/ZmSAUR48,and ZmSAUR60/ZmSAUR22)in maize are retained as duplicates.While,in the closest relative sorghum,of 24 duplication events,only three pairs(SbSAUR4/SbSAUR22,SbSAUR38/SbSAUR43,and SbSAUR35/SbSAUR66)are retained as duplicates(Figure S2).In addition,we also estimated the evolutionary dates of duplicated ZmSAUR genes using Ksas the proxy for time(Table 3).Our study showed that duplication events for maize 9 of 28 pairs occurred within the past 10.21–16.64 million years(Table 3).This period is consistent with the time when the subsequent large-scale genome duplication event is thought to have occurred in maize(Gaut and Doebley 1997).It is clear that gene duplication for most of the ZmSAURs is due to the second large-scale duplication event.Therefore,maize holds more numbers of duplicated SAUR genes than sorghum.Interestingly,we also identified several tandemly clustered SAUR members in maize,such as ZmSAUR8–12 cluster,ZmSAUR25–33 cluster,and ZmSAUR65–68 cluster(Figure 2),suggesting that tandem duplication may also be a factor underlying the genesis of family genes.Taken together,segmental duplication and tandem duplications contribute to expansion of the SAUR gene family in maize.
Synteny analysis is a useful tool for establishing both orthology relationships and functional linkages between genes.The application of synteny analysis to SAUR genes has not been applied previously.In this study,we compared the assembled genomes of maize and sorghum species for regions of conserved synteny and found that the synteny relationship widely existed between maize and sorghum genomes(Figure 3).As an example,one SAUR gene cluster at the bottom of chromosome 2 of both genomes showed conserved synteny in maize and sorghum(Figure 3),which indicated that this region has remained partially conserved since their last common ancestor.This analysis cannot only provide remarkable insight into the evolutionary paths of SAURs among maize and sorghum,but also provide careful dissection of these syntenic networks and comparisons among these species.

Auxin-responsive cis-elements in the promoters of ZmSAUR genes
To elucidate the possible regulatory mechanism of ZmSAUR genes in exogenous auxin stimuli,we identified putative auxinresponsive cis-elements in the 1000 bp promoter regions upstream of the transcription start site(TSS)of the ZmSAUR genes(Table S1).Seven types of cis-elements,which are directly related to the Dof protein binding(S000273)(Baumann et al.1999),NDE element(S000360 and S000370)(Xu et al.1997),ARF binding(S000270)(Goda et al.2004),ASF-1 binding(S000024)(Redman et al.2002),AuxRE(S000026)(Ballas et al.1993),and TGA-box(S000234)(Liu et al.1994),were detected by searching the promoter sequence against the PLACE database(Higo et al.1999).Surprisingly,we found that except for ZmSAUR11,ZmSAUR43,ZmSAUR45,ZmSAUR59,and ZmSAUR64,all other ZmSAUR genes contained at least one of the putative seven types of cis-elements in their promoter regions(Figure 4).Moreover,most members were enriched with multiple cis-elements in their promoters.
Variable selective pressures among amino acid sites
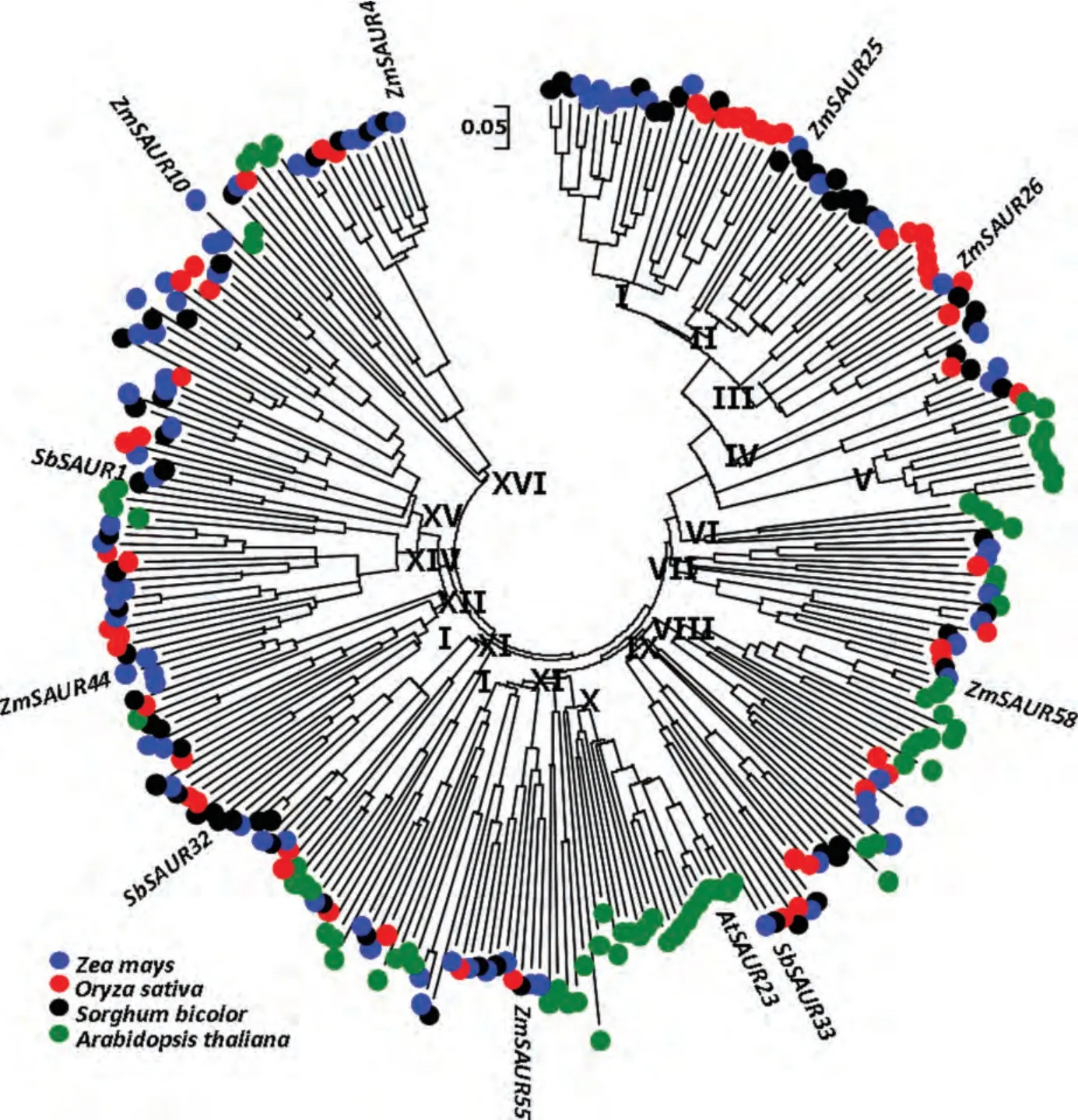
Figure 1.Unrooted Neighbor-joining phylogenetic tree of small auxin-up RNAs(SAURs)in four plantsThe tree is reconstructed using SAUR sequences in Arabidopsis thaliana(green),Oryza sativa(red),S.bicolor(black),and Zea mays(blue).Evolutionary distances are computed using the p-distance method and are in units of the number of amino acid substitutions per site.Some SAURs are also labeled in it.Please visit the online version about the color pictures in this paper.
