Development of a stable SCAR marker for rapid identi fication of Ganoderma lucidum Hunong 5 cultivar using DNA pooling method and inter-simple sequence repeat markers
CHAO Wen-zheng, TANG Chuan-hong,, ZHANG Jing-song, YU Ling Honda Yoichi
1 School of Perfume and Aroma Technology, Shanghai Institute of Technology, Shanghai 201418, P.R.China
2 Key Laboratory of Edible Fungus Resources and Utilization (South), Ministry of Agriculture/National Engineering Research Center of Edible Fungi/National R&D Center for Edible Fungi Processing/Key Laboratory of Agriculture Genetics and Breeding of Shanghai, Institute of Edible Fungi, Shanghai Academy of Agricultural Sciences, Shanghai 201403, P.R.China
3 Laboratory of Forest Biochemistry, Graduate School of Agriculture, Kyoto University, Kyoto 6068502, Japan
RESEARCH ARTICLE
Development of a stable SCAR marker for rapid identi fication of Ganoderma lucidum Hunong 5 cultivar using DNA pooling method and inter-simple sequence repeat markers
CHAO Wen-zheng1,2, TANG Chuan-hong2,3, ZHANG Jing-song2, YU Ling1, Honda Yoichi3
1 School of Perfume and Aroma Technology, Shanghai Institute of Technology, Shanghai 201418, P.R.China
2 Key Laboratory of Edible Fungus Resources and Utilization (South), Ministry of Agriculture/National Engineering Research Center of Edible Fungi/National R&D Center for Edible Fungi Processing/Key Laboratory of Agriculture Genetics and Breeding of Shanghai, Institute of Edible Fungi, Shanghai Academy of Agricultural Sciences, Shanghai 201403, P.R.China
3 Laboratory of Forest Biochemistry, Graduate School of Agriculture, Kyoto University, Kyoto 6068502, Japan
The cultivar Ganoderma lucidum Hunong 5 was obtained using cross-breeding. Hunong 5 has high commercial value due to its high polysaccharide and triterpene content. This is the first report of using a DNA pooling method to develop a stable sequence characterized ampli fied region (SCAR) marker for rapid identi fication of the G. lucidum Hunong 5 cultivar.The SCAR marker was developed by first generating and sequencing a distinctive inter simple sequence repeat (ISSR)fragment (882 bp) from G. lucidum Hunong 5 cultivar. A stable SCAR primer pair GLH5F/GLH5R were obtained to identify the cultivar and the SCAR marker is a DNA fragment of 773 bp.
DNA pooling, Ganoderma lucidum, Hunong 5 cultivar, ISSR marker, SCAR marker
1. lntroduction
Ganoderma lucidum, one of the most important and popular medicinal mushrooms, has been used in Asian countries for centuries as a health-preserving and therapeutic agent to increase human vitality and longevity (Sliva 2006; Xu et al.2010a, b). Polysaccharides and ganoderic acids (GAs) are two major bioactive constituents of G. lucidum (Fang and Zhong 2002; Boh et al. 2007; Xu et al. 2010a, b; Wang et al.2012). Some studies have shown that GAs have biological activities such as anti-tumor and anti-HIV-1 activities(Elmekkawy et al. 1998; Wu et al. 2001). With the dramatic growth in the ratio of elderly people in China, more attention is being paid to health. This focus has contributed to the rapid increase in demand for products containing G. lucidum.
Arti ficial cultivation is the main source of raw materials of G. lucidum. The varieties of G. lucidum are one of the most important factors in cultivation. After extensive cross-breeding work, we obtained the Hunong 5 cultivar of G. lucidum. This cultivar has high polysaccharide and triterpene content and was certi ficated by the Shanghai Committee for Appraisal of Crop Varieties, China. However,the strains of G. lucidum can be easily obtained by other companies through tissue isolation. At present, over 100 Ganoderma strains have been cultivated throughout China,although their quality is uneven. The G. lucidum Hunong 5 cultivar can be identi fied using the cultivation experiments according to the “Guidelines for the conduct of tests for distinctness, uniformity and stability of Lingzhi G. lucidum(Curtis) P. Karst.” of Shanghai Institute of Technology,China; however, it is time-consuming and costly. Therefore,development of an inexpensive and reliable method for rapid identi fication of the Hunong 5 cultivar to protect our intellectual property is needed.
