Molecular characterization of chalcone isomerase (CHI) regulating flower color in herbaceous peony (Paeonia lacti flora Pall.)
WU Yan-qing, ZHU Meng-yuan, JlANG Yu, ZHAO Da-qiu, TAO Jun
Jiangsu Key Laboratory of Crop Genetics and Physiology, College of Horticulture and Plant Protection, Yangzhou University,Yangzhou 225009, P.R.China
RESEARCH ARTICLE
Molecular characterization of chalcone isomerase (CHI) regulating flower color in herbaceous peony (Paeonia lacti flora Pall.)
WU Yan-qing, ZHU Meng-yuan, JlANG Yu, ZHAO Da-qiu, TAO Jun
Jiangsu Key Laboratory of Crop Genetics and Physiology, College of Horticulture and Plant Protection, Yangzhou University,Yangzhou 225009, P.R.China
Chalcone isomerase (CHI) is a key enzyme that converts yellow chalcone to colorless naringenin, playing an important regulatory role in color formation of ornamental flowers. We determined the coding sequence of CHI in herbaceous peony using rapid-ampli fication of cDNA ends (RACE) technology, and subsequently detected the expression pattern of CHI in the inner and outer petals at different developmental stages using qRT-PCR. We cloned the upstream promoter sequences of CHI using genome walking technology and predicted the location of CpG islands and 5´ truncation. In addition, we constructed five dual-luciferase reporter gene carriers and detected the promoter activities of different fragments. Our results showed that the full-length cDNA sequence of CHI was 898 bp, and the 5´-upstream core promoter was located at –1 651 to–2 050 bp region, where contained one CpG island (–1 897 to –2 010 bp) and several important binding sites of transcription factor, such as Sp1, serum response factor (SRF), activating protein (AP)-2alpha and CCAAT/enhancer binding protein (C/EBP)alpha. Expression results showed that the expression of CHI at different developmental stages was generally higher in inner petals than those in outer petals, and the maximum at the bud stage (S1). Thus, this study will provide theoretical basis for an in-depth study of CHI gene function and expression regulation.
herbaceous peony, CHI gene, cloning, promoter, transcriptional activity
1. lntroduction
Flower color affects the ornamental value of plants and directly associates with its commercial development value, so those ornamental plants with novel colors have more market prospects. Paeonia lacti flora Pall. is a traditional famous flower in China. However, although there are rich resources in herbaceous peony cultivars with diverse colors such as pink, red, purple and other cultivars, there is only one yellow cultivar. Therefore, breeding yellow peony cultivars with novel colors has vital signi ficance and market prospects.
Currently, conventional cultivation methods cannot fundamentally solve the de ficiency of yellow herbaceous peony cultivars, suggesting the urgency to start from studying the molecular mechanism of formation and regulation of herbaceous peony color and finally improve color by molecular designing, to promote the sustainable development of the flower industry. Research has found that carotenoids and flavonoids are two categories of pigments mainly involved in yellow color formation. Yellow color formation is associated with the chalcone isomerase (CHI) gene (Zhou et al.2009; Zhao et al. 2014), as a key enzyme catalyzing the formation of colorless naringenin from yellow chalcone. The expression level of CHI directly affects the accumulation of yellow chalcone, a colorless phenotype or light yellow anthoxanthin and red anthocyanin. Therefore, CHI gene plays a very important role in the improvement of yellow flowers (Zhao et al. 2012). The important regulatory role of CHI in color formation has attracted the attention of many researchers. Initially, the researchers isolated the cDNA sequence of CHI from bean (Mehdy and Lamb 1987),Glycine max (Chen et al. 2012), Antirrhinum majus (Sparvoli et al. 1994), and Callistephus chinensis (Li et al. 2006),respectively. Subsequently, CHI from ornamental plants and herbs also gained attention, such as, the full-length cDNA (725 bp) of CHI in Lonicera japonica was cloned successfully (Qiao 2012); the full-length CHI gene (1 163 bp) in Camellia sinensis was cloned with expressed sequence tag(EST) sequencing technology and T4RNA ligase-mediated 5´rapid-ampli fication of cDNA ends (RACE) technology (Ma et al. 2007). However, research on herbaceous peony CHI is still in its early stages, as we are still unclear about its coding sequence, expression pattern and regulatory mechanism of upstream promoter regions affecting gene expression.