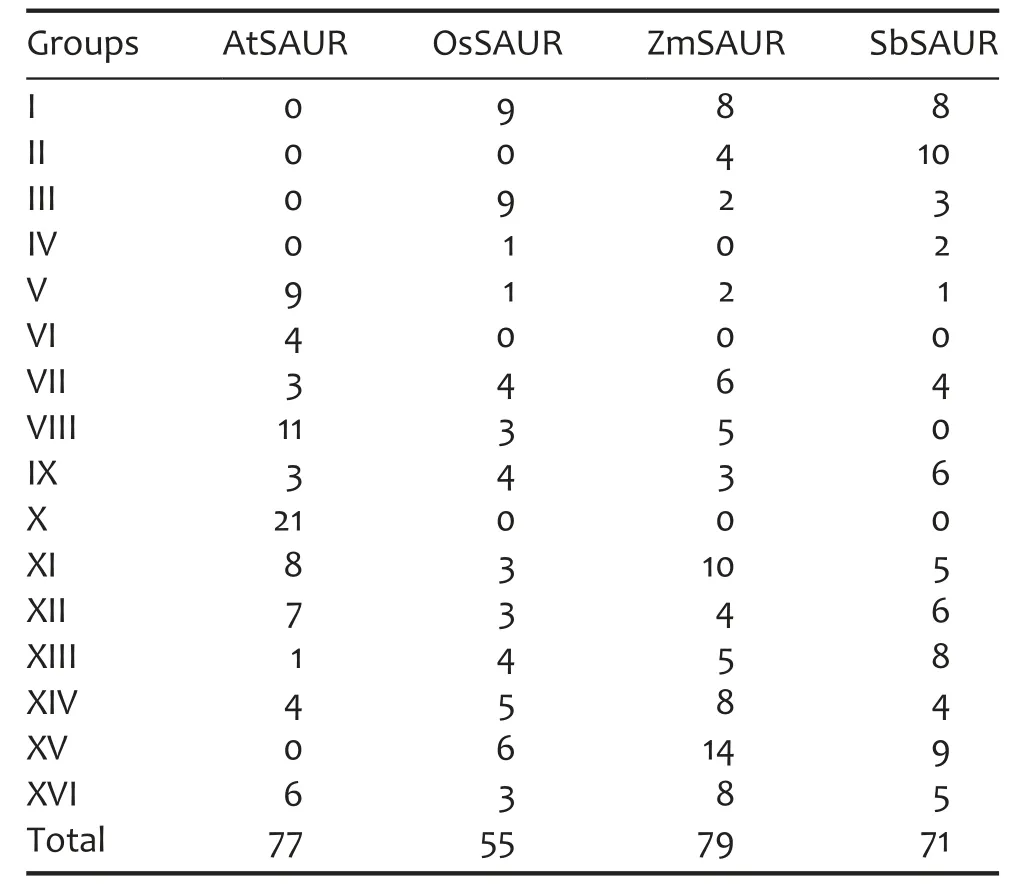
Table 2.Number of small auxin-up RNA(SAUR)genes of Arabidopsis,rice,maize,and sorghum in Groups I–XVI
Ratios of non-synonymous(Ka)versus synonymous(Ks)mutations(Ka/Ks)can measure selection pressure on amino acid substitutions.A Ka/Ksratio greater than 1 suggests positive selection and a ratio less than 1 suggests purifying selection(Hurst 2002).As we know,amino acids in a protein are usually expected to be under different selective pressures and to have different underlying Ka/Ksratios.To analyze the positive or negative selection of specific amino acid sites within the fulllength sequences of the SAUR proteins in the different groups,the Ka/Ksratios were calculated with the Selecton Server(http://selecton.tau.ac.il/)(Stern et al.2007).In this study,four of the evolutionary models(M8(ωs≥1),M8a(ωs=1),M7(beta),and M5(gamma))were used to perform the tests.The selection models M8a and M7 do not indicate the presence of positively selected sites,whereas the M8 and M5 models indicate some sites(Table 4).Moreover,statistical significance of positive selection has been tested for the identified positively selected sites.The results show that the Ka/Ksratios of the sequences from different SAUR groups are significantly different(Table 4).Such as,higher Ka/Ksratios existed in Groups I and IX,indicating a higher evolutionary rate or sitespecific selective relaxation within members of the same group.Despite the differences in Ka/Ksvalues,all the estimated Ka/Ksvalues are substantially lower than 1,suggesting the SAUR sequences within each of the groups are under strong purifying selection pressure.From Table 4,we also found that,while most of the protein sequences are subject to constant purifying selection,a few sites undergo positive selection.An example of detailed distribution of the positive-selection sites in Group VIII sequences as predicted by the M8 model shown in Figure 5.Further analyses indicate that two of the eight positive selection sites in Group VIII are located in α-helices(α1 for sites 6 and 7).Another five predicted positive-selection sites are located in β-strands(β1 for sites 42,43,and 44,β2 for site 46 and β5 for site 94).Site 132 is located in one β-turn(Figure 5).Most of these sites exist in the N-terminal and C-terminal of SAUR amino acid residues.These observations suggest that positive selection pressure on these residues might have changed the protein structure,thus accelerated functional divergence.Therefore,because of the different evolutionary rates predicted at some amino acid sites,SAUR genes may be significantly divergent from each other in their functions.During long periods of evolution,different evolutionary rates at specific amino acid sites can spur SAUR family genes to evolve new functions after divergence.

Figure 2.Gene locations and genomic duplication in maizeSeventy-nine small auxin-up RNA(SAUR)genes are mapped to the maize 10 chromosomes.Paralogous regions in the putative ancestral constituents of the maize genome are depicted using SyMAP v3.4(Soderlund et al.2011)and are shown on one side of each chromosome.The same colors on both each side of a chromosome region and maize chromosome color key indicate the same paralogous regions in the genome.
Expression analysis of ZmSAUR genes
Expression profiling is a useful tool for understanding gene function(Cao 2012;Zou et al.2013).A comprehensive expression analysis was performed using publicly available microarray data for maize.Firstly,to identify their potential roles in plant growth and development,we tested the temporal-and spatial-specific expression patterns of ZmSAUR genes.In this case,the gene expression levels of 60 distinct tissues representing 11 major organ systems were mined through microarray data.Organ systems included the germinating seed,primary root,whole seeding,stem and shoot apical meristem(SAM),cob,internodes,silk,tassel and anthers,leaf,husk,and seed.We detected 75 probes assigned to 75 corresponding ZmSAUR transcripts.The remaining four transcripts with no detectable expression signal are ZmSAUR57,ZmSAUR66,ZmSAUR68,and ZmSAUR70.From Figure 6,it can be seen that all of the 75 detected transcripts produced by 75 genes are expressed with distinct levels and in different tissues,suggesting that they may be involved in various biological processes.Such as,most members of clade i displayed high expression levels in silks,but showed low expression levels in the embryo and endosperm,implying that ZmSAURs in this clade may play an important role in silks development(Figure 6).We also noticed that most members of maize SAUR genes in clade ii were expressed at higher levels in leaf tissue or organs,implying that ZmSAUR genes in this clade may be involved in the growth and development of leaf in maize.While,comparatively speaking,these genes in clade ii showed lower expression levels in seed than that in other organs(Figure 6).Some ZmSAURs that are highly expressed in some specific organs were also found.For example,ZmSAUR6 and ZmSAUR38 displayed unusually high expression levels in the whole period of seed growth,suggesting that they may be involved in seed development.ZmSAUR43 and ZmSAUR63 may play an important role in anthers due to the highest expression levels.ZmSAUR31 showed higher expression levels in coleoptile primary root,etc.indicating that it might be involved in the growth and development of coleoptile primary root in maize.