PCR-based molecular markers including restriction fragment length polymorphism (RFLP) (Kulkarni et al.1991), simple sequence repeats (Zietkiewicz et al. 1994),random ampli fied polymorphic DNA (RAPD) (Hseu et al.1996), inter-retrotransposon ampli fied polymorphism (Murata et al. 2005), sequence-related ampli fied polymorphism(SRAP) (Sun et al. 2006), inter simple sequence repeats(ISSR) (Wang et al. 2002; Zhang et al. 2007), and ampli fied fragment length polymorphism (AFLP) (Zheng et al. 2009;Nusaibah et al. 2010) have been widely used to detect genetic diversity in mushrooms. Although useful, these methods have some drawbacks, e.g., they cannot be used to identify a speci fic strain. Thus, based on the aforementioned markers,speci fic DNA fragments could be converted into sequence characterized ampli fied region (SCAR) markers (Albani et al.2004; Qin et al. 2006; Li et al. 2008; Su et al. 2008) to rapidly and precisely distinguish different strains. However, it is still time-consuming and dif ficult to attain a strain-speci fic DNA fragment from one strain in a large number of Ganoderma strains cultivated in China.
In contrast, DNA pooling is a reliable genotyping method(Darvasi and Soller 1994) in which equal amounts of DNA from a large number of individual samples are pooled and the single nucleotide polymorphism (SNP) allele frequencies are estimated (Hoogendoorn et al. 2000; Lindroos et al.2002; Sham et al. 2002; Silva et al. 2012). DNA pooling is an ef ficient and labor-saving method that has been used successfully in quantitative trait loci (QTLs) association studies in animals and also been used effectively and ef ficiently as a rapid screen for allelic association in casecontrol studies. DNA samples with genomic DNAs that have high similarity based on genetic diversity analysis are pooled into one DNA pool in equal amounts. In addition, 49 DNA pools have been established according to a previous genetic diversity analysis of Ganoderma strains in China(unpublished data).
The objective of this work was to develop a SCAR marker for a rapid, precise, and effective identi fication of the G. lucidum Hunong 5 cultivar using a DNA pooling method and ISSR markers. This is the first report where a strainspeci fic ISSR marker was converted into a SCAR marker using a DNA pooling method in fungi.
2. Materials and methods
2.1. Ganoderma strains
A total of 152 Ganoderma strains were used to develop SCAR markers. All of the tested strains were stored in the Institute of Edible Fungi, Shanghai Academy of Agricultural Sciences, China.
2.2. DNA extraction
Ganoderma strains were transferred to potato dextrose broth medium (BD) (Franklin Lakes, NJ, USA) for 7–10 days of culturing by liquid shaking at 26–28°C. Genomic DNA was extracted from fresh mycelia using a modi fied LETS method(Chen et al. 1999).
As a positive control, DNA samples were ampli fied with the primers ITS1F (Gardes and Bruns 1993) and ITS4(White et al. 1990) to ensure the quality and integrity of extracted DNA, and to minimize the risk of obtaining falsenegative results. The PCR conditions were as follows: initial denaturation at 94°C for 2 min, followed by 28 cycles of 15 s at 94°C, 30 s at 62°C, 1 min at 72°C, and a final extension of 5 min at 72°C. Then, the ampli fied PCR products were mixed with 1.0 μL DNA loading buffer and analyzed by agarose (1.5%, w/v) gel electrophoresis in a 1×TAE buffer at 140 V. Bands were visualized with ethidium bromide.
2.3. Enterobacterial repetitive intergenic consensus(ERlC)-PCR ampli fication
The purity and concentration of all genomic DNA samples were detected using a Synergy™ HT multidetection microplate reader (Bio-Tek, Winooski, VT,USA). All DNA samples were then diluted with 1×TE buffer to make a final concentration of 40 ng μL-1, and finally stored at –20°C. The ERIC-PCR primers are ERIC1R: 5´-ATGTAAGCTCCTGGGGTTCAC-3´, ERIC2:5´-AAGTAAGTGACTGGGGTGAGCG-3´, synthesized by Sangon Biotech (Shanghai) Co., Ltd. (China), and were used to ampli fied the genomic DNA of all strains. ERIC-PCR was performed in a 25-μL reaction system consisting of 1×PCR buffer (2.5 μL) (TaKaRa Bio Inc., Otsu, Japan), 2.0 mmol L-1MgCl2(TaKaRa), 0.2 mmol L-1dNTPs, 1.3 U Taq DNA polymerase, 0.08 μmol L-1primers, and 100 ng DNA template.The PCR conditions were as follows: initial denaturation at 95°C for 7 min followed by 30 cycles of 1 min at 95°C,1 min at 52°C, 8 min at 65°C, and a final extension of 16 min at 65°C. Then, the ampli fied PCR products were mixed with 1.0 μL DNA loading buffer and analyzed by agarose (1.5%, w/v) gel electrophoresis in 1×TAE buffer at 140 V and bands were visualized with ethidium bromide.