In this study, we firstly cloned the cDNA sequence of herbaceous peony CHI gene with RACE technology and revealed the expression pattern of CHI gene in inner and outer petals of Xiangyangqihua (pink outer petals, and yellow inner petals) and Liantai (pink outer petals, and milk white inner petals) at different developmental stages (bud stage (S1), initial bloom stage (S2), blooming stage (S3),and decline stage (S4)) using qRT-PCR. In addition, we further cloned the upstream promoter region of CH1 using genome walking technology, analyzed the promoter region,explored its transcriptional regulatory mechanism through 5´ end truncation treatment by directionally cloning different promoter fragments and detecting promoter activity using a dual luciferase detection system. In this study, we further clari fied the key regulatory regions and candidate transcription factor binding sites of CHI promoter, which provides certain guidance for further study of CHI gene function.
2. Materials and methods
2.1. Plant materials
Petals of herbaceous peony cultivars Xiangyangqihua(anemone type) and Liantai (anemone type) at different developmental stages, S1, S2, S3, and S4, were selected as the experimental materials (Fig. 1). Xiangyangqihua has pink outer petals and yellow inner petals, while Liantai has pink outer petals and milk white inner petals, and their flower color gradually becomes light with the extension of stages from S1 to S4. Samples were collected from the germplasm nursery of herbaceous peony at the School of Horticulture and Plant Protection, Yangzhou University, China from April to July 2015. All materials were immediately frozen in liquid nitrogen after collection, brought back to the laboratory, and stored at –80°C for later use.

Fig. 1 Petal changes at different stages of herbaceous peony.A, Xiangyangqihua. B, Liantai. S1, S2, S3, S4 are the bud,initial bloom, blooming and decline stages, respectively.
2.2. Reagents
3´- and 5´-Full RACE Core Set Kits, SMARTerTMRACE cDNA Ampli fication Kit, Genome Walking Kit, and MiniBEST Agarose Gel DNA Extraction Kit were purchased from TaKaRa(Dalian, China). pEASYTM-T5 Zero Cloning Kit, HEK 293T and DH5α competent cells were bought from TransGen Biotech (Beijing, China). Lipofectamine®3000 Transfection Reagent came from Thermo Fisher Scienti fic (Waltham, MA).Dual-Luciferase Assay System was bought from Promega Promega (Madison, WI, USA). Primers were synthesized by Sangon Biotech (Shanghai, China).
2.3. Primers design
Partial mRNA sequence of CHI gene in herbaceous peony was obtained from previous transcriptome sequencing(Zhao et al. 2014). Speci fic RACE ampli fication primers for 5´- and 3´-end sequences were designed according to the instructions of 3´- and 5´-Full RACE Core Set Kits.Fluorescence quantitative primers were designed after obtaining the full-length coding sequence of CHI by Primer 3.0 Software (http://primer3.ut.ee/). Speci fic primers were designed according to the Genome Walking Kit for genome walking twice. All primers’ information was shown in Table 1.
2.4. 3´ and 5´ RACE
Total RNA was isolated from petals using the RNA Isolation Kit (Invitrogen Life Technologies, China) according to themanufacturer’s instructions. The 1st-strand cDNA was synthesized from 1.0 μg RNA as template using SMARTerTMRACE cDNA Ampli fication Kit (TaKaRa, Dalian, China),followed by two rounds of PCR for ampli fication of 5´ and 3´ ends, according to manufacturers’ protocols. The PCR conditions were 94°C for 3 min for pre-denaturing, then 20 cycles at 94°C for 30 s, 50°C for 30 s, 72°C for 2 min and final extension at 72°C for 10 min. PCR products were separated by electrophoresis on a 1.0% agarose gel and sequenced by Sangon Biotech using a ABI PRISM 377 DNA sequencer (USA).