Quite a few members of the SAUR family have been demonstrated to be regulated by IAA in plants.However,few SAUR genes responses to IAA treatment were reported in maize.For this purpose,we investigated the expression patterns of maize SAUR genes under IAA treatment.The results are given in Figure 7 and the primer sequence in Table S2.The relative quantification method was used to evaluate the quantitative variation between replicates.Among the 10 ZmSAUR genes detected,six and four members were upregulated during IAA treatment in shoot and root,respectively(Figure 7).ZmSAUR23 and ZmSAUR78 are significantly upregulated in the shoot and root 10 min after IAA treatment,respectively,suggesting that these genes are more likely to play critical roles in regulating IAA response in maize shoot and root,respectively.

Table 3.Inference of duplication time of small auxin-up RNA(SAUR)paralogous pairs in maize
DISCUSSION
SAUR gene structure and intron functions
Previous researchers have identified and annotated some SAUR genes in Arabidopsis,rice,Physcomitrella,sorghum,and Solanaceae(Hagen and Guilfoyle 2002;Jain et al.2006;Rensing et al.2008;Wang et al.2010;Wu et al.2012).In this study,we identified 79 SAUR genes in maize.All the members were predicted to encode the SAUR domains.Interestingly,most of the ZmSAURs are intronless(Table 1).This phenomenon also exists in the SAUR genes of other species(Jain et al.2006;Rensing et al.2008;Wang et al.2010;Wu et al.2012).Increasing evidence has revealed the potential function of introns,such as a signal for mRNA export,and alternative splicing exon shuffling(Fedorova and Fodorav 2003;Roy and Gilbert 2006).Moreover,intragenic recombination was also connected with introns(Le Hir et al.2003).Like SAUR,other intronless genes,such as non-chemosensory G-protein-coupled receptors(Takeda et al.2002),olfactory receptor genes(Zhang and Firestein 2002),F-box protein family(Gagne et al.2002),pentatricopeptide repeat family genes(Lurin et al.2004),and DEAD box RNA helicases(Aubourg et al.1999),usually evolve rapidly either by gene duplication or by reverse transcription/integration.In addition,many introns also have the ability to promote gene expression through a process termed intronmediated enhancement(IME)(Mascarenhas et al.1990).It has been shown that enhancing introns typically located near the promoter and are compositionally distinct from downstream introns.We also calculated the IME signals in six ZmSAUR genes containing introns by IMEter v2.0(Parra et al.2011),software for calculating whether an intron is likely to enhance gene expression.Our results showed that two introns of ZmSAUR10 and ZmSAUR13 genes obtain a higher score,implying that these introns are expected to enhance expression(Table 1).Although the results require additional experiments to be validated,more and more studies have indicated that introns may play an important role in genetic evolution.
Subcellular localization of ZmSAUR proteins
We also found that some SAUR proteins were predicted to localize in the mitochondria or chloroplasts(Table 1).As we know,auxin plays a critical role in plant growth and development by modulating the expression of genes involved in several physiological processes.As energy organelles,plant mitochondria and chloroplasts are capable of generating ATP by either oxidative or photosynthetic phosphorylation,respectively.Several transporters localized at the membrane of both organelles serve to interconnect the metabolism between the chloroplasts,the mitochondria,and the surrounding cytosol(Flügge et al.2011).One question is whether the auxin is also involved in the energy transfer process.Research by Mockevičiūtė et al.(2006)shed some light on this problem,which indicated that some IAA-ABP(auxin-binding protein)complexes might be located in the mitochondria and chloroplast.It is likely that the auxin may be involved in the energy transfer process occurring for the first time on the membrane of both organelles.As one early auxinresponsive gene family,these SAUR proteins might also be involved in this process.Previous studies have examined the localization of some SAUR proteins to the nucleus(Knauss et al.2003;Park et al.2007),cytoplasm(Kant et al.2009),or plasma membrane(Spartz et al.2012).Our results for ZmSAUR protein localization indicate that more than half of the ZmSAUR proteins can be predicted to localize to the mitochondria or chloroplasts(Table 1).This phenomenon has also been found in SlSAUR proteins,that is,more than a third of SlSAUR proteins can be predicted to localize at these organelles(Wu et al.2012).In addition,we also searched the CoreMitoP(an experimentally confirmed mitochondria proteins database)(Cui et al.2011),and found that over 67.5%of the Arabidopsis SAUR proteins can be detected in this database.Therefore,subcellular localization may be another process needing to be further examined in SAUR proteins.Furthermore,additional experiments and methodical approaches will be carried out for elucidating these SAUR localizations and their possible role in response to auxin signals and the mechanism of energy transfer.

Figure 3.Synteny analyses between maize and sorghum genomesConservation of synteny generated from the SyMAP v3.4(Soderlund et al.2011)was analyzed in the maize and sorghum genomes(A).Red asterisks mean the two segments analyzed in(B).Two conserved segments held some small auxin-up RNA(SAUR)genes from the chromosome 2(197.000–197.149 Mb)of the maize genome and the chromosome 2(66.616–66.654 Mb)of sorghum genome were also shown in(B).
Species-specific expansion of SAUR genes
Phylogenetic analyses can allow us to identify evolutionarily conservative and divergent SAUR genes.Remarkably,Groups VI and X do not include any rice,sorghum,or maize proteins but contain only proteins from Arabidopsis(Table 2;Figure 1).Likewise,Groups I,II,III,IV,and XV do not include any Arabidopsis SAUR proteins but contain only proteins from monocots(Table 2;Figure 1).It is possible that these groups have evolved after the monocot-dicot divergence and that they have specialized roles in monocots or dicots.In addition,we also found that some SAUR genes were suggested to form species-specific clades or sub-clades after the divergence of these species(Table 2;Figures 1 and S1).In general,genes encoding proteins are required largely as components of an organism’s morphological structures or involved in recognition and binding of pathogens and withstanding environmental stress are often subject to species or lineage-specific expansion(Kondrashov et al.2002;Lespinet et al.2002).These expansions may provide the raw material for generating the diversity required to counter rapidly changing pathogens and to respond to other variable environmental factors(Lespinet et al.2002).Therefore,species or lineage-specific expansion appears to be a common means of generating additional specificities in signaling pathways.In plant,rapid gene expansion has been reported in multigene families involved in stress-responsive(Hanada et al.2008;Lehti-Shiu et al.2009).Our work demonstrates that SAUR genes involved in early auxin-response can also increase rapidly in copy number in some clades during speciation.To our knowledge,a species-specific gene cluster has not been reported for this gene family.Given the important roles of SAURs in numerous physiological processes of the plant,it is likely that SAUR duplicates retained for some clades increase the capacity of signal transduction.It is difficult to explain why certain duplicates were retained or lost.Further molecular genetic or biochemical analyses of members within a subfamily may shed light on their relative contributions in the SAUR family.

Figure 4.Distribution of major auxin-responsive cis-elements in the promoter sequences of the 79 ZmSAUR genesSeven putative cis-elements are represented by different symbols as indicated.