2.4. Genetic diversity analysis of Ganoderma strains
Strain polymorphisms were represented by the molecular weight of the DNA bands. Bands with the same molecular weight were regarded as having the identical character.Ampli fied fragments were scored as 1 for presence and 0 for absence. Similarities between strains were analyzed by the unweighted pair group method with arithmetic mean(UPGMA) in NTSYSpc Software (version 2.11a).
2.5. Construction of DNA pools
According to the previous genetic diversity analysis of Ganoderma strains in China, 49 DNA pools were established (Pool 49 contained Hunong 5 only) (Table 1).DNA samples with high similarity based on genetic diversity analysis were pooled into a DNA pool according to equal volume to make a final concentration of 40 ng μL-1and then stored at –20°C.
2.6. lSSR ampli fication
ISSR PCR primer (5´-(GAA)6-3´) was synthesized by Sangon Biotech (Shanghai) Co., Ltd. (China). ISSR ampli fications were performed in a 25-μL reaction system consisting of 1× PCR buffer (2.5 μL) (TaKaRa), 2.0 mmol L–1MgCl2(TaKaRa), 0.2 mmol L–1dNTPs, 0.4 μmol L–1primers,1.5 U Taq DNA polymerase, and 40 ng DNA template. The PCR conditions were as follows: initial denaturation at 94°C for 3 min, followed by 35 cycles of 1 min at 94°C, 1 min at 52°C, 1 min at 72°C, and a final extension of 5 min at 72°C.
2.7. Cloning and sequencing of the speci fic lSSR DNA fragment
The speci fic genomic DNA fragment of G. lucidum Hunong 5 cultivar was obtained using the ISSR-PCR method and puri fied using an Axygen®AxyPrepTMDNA Gel Extraction Kit (Corning Inc., Corning, NY, USA). The DNA fragment was ligated to a pGEM-T vector and sequenced by Sangon Biotech (Shanghai) Co., Ltd., China.
2.8. Design of primers and analysis of SCAR markers
Primer pairs (forward and reverse) were designed based on the sequence of the speci fic DNA fragment using Primer Premier 6.0 Software (PREMIER Biosoft Intl., Palo Alto, CA,USA). All of the 49 DNA pools were used to test the primerpairs to ensure that the primer pair accurately identi fied the corresponding strain. SCAR-PCR was performed in a 25 μL reaction system consisting of 1×PCR buffer(2.5 μL) (TaKaRa), 2.0 mmol L–1MgCl2(TaKaRa), 0.2 mmol L–1dNTPs, 0.4 μmol L–1primers, 1.5 U Taq DNA polymerase,and a 40-ng DNA template. The PCR conditions were as follows: initial denaturation at 94°C for 3 min, followed by 40 cycles of 1 min at 94°C, 1 min at 52°C, 1 min at 72°C,and a final extension of 5 min at 72°C. Then, the ampli fied PCR products were mixed with 1.0 μL DNA loading buffer and analyzed by agarose (1.5%, w/v) gel electrophoresis in a 1×TAE buffer at 140 V and bands were visualized with ethidium bromide.

Table 1 DNA pooling
3. Results
3.1. Test of extracted DNA through internal transcribed spacer (lTS)-PCR
Clear bands were ampli fied from 152 DNA samples (Fig. 1),which shows that all extracted DNA had good quality and integrity. Therefore, the possibility of false-negative results due to abnormal DNA was ruled out.
3.2. ERIC-PCR
A total of 151 strains of Ganoderma (without Hunong 5)and 3 non-Ganoderma strains (strains 28, 87, and 110)were ampli fied with the ERIC primers, and a total of 105 PCR products were obtained (data not shown). Fig. 2 is the ERIC fingerprint of strains 1–38, showing that the number of bands with each strain ranged from 3 to 17 and the molecular weight ranged from 160 to 5 000 bp. The electrophoresis patterns showed a high DNA polymorphism.