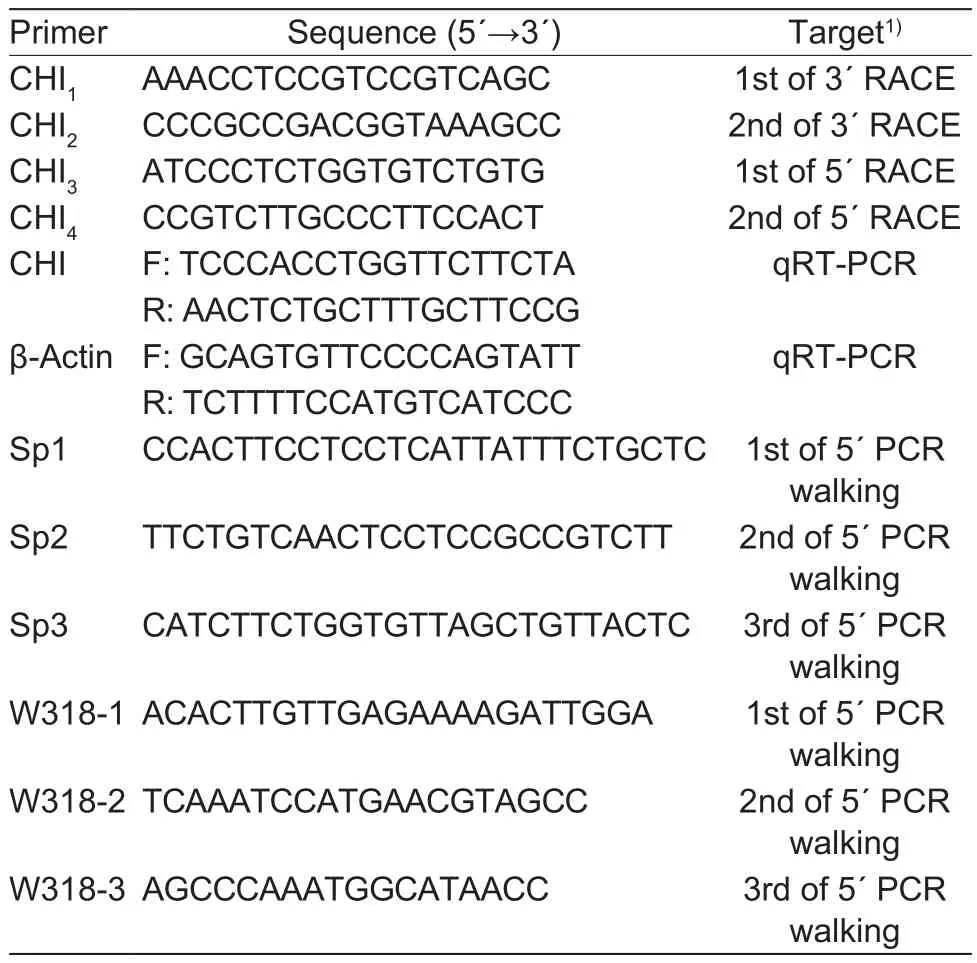
Table 1 Chalcone isomerase (CHI) gene cloning and quantitative primer information
2.5. Differential expression analysis of CHI gene between inner and outer petals at different developmental stages
Inner and outer petals of herbaceous peony cultivars Xiangyangqihua and Liantai at four developmental stages(S1, S2, S3, and S4) were selected as the experimental materials. Total RNA was isolated from the petals using the RNA Isolation Kit according to manufacturer’s instructions.cDNA was synthesized from 1.0 μg total RNA followed by PCR using a RT-PCR Kit (TaKaRa) according to the manufacturer’s instructions. The qRT-PCR reaction mix included 12.5 μL SYBR®Premix Ex TaqTM(2×), 0.5 μL PCR primers(10 pmol mL–1), 0.5 μL Rox reference dye II (50×) and 2 μL cDNA (100 ng μL–1) as a template. The volume was made up to 25 μL with double distilled water. The PCR conditions were 94°C for 5 min for pre-denaturing, then 40 cycles for 95°C for 15 s, 50°C for 15 s, 72°C for 40 s and final extension at 72°C for 10 min. The comparative CTmethod (2–ΔΔCTmethod) (Yuan et al. 2006) was used to analyze the relative expression level of CHI. β-Actin was used as an internal standard for qRT-PCR.
2.6. Genome walking of 5´-upstream sequence
For the 1st-round of PCR walking, PCR was performed using three speci fic sequences (Sp1, Sp2 and Sp3) and genomic DNA as a template. The PCR system and conditions followed the instructions of the Genome Walking Kit (TaKaRa).Thermal asymmetric interlaced PCR (TAIL-PCR) was run three times resulting in the 1st, 2nd and 3rd PCR products.Approximately 5 μL of PCR products were separated on a 1% agarose gel. The DNA was recovered from the target band using a gel extraction kit for sequencing. For the 2nd-round PCR walking, PCR was performed using the speci fic sequences (W318-1, W318-2 and W318-3) (Table 1) to clone about 2 500 bp of 5´-upstream sequences of the CHI gene.