Table 4.Likelihood values and parameter estimates for ZmSAUR genes

Table 4.(Continued)
Genome duplication and SAUR evolution
Genome-wide duplication events,gene loss,and local rearrangements have created the present complexities of the genome(Blanc and Wolfe 2004;Cao et al.2010,2011;Lucas et al.2013;Shang et al.2013).To further investigate the relationship between genetic divergence within the SAUR family and gene duplication and loss in the maize and sorghum genomes,we determined the chromosomal location of each SAUR gene.As described above,these estimates of proportions of duplicated SAUR loci suggest that a larger proportion of the SAUR are duplicated in the maize genome(12 of 28 pairs)than in the sorghum genome(3 of 24 pairs).Maize has experienced two genome amplification events(ancestral tetraploidy and a large-scale duplication event)(Gaut and Doebley 1997).That is,maize genome duplication happened after divergence from sorghum.Compared with sorghum,it may be the reason why maize retained so many duplicated SAUR genes.Further analyses using evolutionary dates of the duplicated ZmSAUR genes also confirmed it(Table 3).We also found that most of these species-specific SAURs are located in clusters(Jain et al.2006).That is,tandem gene duplications have clearly contributed to the expansion of SAUR gene families within these species or lineage-specific clades.Based on analysis of orthologous groups of genes in some model plants,genes expanded via tandem duplication tend to be involved in responses to environmental stimuli(Hanada et al.2008;Cai et al.2013).It is likely that these tandem gene arrangements would therefore be consistent with functions of SAUR related to stress adaptation.Therefore,segmental duplication and tandem duplication both might contribute to the expansion of ZmSAURs.Like maize SAUR genes,the expansion of most of the SAUR genes can also be accounted for by the segmental duplication and tandem duplication in Arabidopsis,rice,and Solanaceae(Hagen and Guilfoyle 2002;Jain et al.2006;Wu et al.2012).It suggests that SAURs might experience similar evolutionary mechanisms in the process of gene expansion in these species.
Auxin-responsive cis-elements in ZmSAURs
Furthermore,we also identified the genomic structure of SAUR genes in maize.Some putative auxin-responsive cis-elements were identified in the promoter regions of SAUR genes in maize,suggesting that these SAURs might be involved in auxin signal transduction pathway.Through comparing the distribution of the seven regulatory elements in the promoter regions,all sister pairs of ZmSAUR genes were found to exhibit significant differences in their promoter sequences,suggesting that the duplicated genes may not have some regulatory features in common,but rather under similar regulatory pathways in some respects(Zhao et al.2011).For example,each of the duplicated genes contained at least one auxin-responsive cis-element in their promoter regions.Through massively parallel signature sequencing in Arabidopsis,Haberer et al.(2004)revealed that more than two-thirds of the duplicated genes exhibit divergence in their expression characteristics.Significant differences in promoter sequences result in the expression divergence between duplicated genes,which might represent an important evolutionary mechanism of neofunctionalization or subfunctionalization(Prince and Pickett 2002).

Figure 5.Distribution of positive selection sites of Group VIII small auxin-up RNA(SAUR)members predicted by M8 modelThe predicted tertiary structure(alpha helix,beta strand,and turn)(A)and partial alignment results(B)are shown.Seven potential positive selection sites are marked with a star in the predicted tertiary structure and one site(94)is shown with gray background in alignment results.
Differential expression of ZmSAUR genes
Since expression profiling can provide important clues for gene function,we examined the expression of ZmSAUR genes in 60 distinct tissues of maize development by microarray data.The expression profiles reveal variations in the expression of ZmSAURs in different tissues.In general,most members of ZmSAURs were expressed at higher levels in root and leaf tissues or organs,and showed lower expression levels in endosperm and seed(Figure 6).For plants,compared with some of the dormant parts(such as endosperm and seed),the growing parts(such as root and leaf)often can produce a large amount of auxin to maintain the needs of plant growth.Furthermore,as we know,SAUR is one of three different early auxin-responsive gene families.Therefore,it might be the reason why most ZmSAURs were expressed at higher levels in root and leaf,but not in endosperm or seed.While,some ZmSAUR genes seem not to follow this trend.Such as,expression levels of ZmSAUR6 and ZmSAUR38 were higher in the whole period of seed growth,suggesting that they could play key roles in the maize seed development.In addition,we also identified expression profiles of 10 ZmSAUR genes response to IAA treatment by quantitative reverse transcription polymerase chain reaction(qRT-PCR)analyses.Among these 10 ZmSAUR genes detected,six and three members were upregulated during IAA treatment in shoot and root,respectively(Figure 7),implying that these genes are more likely to play critical roles in regulating IAA response in maize shoot or root.
In conclusion,this study provided a comparative genomic analysis addressing phylogeny,chromosomal location,synteny analysis,auxin-responsive cis-elements,selective pressures,and expression profiling of the SAUR gene family in maize.Phylogenetic analyses revealed 16 groups in the SAUR family.SAUR genes were non-randomly distributed across the maize chromosomes,and a high proportion of the SAUR genes might be derived from segmental duplication and tandem duplications.Synteny analysis established orthology relationships between SAUR genes in maize and sorghum genomes.Some auxin-responsive cis-elements were also found in the upstream sequence of ZmSAURs.An additional selection analysis suggested that significant site-specific selective constraints might have acted on most SAUR paralogs after gene duplication,leading to subgroup-specific functional evolution.Furthermore,comprehensive analysis of the expression profiles provided insights into possible functional divergence among members of the SAUR gene family.These data may provide valuable information for future functional investigations of this gene family.
MATERIALS AND METHODS
Sequence retrieval and characterization analysis
To identify potential members of the SAUR gene family in maize(Zea mays subsp.mays),we performed multiple database searches.Rice and sorghum SAUR sequences(Jain et al.2006;Wang et al.2010)were retrieved and used as queries in BLAST searches against the maize B73 genome sequence(http://www.maizesequence.org/)(Schnable et al.2009).ProtParam tool(http://web.expasy.org/protparam)was used to analyze the physicochemical parameters of SAUR proteins.Subcellular localization prediction of each of these family genes was carried out using the CELLO v2.5 server(http://cello.life.nctu.edu.tw/)(Yu et al.2004)and the TargetP 1.1 server(Emanuelsson et al.2000).IME signals of introns were calculated by IMEter v2.0(Parra et al.2011).