3.3. Clustering analysis
The dendrogram of all 154 strains (Fig. 3), which was based on the data of the ERIC-PCR fingerprints, showed that the similarity coef ficients spread from 0.60 to 1.00.The dendrogram also indicated that these strains could be divided into four distinct groups at the low level of 0.620:(1) Oligoporus gurrualntus (Pk.) Gilbn.et. Ryv. (strain 28);(2) Xuezhi (common name) (strains 87 and 110); (3) strain 117 and 165 were af filiated with subgen. Elfvingia based on morphological classi fications (Zhao 1989); (4) the other strains were af filiated with subgen. Ganoderma based on morphological classi fications (Zhao et al. 1989).
Ganoderma sp. from China (strain 104) and G. lucidum from India (strain 128) had a close relationship, but they were distantly related to the G. lucidum strains from America and Canada. Moncalvo et al. (1995) thought that these geographically separated strains should not be treated as a single species. Overall, 151 strains of Ganoderma were divided into 48 groups at the high similarity level of 0.83.
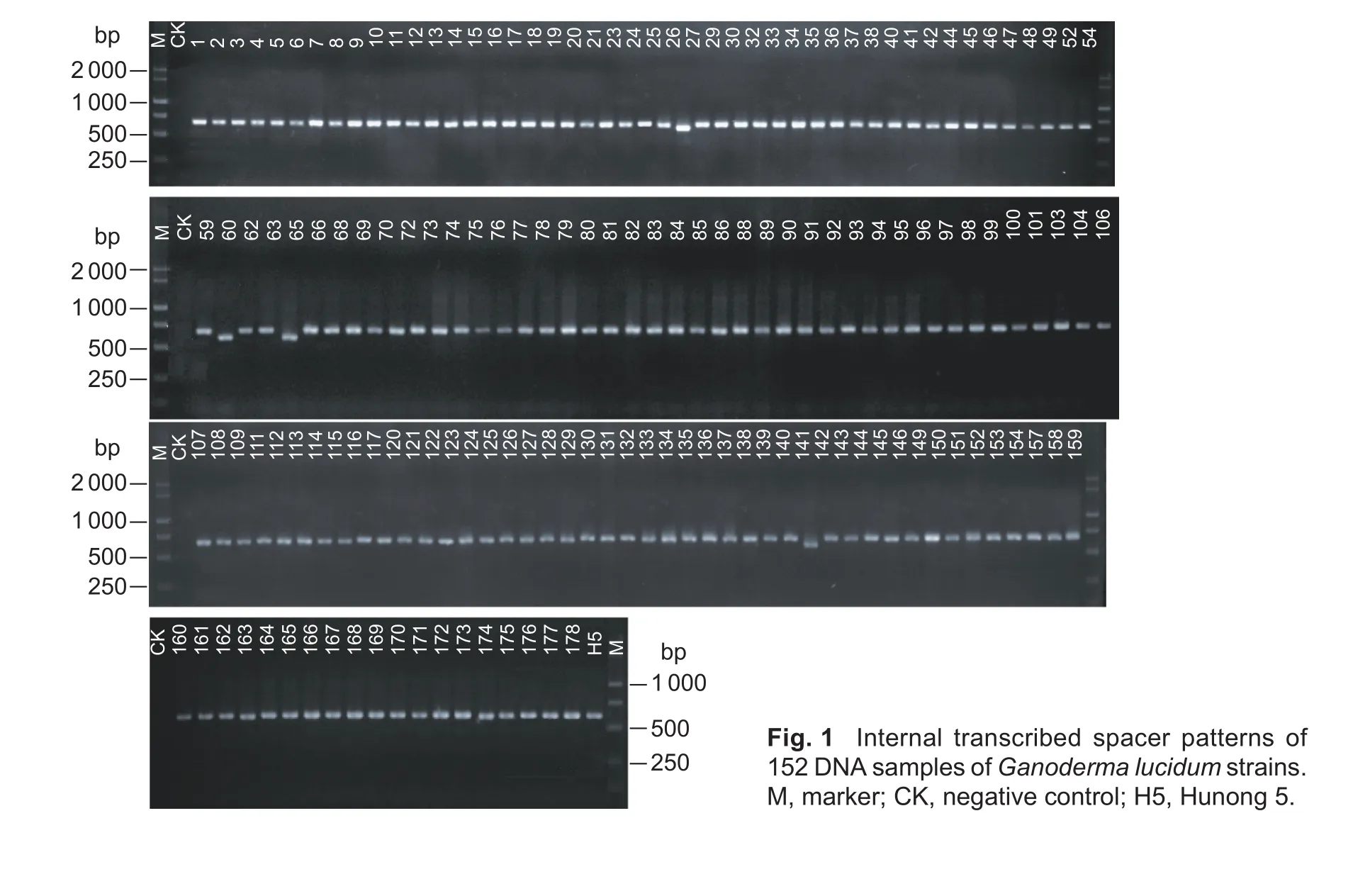
Fig. 1 Internal transcribed spacer patterns of 152 DNA samples of Ganoderma lucidum strains.M, marker; CK, negative control; H5, Hunong 5.