2.7. CpG island prediction and core promoter determination
After obtaining 2 500 bp of 5´-upstream sequence of the CHI gene, we predicted the location of CpG islands using the MethPrimer Software (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi), then truncated per 400 bp according to the 2 000 bp of 5´-upstream sequence. We subsequently designed ampli fication primers (Table 2) and cloned sequences containing different fragments of the promoter region into pGL4.17 (Amp+) expression plasmid. The re-constructed plasmid was extracted with endotoxin-free plasmid extraction kit, and plasmid concentration was measured. HEK 293T cells were seeded into 24-well plates at 1×105cells per well with 500 μL DMEM medium (10% FBS), and cultured at 37°C overnight. Cells were transfected with Lipofectamine 3000(Thermo Fisher Scienti fic, England), medium discarded 24 h after transfection and washed once with PBS. The cells were collected and the promoter activity was analyzed with the Dual-Luciferase Assay System (Promega, UK) on Promega Glomax 2020.
2.8. Transcription factor binding sites of core promoter region
The core promoter region of herbaceous peony CHI gene was determined based on 5´-end truncation and promoter dual-luciferase activity detection, and the transcription factor binding sites of its promoter region was predicted using AliBaba software (http://www.gene-regulation.com/pub/programs/alibaba2/index.html?).
3. Results
3.1. RACE cloning of herbaceous peony CHI coding region
We ampli fied the 3´- and 5´-end cDNA fragments of CHI with RACE using RNA extracted from petals of herbaceous peony as a template. We obtained one band of approximately 800 bp in the ampli fication of the 3´ end (Fig. 2-A),and one band of approximately 300 bp in the ampli fication of the 5´ end (Fig. 2-B); their sizes were consistent with expected fragment lengths. Sequencing and splicing results (Fig. 2-C) showed that the CHI full-length cDNA sequence is 898 bp, the 5´-untranslated regions (UTR) is 194 bp with one complete open reading frame (ORF)(654 bp), one 3´-UTR (50 bp) and one poly A+tail (12 bp).

Table 2 Ampli fication primers for different regions of chalcone isomerase (CHI) upstream sequence

Fig. 2 Rapid-ampli fication of cDNA ends (RACE) cloning and sequence analysis of chalcone isomerase (CHI) gene coding region of herbaceous peony. A and B, the RACE ampli fication fragment of 3´ and 5´ ends, respectively. M, DL2000 marker. C, the nucleotide sequence and deduced amino acid sequence of herbaceous peony CHI gene. The box indicates initiation codon.* indicates termination codon. The sequence underlined shows the poly adenosine acidi fication signal.
This gene sequence has been submitted to GenBank database, whose accession number is JN119872.
3.2. Expression pattern analysis of herbaceous peony CHI gene
We detected the expression level of CHI in the inner and outer petals of Xiangyangqihua and Liantai at different developmental stages (S1-S4) using qRT-PCR. Our results showed, the expression in Xiangyangqihua was signi ficantly higher in the inner petals than that in the outer petals at S2 (P<0.05, Fig. 3-A), signi ficantly lower in the inner petals at S3 than those at S1 and S2, and obviously higher in the outer petals at S1 than those at S2 and S3(P<0.05, Fig. 3-C). The expression in Liantai was signi ficantly higher in the inner petals than that in the outer petals at S1 (P<0.05, Fig. 3-B), signi ficantly higher in the inner petals at S1 than those at S2 and S4, and obviously higher in the inner petals at S3 than that at S2 (P<0.05, Fig. 3-D).Overall, CHI gene expression was generally higher in the inner petals than that in the outer petals, and generally higher at S1 than those at other stages.
3.3. Walking results of 5´-upstream sequence of CHI
After obtaining the cDNA sequence of CHI, we further obtained the 5´-upstream sequence of approximately 2 500 bp by two-step walking ampli fication (Fig. 4-A). Combined with sequencing (Fig. 4-C) and prediction of the CpG island region with MethPrimer software (Fig. 4-B), we found that the upstream sequence of CHI had two CpG islands, located at –657 to –803 bp and –1 897 to –2 010 bp.