Phylogenetic analyses of the SAUR gene family in maize,Arabidopsis,rice,and sorghum
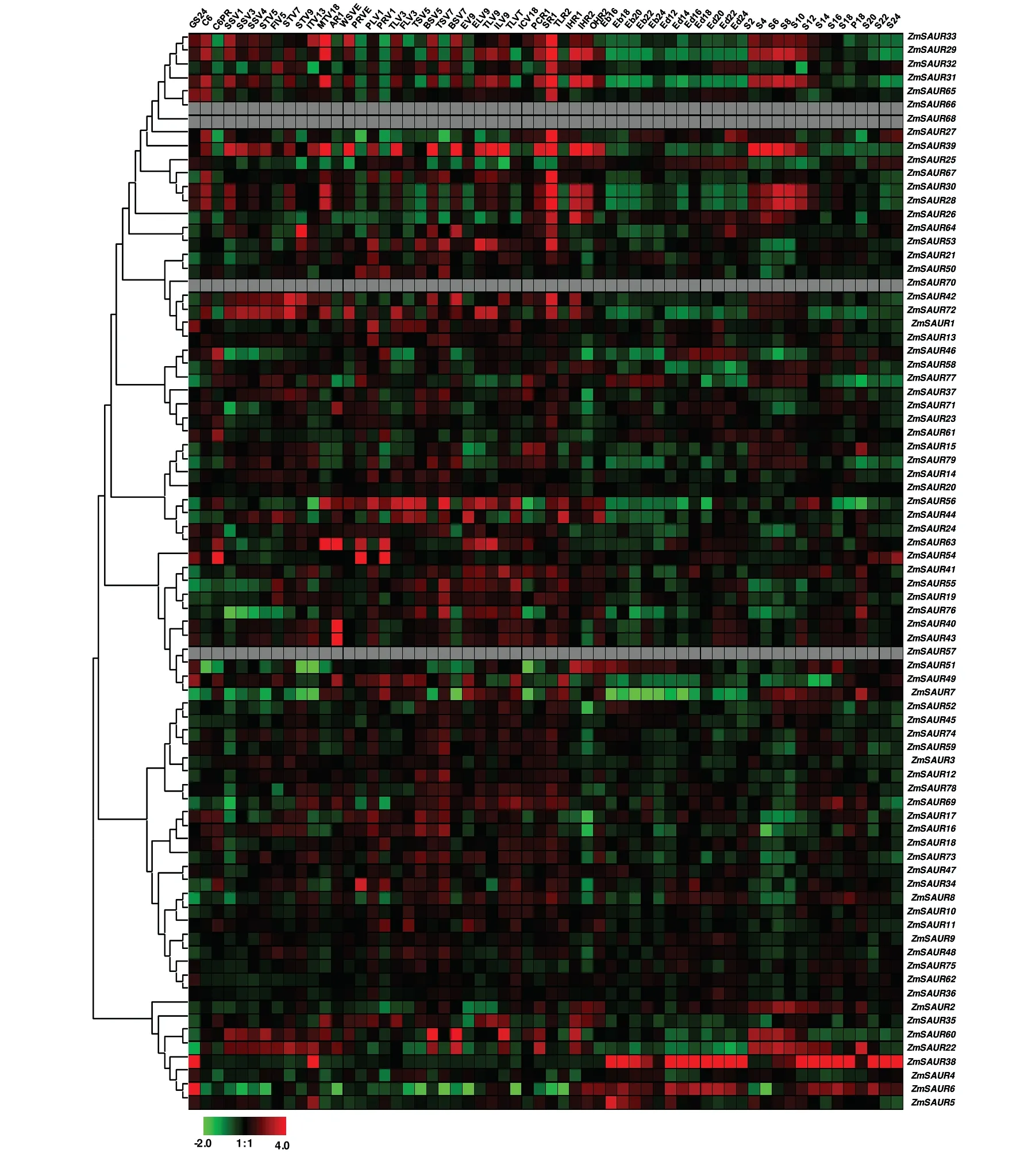
Figure 6.Expression profiles of the maize small auxin-up RNA(SAUR)genesDynamic expression profiles of SAUR genes for 11 different development tissues or organ systems through microarray data.As examples,two clades(i and ii)are also labeled in here.The abbreviation represents specific developmental stages:GS24,Germinating Seed 24 h;C6,Coteoptile 6DAS GH;C6PR,Coteoptile 6DAS Primary Root;SSV1,Stem and SAM(V1);SSV3,Stem and SAM(V3);SSV4,Stem and SAM(V4);STV5,Shoot tip(V5);FIV5,First Internode(V5);FIV7,First Internode(V7);FIV9,Fourth Internode(V9);ITV13,Immature Tassel(V13);MTV18,Meiotic Tassel(V18);AR1,Anthers(R1);WSVE,Whole Seedling(VE);PRVE,Primary Root(VE);PLV1,Pooled Leaves(V1);PRV1,Primary Root(V1);TLV3,Topmost Leaf(V3);FLV3,First Leaf(V3);TSV5,Tip of Stage 2 leaf(V5);BSV5,Base of Stage 2 leaf(V5);TSV7,Tip of Stage 2 leaf(V7);BSV7,Base of Stage 2 leaf(V7);EV9,Eighth Leaf(V9);ELV9,Eleventh Leaf(V9);TLV9,Thirteenth Leaf(V9);ILV9,Immature Leaf(V9);TLVT,Thirteenth Leaf(VT);ICV18,Immature Cob(V18);PCR1,Pre-pollination Cob(R1);SR1,Silks(R1);TLR2,Thirteenth Leaf(R2);IHR1,Innermost Husk(R1);IHR2,Innermost Husk(R2);OHR2,Outer Husk(R2);Eb16,Embryo 16DAP;Eb18,Embryo 18DAP;Eb20,Embryo 20DAP;Eb22,Embryo 22DAP;Eb24,Embryo 24DAP;Ed12,Endosperm 12DAP;Ed14,Endosperm 14DAP;Ed16,Endosperm 16DAP;Ed18,Endosperm 18DAP;Ed20,Endosperm 20DAP;Ed22,Endosperm 22DAP;Ed24,Endosperm 24DAP;S2,Seed 2DAP;S4,Seed 4DAP;S6,Seed 6DAP;S8,Seed 8DAP;S10,Seed 10DAP;S12,Seed 12DAP;S14,Seed 14DAP;S16,Seed 16DAP;S18,Seed 18DAP;P18,Pericarp 18DAP;S20,Seed 20DAP;S22,Seed 22DAP;S24,Seed 24DAP.
Multiple sequence alignments of full-length protein sequences were performed using MUSCLE 3.52(Edgar 2004),followed by manual comparisons and refinement.Phylogenetic analyses of the SAUR proteins based on amino acid sequences were carried out using the NJ method in MEGA v5(Tamura et al.2011).NJ analyses were done using p-distance methods,pairwise deletion of gaps,and default assumptions that the substitution patterns among lineages and substitution rates among sites are homogeneous.Support for each node was tested with 1 000 bootstrap replicates.
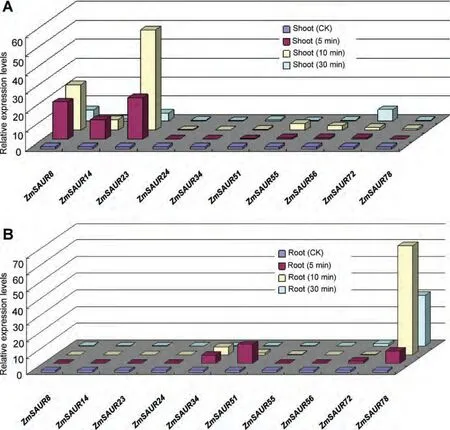
Figure 7.Expression profiles of 10 ZmSAUR genes in response to IAA treatmentQuantitative reverse transcription polymerase chain reaction(qRT-PCR)analyses were used to assess ZmSAUR transcript levels in shoots(A)and roots(B)sampled at 5,10,and 30 min after spraying 10 μM IAA in 1 week maize seedlings.Control(CK)seedlings were grown with normal irrigation.The mean of three experiments stands for their relative expression levels.
Chromosomal location and gene structure of the SAUR genes
Chromosomal locations of the SAUR genes were determined using their annotation information on MaizeSequence(http://www.maizesequence.org)(Schnable et al.2009)for maize and the reference(Wang et al.2010)for sorghum.Gene intron–extron structure information was also collected for genome annotations of maize from MaizeSequence.
Inference of duplication time
MEGA v5(Tamura et al.2011)was used to perform the pairwise alignment of nucleotide sequences of the SAUR paralogs with ClustalW(codons).K-Estimator 6.0 program(Comeron 1999)was used to estimate the Kaand Ksvalues of paralogous genes.Estimates of the evolutionary rates are useful for explaining patterns of macroevolution because Kscan be used as a proxy for time when estimating dates of segmental duplication events.Ksvalue was calculated for each of gene pair and then used to calculate the approximate date of the duplication event(T=Ks/2λ),assuming clock-like rates(λ)of 6.5×10-9synonymous/substitution site/year for maize(Gaut et al.1996).
Upstream sequence analysis of ZmSAUR genes
Firstly,we used expressed sequence tag(EST)information to define the TSS of each ZmSAUR gene.Once the TSS starting positions of ZmSAUR genes were determined on the maize genome,1 000 bp genomic sequences upstream of the TSS were acquired from maize B73 genomic database.PLACE(http://www.dna.affrc.go.jp/PLACE/signalscan.html)(Higo et al.1999),a database of plant cis-acting regulatory DNA elements,was used for searching auxin-responsive elements in the promoter regions of the ZmSAUR genes.