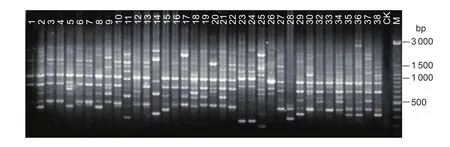
Fig. 2 Enterobacterial repetitive intergenic consensus-PCR fingerprints. CK, negative control; M, marker.
3.4. Selecting and cloning of the speci fic lSSR fragment to G. lucidum Hunong 5 cultivar
ISSR-PCR was performed on 49 DNA pools using the primer 5´-(GAA)6-3´. A single bright band (Fig. 4) matched a sequence that was speci fic to Pool 49, that was strain G. lucidum Hunong 5 cultivar. Therefore, the band was selected for conversion into a strain-speci fic SCAR marker.First, it was puri fied with the aforementioned DNA gel extraction kit, and then cloned and sequenced, which revealed the actual length of the band to be 882 bp.
3.5. Design of primer pairs and development of SCAR marker
The speci fic ISSR DNA fragment of G. lucidum Hunong 5 cultivar was cloned and sequenced. The DNA sequence was submitted to GenBank with the accession no. FY747522.
A speci fic SCAR primer pair was designed and synthesized based on this sequence with Primer Premier 6.0 Software. The sequences of the SCAR primer pair were:GLH5-F, 5´-GACTTTCTCGATGTACTT-3´, and GLH5-R,5´-CTGGTCCAATGCGTCCCT-3´. The SCAR primer pair was tested in 49 DNA pools, and a single bright, distinct, and easily identi fiable band of 773 bp (Fig. 5) was only present in Pool 49. Pool 49 consisted of G. lucidum Hunong 5 cultivar only; therefore, the SCAR markers designed in this study can be used for identi fication of the G. lucidum Hunong 5 cultivar.
4. Discussion
The natural products developed from G. lucidum are being increasingly favored by consumers around the world (Gao et al. 2004; Noguchi et al. 2008; Zhang et al. 2008; Zhao et al. 2012; Suprasert et al. 2014). Different G. lucidum varieties have different active compounds, so in order to manufacture a speci fic product, it is necessary to produce it from corresponding raw material with consistent properties. At present, certain products produced from different G. lucidum raw materials may have different amounts of active compounds, affecting product quality and uniformity(Lin et al. 2001; Fu et al. 2009; Wu et al. 2009; Xiao et al.2009). In addition, the breeding level and ef ficiency of identifying G. lucidum is seriously restricted due to lack of protection for intellectual property rights of G. lucidum cultivars. Therefore, development of an accurate, rapid,and ef ficient way to identify G. lucidum varieties is needed.In this study, a strain-speci fic SCAR marker was proven to be a simple, reliable, and effective method for rapid identification of a particular G. lucidum strain. This marker can be derived from any PCR-based markers, such as RAPD(Chen et al. 2015), ISSR (Merlera et al. 2015), SRAP (Li et al.2007), and AFLP (Che et al. 2003). Compared to RFLP and RAPD, many more polymorphic fragments can be revealed using ISSR, and ISSR is more reproducible and reliable than RAPD (Wang et al. 2002). The pooling technique in genetic association studies reduces the number of required genotyping reactions, and thus is cost-effective (Cui et al.2005; Piergiorge et al. 2014; Ashfaqur et al. 2015). The DNA pooling method has been widely applied in studies of disease mechanisms and animal breeding due to its advantages of saving time, labor, and cost. We successfully used the DNA pooling method to develop a simple, reliable, and effective SCAR marker for identi fication of the G. lucidum Hunong 5 cultivar. This work lays a foundation for further utilization of SCAR markers in other mushrooms, including other Ganoderma cultivars.