3.4. Promoter activity detection and prediction of transcription factor binding sites
Due to the CpG island region located upstream of the ATG codon at 2 000 bp, we divided the upstream 2 000 bp into five segments according to the position of the CpG island after 5´ end truncation (per 400 bp). Five segments were named as P870 (–51 to –450 bp), P869 (–451 to –850 bp),P868 (–851 to –1 250 bp), P867 (–1 251 to –1 650 bp),P866 (–1 651 to –2 050 bp), respectively, then cloned into pGL4.17 plasmids and transfected into HEK 293T cells.Dual luciferase activity detection results showed that the constructed recombinant plasmid pGL-P866 had obvious promoter activity, while others existed almost without any activity (Fig. 5). Finally, we determined the core promoter region of CHI located at –1 651 bp to –2 050 bp. According to the CpG island location and activity detection results, we further predicted its transcription factor binding sites using AliBaba software and found the presence of important transcription factors such as Sp1, serum response factor (SRF),activating protein (AP)-2alpha and CCAAT/enhancer binding protein (C/EBP)alpha (Fig. 6).

Fig. 3 Expression pattern analysis of herbaceous peony chalcone isomerase (CHI) gene. A and C, CHI expression analysis in the inner and outer petals of Xiangyangqihua at different developmental stages. B and D, CHI expression analysis in the inner and outer petals of Liantai at different developmental stages. S1, S2, S3, and S4 are the bud, initial bloom, blooming and decline stages, respectively. Each image shows mean±SE (n=3) from petal tissues. *, signi ficant at P<0.05.

Fig. 4 5´-Upstream sequence of herbaceous peony chalcone isomerase (CHI) gene. A, the genome walking result. M, DL2000 marker; 1, the 1st walking; 2, the 2nd walking. B, CpG island prediction. Blue regions indicate CpG. F1-F5 and R1-R5 indicate the forward and reverse primer, respectively. MSP, methylation speci fic PCR. C, CHI upstream sequence (2 500 bp) after walking.ATG initiation codon was included.

Fig. 5 Dual luciferase activity detection of chalcone isomerase(CHI) promoter region. Bars mean SE.
4. Discussion
Chalcone isomerase (CHI) is the key enzyme related to flavonoid synthesis, and is also one of the enzymes required in biosynthesis of flavonoid pigments. Studies have shown that decreased expression or activity de ficiency of CHI will severely hamper the flavonoid biosynthetic pathway in many plants, leading to signi ficantly reduced anthocyanin and flavonoid contents (van Tunen et al. 1988; Kim et al. 2004),while CHI over-expression can increase flavonoid content(Muir et al. 2001). The importance of CHI in biosynthesis of flavonoids has attracted the attention of many scholars in recent years, meanwhile its application mainly focused on improving the flower color or improving medicinal component of ornamental plants (Wu et al. 2008). Muir et al. (2001)transferred the petunia CHI gene into tomato and obtained transgenic tomato with signi ficantly increased flavonoid content. Nishihara et al. (2005) silenced CHI gene expression in Solanum lycopersicum and found that the chalcone content was signi ficantly increased in petals of tobacco. Bednar and Hadcock (1988) found that CHI mutation in Petunia could increase chalcone content in pollen, thus leading to the formation of yellow or green petals. However, there is still a lack of research aiming at CHI gene of herbaceous peony, whose sequence information and expression pattern are unclear yet.

Fig. 6 Transcription factor binding sites predicted at chalcone isomerase (CHI) core active promoter region. Sp1, RAR-alpha,NF-kappaB, SRF, T3R-alpha, NF-1, AP-2alpha, GATA-1, and C/EBPalpha are all transcription factors. SRF, serum response factor; AP, activating protein; C/EBP, CCAAT/enhancer binding protein.