Site-specific selection assessment and testing
We used Selecton Server(http://selecton.tau.ac.il/),a Bayesian inference approach for evolutionary models(Stern et al.2007),to calculate site-specific positive and purifying selection.Ka/Ksvalues are used to estimate two types of substitutions events by calculating the synonymous rate(Ks)and the nonsynonymous rate(Ka),at each codon site.This server implements several evolutionary models that describe in probabilistic terms how the characters evolve.In this study,four of the evolutionary models(M8(ωs≥1),M8a(ωs=1),M7(beta),and M5(gamma))were used.Each of the models uses different biological assumptions and the model that best fits the data can be selected.These models all assume a statistical distribution to account for heterogeneous Ka/Ksvalues among sites.The distributions are approximated using eight discrete categories and the Ka/Ksvalues are computed by calculating the expectation of the posterior distribution(Stern et al.2007).
Protein tertiary structure model-building and visualization analysis
The tertiary structure prediction and visualization analysis of SAUR proteins were performed with SAM server(Karplus et al.2005)and NOC 3.01(http://noch.sourceforge.net/),respectively.
Microarray-based expression analysis
Genome-wide microarray data of maize published by Sekhon et al.(2011)were obtained from the National Center for Biotechnology Information(NCBI)Gene Expression Omnibus(GEO)with Accession Numbers GSE27004.Expression data were gene-wise normalized and hierarchically clustered based on Pearson coefficients with average linkage in the Genesis(v 1.7.6)program(Sturn et al.2002).
Plant materials and exogenous hormone treatment
In this study,1-week-old maize seedlings were used to examine the expression patterns of SAUR genes under hormone stress.Plants were grown in a plant growth chamber at 23±1°C with a 14-h light/10-h dark photoperiod.For IAA treatment,the seedlings were transferred in the solution with 10 μM IAA and then sampled at 5,10,and 30 min.Control(CK)seedlings were grown at 23±1°C with normal irrigation.Three replicates were conducted for each sample.
RNA isolation and qRT-PCR analysis
Total RNA was extracted from by using the Trizol total RNA extraction kit(Sangon,Shanghai,China,SK1321)and was treated with RNase-free DNase-I to remove genomic DNA.Reverse transcription was performed with total RNA(2 μg)using M-MLV(TakaRa,Dalian,China).The cDNA samples were diluted to 8 ng/μL.Triplicate quantitative assays were performed on 1 μL of each cDNA dilution using SYBR Green Master Mix(TakaRa)with an ABI 7500 sequence detection system,according to the manufacturer’s protocol.The mean of three experiments stands for their relative expression levels.The gene-specific primers were synthesized in Sangon.Ten ZmSAUR genes were randomly selected for qRT-PCR analysis from different major branches of the phylogenetic tree.Expression level of the maize Actin 1(GRMZM2G126010)gene was used as the endogenous control.The relative expression level was calculated as 2-ΔΔCTmethod(Livak and Schmittgen 2001).Their gene-specific primers are given in Table S2.
ACKNOWLEDGEMENTS
This project is supported by grants from the National Science Foundation of China(31100923),the National Science Foundation of Jiangsu Province(BK2011467)and Jiangsu University“Youth Backbone Teacher Training Project” in 2012 to J.C.
Aubourg S,Kreis M,Lecharny A(1999)The DEAD box RNA helicase family in Arabidopsis thaliana.Nucleic Acids Res 27:628–636
Ballas N,Wong LM,Theologis A(1993)Identification of the auxinresponsive element,AuxRE,in the primary indoleacetic acidinducible gene,PS-IAA4/5,of pea(Pisum sativum).J Mol Biol 233:580–596
Baumann K,De Paolis A,Costantino P,Gualberti G(1999)The DNA binding site of the Dof protein NtBBF1 is essential for tissuespecific and auxin-regulated expression of the rolB oncogene in plants.Plant Cell 11:323–334
Blanc G,Wolfe KH(2004)Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes.Plant Cell 16:1667–1678
Cai X,Zhang Y,Zhang C,Zhang T,Hu T,Ye J,Zhang J,Wang T,Li H,Ye Z(2013)Genome-wide analysis of plant-specific Dof transcription factor family in tomato.J Integr Plant Biol 55:552–566
Cao J(2012)The pectin lyases in Arabidopsis thaliana:Evolution,selection and expression profiles.PLoS ONE 7:e46944
Cao J,Huang J,Yang Y,Hu X(2011)Analyses of the oligopeptide transporter gene family in poplar and grape.BMC Genomics 12:465
Cao J,Shi F(2012)Evolution of the RALF gene family in plants:Gene duplication and selection patterns.Evol Bioinfor Online 8:271–292
Cao J,Shi F,Liu X,Huang G,Zhou M(2010)Phylogenetic analysis and evolution of aromatic amino acid hydroxylase.FEBS Lett 584:4775–4782
Chae K,Isaacs CG,Reeves PH,Maloney GS,Muday GK,Nagpal P,Reed JW(2012)Arabidopsis small auxin-up RNA63 promotes hypocotyl and stamen filament elongation.Plant J 71:684–697
Comeron JM(1999)K-estimator:Calculation of the number of nucleotide substitutions per site and the confidence intervals.Bioinformatics 15:763–764
Cui J,Liu J,Li Y,Shi T(2011)Integrative identification of Arabidopsis mitochondrial proteome and its function exploitation through protein interaction network.PLoS ONE 6:e16022
Edgar RC(2004)MUSCLE:Multiple sequence alignment with high accuracy and high throughput.Nucleic Acids Res 32:1792–1997
Emanuelsson O,Nielsen H,Brunak S,von Heijne G(2000)Predicting subcellular localization of proteins based on their N-terminal amino acid sequence.J Mol Biol 300:1005–1016
Esmon CA,Tinsley AG,Ljung K,Sandberg G,Hearne LB,Liscum E(2006)A gradient of auxin and auxin-dependent transcription precedes tropic growth responses.Proc Natl Acad Sci USA 103:236–241
Fedorova L,Fedorov A(2003)Introns in gene evolution.Genetica 118:123–131
Flügge UI,Häusler RE,Ludewig F,Gierth M(2011)The role of transporters in supplying energy to plant plastids.J Exp Bot 62:2381–2392
Franco A,Gee MA,Guilfoyle TJ(1990)Induction and superinduction of auxin-responsive genes with auxin and protein synthesis inhibitors.J Biol Chem 265:15845–15849
Gagne JM,Downes BP,Shiu SH,Durski AM,Vierstra RD(2002)The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis.Proc Natl Acad Sci USA 99:11519–11524
Gaut BS,Doebley JF(1997)DNA sequence evidence for the segmental allotetraploid origin of maize.Proc Natl Acad Sci USA 94:6809–6814