For the first time, we obtained the full-length cDNA coding sequence of herbaceous peony CHI gene using RACE cloning and sequencing technology, providing the basis for exploring the function and mechanism of CHI through the method of RNA interference, over-expression and gene knockout in the future. Subsequently, we observed the expression pattern of CHI in this study, using Xiangyangqihua and Liantai as the experimental materials. We detected the differential expression of CHI between inner and outer petals of two herbaceous peony cultivars at different developmental stages with qRT-PCR. Results showed that CHI expression level was generally higher in inner petals than outer petals at different stages. The expression quantity was signi ficantly higher in inner petals than outer petals of Xiangyangqihua at S2 and signi ficantly higher in inner petals than outer petals of Liantai at S1; this differential expression may be related to the different colors between inner and outer petals of Xiangyangqihua (pink outer petals, yellow inner petals) and Liantai (pink outer petals, milk white inner petals). Additionally, there also existed a certain signi ficant difference in expression levels at four different stages, which may be due to the fact that the petal color of herbaceous peony gradually becomes lighter with the extension of stage(Zhao et al. 2014). The above results further revealed that the expression level of CHI had direct association with the petal color formation of herbaceous peony. Thus, adjusting the CHI expression level is an important approach to change the petal color in herbaceous peony.
Gene expression level is regulated by transcription factors binding to promoter regions to a certain extent.We therefore further obtained the CHI upstream promoter sequences using Genome walking technology. Combined with bioinformatics analysis and promoter activity detection in this study, we determined a core promoter region of herbaceous peony CHI located at -1 651 to-2 050 bp,amongst this promoter there was one CpG island (-1 897 to-2 010 bp) and important transcription factor binding sites such as Sp1, SRF and C/EBPalpha. Sp1 binding to DNA has a transcription-activating effect, and its binding sequences are often distributed in gene promoters or enhancers in the form of multiple copies (Zhu et al. 2003; Isomura et al. 2005).C/EBP is one transcriptional regulation factor of eukaryotic cells found by Nye and Graves (1990). Studies found that C/EBPalpha proteins are mainly involved in lipid metabolism,its expression level being associated with the synthesis of fatty acids and the mature differentiation of liver cells (Wang et al. 2009), but is unreported in plant research. SRF is one main representative member in MADS-box transcription factor family, regulating the initiation and expression of downstream genes mainly through speci fic recognition and combination of corresponding DNA cis-elements (Alvarez et al. 2000).
This study carried out preliminary study on herbaceous peony CHI gene and determined its core promoter regions(-1 651 to -2 050 bp). As a next step, this study will not only provides favorable material for transcriptional regulatory mechanism of CHI gene by methylation modi fication with BiSul fite Amplicon Sequencing (BSAS), but also provides a theoretical basis for improving flower color breeding in herbaceous peony through biological engineering.
Acknowledgements
This work was supported by the Natural Science Fundation of Jiangsu Province, China (14KJB210011), the Opening Project of Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement, China (2014014), and the Natural Science Foundation of Yangzhou, China (YZ2014033).
Alvarez-Buylla E R, Pelaz S, Liljegren S J, Gold S E, Burgeff C, Ditta G S, Ribas-de-Pouplana L, Martínez-Castilla L, Yanofsky M F. 2000. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proceedings of the National Academy of Sciences of the United States of America,9, 5328–5333.
Bednar R A, Hadcock J R. 1988. Puri fication and characterization of chalcone isomerase from soybeans. Journal of Biological Chemistry,263, 9582–9588.
Chen X Q, Zhang L, Xu H N, Chen L M, Li K Z. 2012. Key enzymes in soybean iso flavones biosynthesis and its metabolic engineering. China Biotechnology,32, 133–138.(in Chinese)
Isomura H, Stinski M F, Kudoh A, Daikoku T, Shirata N, Tsurumi T. 2005. Two Sp1/Sp3 binding sites in the major immediateearly proximal enhancer of human cytomegalovirus have a signi ficant role in viral replication. Journal of Virology,79,9597–9607.
Kim S, Jones R, Yoo K S, Pike L M. 2004. Gold color in onions(Allium cepa): A natural mutation of the chalcone isomerase gene resulting in a premature stop codon. Molecular Genetics and Genomics,272, 411–419.
Li F, Jin Z, Qu W, Zhao D, Ma F. 2006. Cloning of a cDNA encoding the Saussurea medusa chalcone isomerase and its expression in transgenic tobacco. Plant Physiology and Biochemistry,44, 455–461.
Ma C L, Zhao L P, Zhang Y L, Chen L. 2007. Molecular cloning and sequence analysis of chalcone isomerase gene of tea plant (Camellia sinensis). Journal of Tea Science,27,127–132.
Mehdy M C, Lamb C J. 1987. Chalcone isomerase cDNA cloning and mRNA induction by fungal elicitor, wounding and infection. EMBO Journal,6, 1527–1533.
Muir S R, Collins G J, Robinson S, Hughes S, Bovy A,De Vos C H R, van Tunen A J, Verhoeyen M E. 2001.Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols.Nature Biotechnology,19, 470–474.
Nishihara M, Nakatsuka T, Yamamura S. 2005. Flavonoid components and flower color change in transgenic tobacco plants by suppression of chalcone isomerase gene. FEBS Letters,579, 6074–6078.
Nye J A, Graves B J. 1990. Alkylation interference identi fies essential DNA contacts for sequence-speci fic binding of the eukaryotic transcription factor C/EBP. Proceedings of the National Academy of Sciences of the United States of America,87, 3992–3996.
Qiao Z Q. 2012. Full-length cloning and sequence analysis of chs, fls, and chi genes of Lonicera japonica. MSc thesis, Central South Forestry University of Science and Technology, China. (in Chinese)
Sparvoli F, Martin C, Scienza A, Gavazzi G, Tonelli C. 1994.Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.). Plant Molecular Biology,24, 743–755.
van Tunen A J, Koes R E, Spelt C E, van der Krol A R, Stuitje A R, Mol J N. 1988. Cloning of the two chalcone flavanone isomerase genes from petunia hybrida: Coordinate, lightregulated and differential expression of flavonoid genes.EMBO Journal,7, 1257–1263.
Wang X, Huang G, Mei S, Qian J, Ji J, Zhang J. 2009. Overexpression of C/EBP-alpha induces apoptosis in cultured rat hepatic stellate cells depending on p53 and peroxisome proliferator-activated receptor-gamma. Biochemical and Biophysical Research Communications,6, 286–291.
Wu B, Zhu Q L, Guo Y L, Sui S Z, Pi W, Li M Y. 2008.Molecular characteristics of chalcone isomerase gene and its application in gene engineering. Plant Physiology Communications,44, 175–181. (in Chinese)
Yuan J S, Reed A, Chen F. 2006. Stewart CN. Statistical analysis of real-time PCR data. BMC Bioinformatics,7, 85.
Zhao D Q, Jiang Y, Ning C L, Meng J S, Lin S S, Ding W, Tao J. 2014. Transcriptome sequencing of a chimaera reveals coordinated expression of anthocyanin biosynthetic genes mediating yellow formation in herbaceous peony (Paeonia lacti flora Pall.). BMC Genomics,15, 689.
Zhao D Q, Tao J, Han C X, Ge J T. 2012. Flower color diversity revealed by differential expression of flavonoid biosynthetic genes and flavonoid accumulation in herbaceous peony(Paeonia lacti flora Pall.). Molecular Biology Reports,39,11263–11275.
Zhou L, Wang Y, Peng Z. 2009. Advances in study on formation mechanism and genetic engineering of yellow flowers.Scientia. Silvae Sinicae,45, 111–119. (in Chinese)
Zhu W G, Srinivasan K, Dai Z, Duan W, Druhan L J, Ding H,Yee L, Villalona-Calero M A, Plass C, Otterson G A. 2003.Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Molecular and Cellular Biology,23, 4056–4065.
18 January, 2017 Accepted 6 March, 2017
Correspondence TAO Jun, Tel: +86-514-87997219, Fax: +86-514-87347537, E-mail: taojun@yzu.edu.cn
© 2018 CAAS. Publishing services by Elsevier B.V. All rights reserved.
10.1016/S2095-3119(16)61628-3
Section editor HUANG San-wen
Managing editor SHI Hong-liang
 Journal of Integrative Agriculture2018年1期
Journal of Integrative Agriculture2018年1期
- Journal of Integrative Agriculture的其它文章
- Climate change and agriculture: Impacts and adaptive responses in Iran
- Development of elite restoring lines by integrating blast resistance and low amylose content using MAS
- Evaluation of stability and yield potential of upland rice genotypes in North and Northeast Thailand
- ldenti fication of the resistance gene to powdery mildew in Chinese wheat landrace Baiyouyantiao
- New clues concerning pigment biosynthesis in green colored fiber provided by proteomics-based analysis
- Constitutive expression of feedback-insensitive cystathionine γ-synthase increases methionine levels in soybean leaves and seeds