Gaut BS,Morton BR,McCaig BC,Clegg MT(1996)Substitution rate comparisons between grasses and palms:Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL.Proc Natl Acad Sci USA 93:10274–10279
Gee MA,Hagen G,Guilfoyle TJ(1991)Tissue-specific and organ-specific expression of soybean auxin-responsive transcripts GH3 and SAURs.Plant Cell 3:419–430
Gil P,Green PJ(1997)Regulatory activity exerted by the SAUR-AC1 promoter region in transgenic plants.Plant Mol Biol 34:803–808
Goda H,Sawa S,Asami T,Fujioka S,Shimada Y,Yoshida S(2004)Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis.Plant Physiol 134:1555–1573
Haberer G,Hindemitt T,Meyers BC,Mayer KF(2004)Transcriptional similarities,dissimilarities,and conservation of cis-elements in duplicated genes of Arabidopsis.Plant Physiol 136:3009–3022
Hagen G,Guilfoyle T(2002)Auxin-responsive gene expression:Genes,promoters and regulatory factors.Plant Mol Biol 49:373–385
Hanada K,Zou C,Lehti-Shiu MD,Shinozaki K,Shiu SH(2008)Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli.Plant Physiol 148:993–1003
Higo K,Ugawa Y,Iwamoto M,Korenaga T(1999)Plant cis-acting regulatory DNA elements(PLACE)database.Nucleic Acids Res 27:297–300
Hoffmann M,Hentrich M,Pollmann S(2011)Auxin-oxylipin crosstalk:Relationship of antagonists.J Integr Plant Biol 53:429–445
Hou K,Wu W,Gan SS(2013)SAUR36,a small auxin-up RNA gene,is involved in the promotion of leaf senescence in Arabidopsis.Plant Physiol 161:1002–1009
Hurst LD(2002)The Ka/Ksratio:Diagnosing the form of sequence evolution.Trends Genet 18:486–487
Jain M,Tyagi AK,Khurana JP(2006)Genome-wide analysis,evolutionary expansion,and expression of early auxin-responsive SAUR gene family in rice(Oryza sativa).Genomics 88:360–371
Kant S,Bi YM,Zhu T,Rothstein SJ(2009)SAUR39,a small auxin-up RNA gene,acts as a negative regulator of auxin synthesis and transport in rice.Plant Physiol 151:691–701
Karplus K,Katzman S,Shackleford G,Koeva M,Draper J,Barnes B,Soriano M,Hughey R(2005)SAM-T04:What is new in proteinstructure prediction for CASP6.Proteins 61(Suppl 7):135–142
Knauss S,Rohrmeier T,Lehle L(2003)The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues.J Biol Chem 278:23936–23943
Kondrashov FA,Rogozin IB,Wolf YI,Koonin EV(2002)Selection in the evolution of gene duplications.Genome Biol 3:RESEARCH0008
Le Hir H,Nott A,Moore MJ(2003)How introns influence and enhance eukaryotic gene expression.Trends Biochem Sci 28:215–220
Lehti-Shiu MD,Zou C,Hanada K,Shiu SH(2009)Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes.Plant Physiol 150:12–26
Lespinet O,Wolf YI,Koonin EV,Aravind L(2002)The role of lineagespecific gene family expansion in the evolution of eukaryotes.Genome Res 12:1048–1059
Liu ZB,Ulmasov T,Shi X,Hagen G,Guilfoyle TJ(1994)Soybean GH3 promoter contains multiple auxin-inducible elements.Plant Cell 6:645–657
Livak KJ,Schmittgen TD(2001)Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method.Methods 25:402–408
Lucas WJ,Groover A,Lichtenberger R,Furuta K,Yadav SR,Helariutta Y,He XQ,Fukuda H,Kang J,Brady SM,Patrick JW,Sperry J,Yoshida A,López-Millán AF,Grusak MA,Kachroo P(2013)The plant vascular system:Evolution,development and functions.J Integr Plant Biol 255:294–388
Lurin C,Andrés C,Aubourg S,Bellaoui M,Bitton F,Bruyère C,Caboche M,Debast C,Gualberto J,Hoffmann B,Lecharny A,Le Ret M,Martin-Magniette ML,Mireau H,Peeters N,Renou JP,Szurek B,Taconnat L,Small I(2004)Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis.Plant Cell 16:2089–2103
Marchler-Bauer A,Anderson JB,Chitsaz F,Derbyshire MK,DeWeese-Scott C,Fong JH,Geer LY,Geer RC,Gonzales NR,Gwadz M,He S,Hurwitz DI,Jackson JD,Ke Z,Lanczycki CJ,Liebert CA,Liu C,Lu F,Lu S,Marchler GH,Mullokandov M,Song JS,Tasneem A,Thanki N,Yamashita RA,Zhang D,Zhang N,Bryant SH(2009)CDD:Specific functional annotation with the Conserved Domain Database.Nucleic Acids Res 37:D205–D210
Mascarenhas D,Mettler IJ,Pierce DA,Lowe HW(1990)Intronmediated enhancement of heterologous gene expression in maize.Plant Mol Biol 15:913–920
McClure BA,Guilfoyle T(1987)Characterization of a class of small auxininducible soybean polyadenylated RNAs.Plant Mol Biol 9:611–623
McClure BA,Hagen G,Brown CS,Gee MA,Guilfoyle TJ(1989)Transcription,organization,and sequence of an auxin-regulated gene cluster in soybean.Plant Cell 1:229–239
Mockevičiūtė R,Anisimovienė N,Merkys A(2006)Comparison of different IAA-ABP complexes formed in kidney bean cell chloroplasts and mitochondria.Biologija 3:102–105
Newman TC,Ohme-Takagi M,Taylor CB,Green PJ(1993)DST sequences,highly conserved among plant SAUR genes,target reporter transcripts for rapid decay in tobacco.Plant Cell 5:701–714
Park JE,Kim YS,Yoon HK,Park CM(2007)Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis.Plant Sci 172:150–157
Park SH,Chung PJ,Juntawong P,Bailey-Serres J,Kim YS,Jung H,Bang SW,Kim YK,Do Choi Y,Kim JK(2012)Posttranscriptional control of photosynthetic mRNA decay under stress conditions requires 3′and 5′untranslated regions and correlates with differential polysome association in rice.Plant Physiol 159:1111–1124
Parra G,Bradnam K,Rose AB,Korf I(2011)Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants.Nucleic Acids Res 39:5328–5337
Prince VE,Pickett FB(2002)Splitting pairs:The diverging fates of duplicated genes.Nat Rev Genet 3:827–837
Punta M,Coggill PC,Eberhardt RY,Mistry J,Tate J,Boursnell C,Pang N,Forslund K,Ceric G,Clements J,Heger A,Holm L,Sonnhammer EL,Eddy SR,Bateman A,Finn RD(2012)The Pfam protein families database.Nucleic Acids Res 40:D290–D301
Reddy VS,Ali GS,Reddy AS(2002)Genes encoding calmodulin-binding proteins in the Arabidopsis genome.J Biol Chem 277:9840–9852
Redman J,Whitcraft J,Johnson C,Arias J(2002)Abiotic and biotic stress differentially stimulate as-1 element activity in Arabidopsis.Plant Cell Rep 21:180–185
Rensing SA,Lang D,Zimmer AD,Terry A,Salamov A,Shapiro H,Nishiyama T,Perroud PF,Lindquist EA,Kamisugi Y,Tanahashi T,Sakakibara K,Fujita T,Oishi K,Shin-I T,Kuroki Y,Toyoda A,Suzuki Y,Hashimoto S,Yamaguchi K,Sugano S,Kohara Y,Fujiyama A,Anterola A,Aoki S,Ashton N,Barbazuk WB,Barker E,Bennetzen JL,Blankenship R,Cho SH,Dutcher SK,Estelle M,Fawcett JA,Gundlach H,Hanada K,Heyl A,Hicks KA,Hughes J,Lohr M,Mayer K,Melkozernov A,Murata T,Nelson DR,Pils B,Prigge M,Reiss B,Renner T,Rombauts S,Rushton PJ,Sanderfoot A,Schween G,Shiu SH,Stueber K,Theodoulou FL,Tu H,Van de Peer Y,Verrier PJ,Waters E,Wood A,Yang L,Cove D,Cuming AC,Hasebe M,Lucas S,Mishler BD,Reski R,Grigoriev IV,Quatrano RS,Boore JL(2008)The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants.Science 319:64–69
Roy SW,Gilbert W(2006)The evolution of spliceosomal introns:Patterns,puzzles and progress.Nat Rev Genet 7:211–221
Schnable PS,Ware D,Fulton RS,Stein JC,Wei F,Pasternak S,Liang C,Zhang J,Fulton L,Graves TA,Minx P,Reily AD,Courtney L,Kruchowski SS,Tomlinson C,Strong C,Delehaunty K,Fronick C,Courtney B,Rock SM,Belter E,Du F,Kim K,Abbott RM,Cotton M,Levy A,Marchetto P,Ochoa K,Jackson SM,Gillam B,Chen W,Yan L,Higginbotham J,Cardenas M,Waligorski J,Applebaum E,Phelps L,Falcone J,Kanchi K,Thane T,Scimone A,Thane N,Henke J,Wang T,Ruppert J,Shah N,Rotter K,Hodges J,Ingenthron E,Cordes M,Kohlberg S,Sgro J,Delgado B,Mead K,Chinwalla A,Leonard S,Crouse K,Collura K,Kudrna D,Currie J,He R,Angelova A,Rajasekar S,Mueller T,Lomeli R,Scara G,Ko A,Delaney K,Wissotski M,Lopez G,Campos D,Braidotti M,Ashley E,Golser W,Kim H,Lee S,Lin J,Dujmic Z,Kim W,Talag J,Zuccolo A,Fan C,Sebastian A,Kramer M,Spiegel L,Nascimento L,Zutavern T,Miller B,Ambroise C,Muller S,Spooner W,Narechania A,Ren L,Wei S,Kumari S,Faga B,Levy MJ,McMahan L,Van Buren P,Vaughn MW,Ying K,Yeh CT,Emrich SJ,Jia Y,Kalyanaraman A,Hsia AP,Barbazuk WB,Baucom RS,Brutnell TP,Carpita NC,Chaparro C,Chia JM,Deragon JM,Estill JC,Fu Y,Jeddeloh JA,Han Y,Lee H,Li P,Lisch DR,Liu S,Liu Z,Nagel DH,McCann MC,SanMiguel P,Myers AM,Nettleton D,Nguyen J,Penning BW,Ponnala L,Schneider KL,Schwartz DC,Sharma A,Soderlund C,Springer NM,Sun Q,Wang H,Waterman M,Westerman R,Wolfgruber TK,Yang L,Yu Y,Zhang L,Zhou S,Zhu Q,Bennetzen JL,Dawe RK,Jiang J,Jiang N,Presting GG,Wessler SR,Aluru S,Martienssen RA,Clifton SW,McCombie WR,Wing RA,Wilson RK(2009)The B73 maize genome:Complexity,diversity and dynamics.Science 326:1112–1115
Sekhon RS,Lin H,Childs KL,Hansey CN,Buell CR,de Leon N,Kaeppler SM(2011)Genome-wide atlas of transcription during maize development.Plant J 66:553–563
Shang H,Li W,Zou C,Yuan Y(2013)Analyses of the NAC transcription factor gene family in Gossypium raimondii Ulbr.:Chromosomal location,structure,phylogeny,and expression patterns.J Integr Plant Biol 55:663–676
Soderlund C,Bomhoff M,Nelson WM(2011)SyMAP v3.4:A turnkey synteny system with application to plant genomes.Nucleic Acids Res 39:e68
Spartz AK,Lee SH,Wenger JP,Gonzalez N,Itoh H,Inzé D,Peer WA,Murphy AS,Overvoorde PJ,Gray WM(2012)The SAUR19 subfamily of small auxin-up RNA genes promote cell expansion.Plant J 70:978–990
Staswick PE,Serban B,Rowe M,Tiryaki I,Maldonado MT,Maldonado MC,Suza W(2005)Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid.Plant Cell 17:616–627
Stern A,Doron-Faigenboim A,Erez E,Martz E,Bacharach E,Pupko T(2007)Selecton 2007:Advanced models for detecting positive and purifying selection using a Bayesian inference approach.Nucleic Acids Res 35:W506–W511
Sturn A,Quackenbush J,Trajanoski Z(2002)Genesis:Cluster analysis of microarray data.Bioinformatics 18:207–208
Takeda S,Kadowaki S,Haga T,Takaesu H,Mitaku S(2002)Identification of G protein-coupled receptor genes from the human genome sequence.FEBS Lett 520:97–101
Tamura K,Peterson D,Peterson N,Stecher G,Nei M,Kumar S(2011)MEGA5:Molecular evolutionary genetics analysis using maximum likelihood evolutionary distance,and maximum parsimony methods.Mol Biol Evol 28:2731–2739
Teale WD,Paponov IA,Palme K(2006)Auxin in action:Signalling,transport and the control of plant growth and development.Nat Rev Mol Cell Biol 7:847–859
Ulmasov T,Murfett J,Hagen G,Guilfoyle TJ(1997)Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements.Plant Cell 9:1963–1971
Wang S,Bai Y,Shen C,Wu Y,Zhang S,Jiang D,Guilfoyle TJ,Chen M,Qi Y(2010)Auxin-related gene families in abiotic stress response in Sorghum bicolor.Funct Integr Genomics 10:533–546
Woodward AW,Bartel B(2005)Auxin:Regulation,action and interaction.Ann Bot 95:707–735
Wu J,Liu S,He Y,Guan X,Zhu X,Cheng L,Wang J,Lu G(2012)Genomewide analysis of SAUR gene family in Solanaceae species.Gene 509:38–50
Xu N,Hagen G,Guilfoyle T(1997)Multiple auxin response modules in the soybean SAUR 15A promoter.Plant Sci 126:193–201
Yang T,Poovaiah BW(2000)Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action.J Biol Chem 275:3137–3143
Yu CS,Lin CJ,Hwang JK(2004)Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions.Protein Sci 13:1402–1406
Zhang X,Firestein S(2002)The olfactory receptor gene superfamily of the mouse.Nat Neurosci 5:124–133
Zhao Y,Zhou Y,Jiang H,Li X,Gan D,Peng X,Zhu S,Cheng B(2011)Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize.PLoS ONE 6:e28488
Zou C,Lu C,Shang H,Jing X,Cheng H,Zhang Y,Song G(2013)Genomewide analysis of the Sus gene family in cotton.J Integr Plant Biol 55:643–653
SUPPORTING INFORMATION
Additional supporting information can be found in the online version of this article:
Figure S1.Neighbor-joining phylogenetic tree of SAURs in Arabidopsis,rice,sorghum,and maize
Figure S2.Gene locations and genomic duplication in sorghum Seventy of 71 SAUR genes are mapped to the Sorghum 9 of 10 chromosomes
SbSAUR71 is localized to unassembled genomic sequence scaffold and thus could not be mapped to any particular chromosome.No SAUR gene was found to localize to the chromosome 5 of the sorghum genome.Paralogous regions in the putative ancestral constituents of the maize genome are depicted using SyMAP v3.4(Soderlund et al.2011)
Table S1.1,000 bp promoter regions upstream of the transcription start site(TSS)of the ZmSAUR genes
Table S2.Primers used in this study
 Journal of Integrative Plant Biology2014年2期
Journal of Integrative Plant Biology2014年2期
- Journal of Integrative Plant Biology的其它文章
- Diversity characterization and association analysis of agronomic traits in a Chinese peanut(Arachis hypogaea L.)mini-core collection
- Characterization of a novel DUF1618 gene family in rice
- Why mosaic?Gene expression profiling of African cassava mosaic virus-infected cassava reveals the effect of chlorophyll degradation on symptom development
- Two hydroxypyruvate reductases encoded by OsHPR1 and OsHPR2 are involved in photorespiratory metabolism in rice