5. Conclusion
Using DNA pooling and ISSR markers, the first stable SCAR marker of the G. lucidum Hunong 5 cultivar was successfully developed. This SCAR marker will play two important roles in development of the Ganoderma industry. First, this marker can be used to rapidly identify the cultivar to obtain suitable G. lucidum raw materials and to provide quality health products and medicines to consumers. Second, this marker can protect the intellectual property of the Hunong 5 cultivar and the rights and interests of the breeders,which promotes breeding additional quality cultivars for the Ganoderma industry. In order to protect intellectual properties of more cultivars and to control the quality of raw materials, we are developing new SCAR markers for other Ganoderma cultivars.

Fig. 3 Dendrogram of cluster analysis based on enterobacterial repetitive intergenic consensus-PCR fingerprinting.

Fig. 4 Inter simple sequence repeat (ISSR) band patterns of the primer 5´-(GAA)6-3´ in 49 DNA pools. CK, negative control;P1-P49, Pool 1-Pool 49 of DNA pool; M, marker.

Fig. 5 Banding patterns of the sequence-characterized ampli fied region (SCAR) primer pair GLH5F/GLH5R in 49 DNA pools. CK,negative control; P1-P49, Pool 1-Pool 49 of DNA pool; M, marker.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (31401933) and the Shanghai Municipal Committee of Agriculture, China (G2014070107).
Albani M C, Battey N H, Wilkinson M J. 2004. The development of ISSR-derived SCAR markers around the SEASONAL FLOWERING LOCUS (SFL) in Fragaria vesca. Theoretical and Applied Genetics,109, 571–579.
Ashfaqur R, Andrew H, Daniel S, John M H. 2015. Allele frequency calibration for SNP based genotyping of DNA pools: A regression based local-global error fusion method.Computers in Biology and Medicine,61, 48–55.
Boh B, Berovic M, Zhang J, Zhi B L. 2007. Ganoderma lucidum and its pharmaceutically active compounds. Biotechnology Annual Review,13, 265–301.
Che K P, Xu Y, Liang C Y, Gong G Y, Weng M L, Zhang H Y, Jin D M, Wang B. 2003. AFLP fingerprint and SCAR markers of watermelon core collection. Acta Botanica Sinica,6, 731–735.
Chen J L, Long Y, Khan M A, Wei C L, Shelly Fu, Fu J J. 2015.Development and signi ficance of RAPD-SCAR markers for identi fication of Litchi chinensis Sonn. by improved RAPD ampli fication and molecular cloning. Electronic Journal of Biotechnology,1, 35–39.
Chen X, Romaine C P, Ospina-Giraldo M D, Royse D J. 1999.A polymerase chain reaction-based test for the identi fication of Trichoderma harzianum, biotypes 2 and 4, responsible for the worldwide green mold epidemic in cultivated Agaricus bisporus. Applied Microbiology & Biotechnology,52, 246–250.
Cui J X, Du H L, Zhang X Q. 2005. Rapidly screening SNPs and estimating allelic frequencies by DNA pooling and sequencing. Acta Genetic Sinica,4, 372–377. (in Chinese)
Darvasi A, Soller M. 1994. Selective DNA pooling for determination of linkage between a molecular marker and a quantitative trait locus. Genetics,138, 1365–1373.
El-Mekkawy S, Meselhy M R, Nakamura N, Tezuka Y, Hattori M, Kakiuchi N, Shimotohno K, Kawahata T, Otaka T.1998. Anti-HIV-1 and anti-HIV-protease substances from Ganoderma lucidum. Phytochemistry,49, 1651–1657.
Fang Q H, Zhong J J. 2002. Effect of initial pH on production of ganoderic acid and polysaccharide by submerged fermentation of Ganoderma lucidum. Process Biochemistry,37, 769–774.
Fu L Z, Wu X Q, Li M Y, Cheng J W, He L, Wu Q Q, Li H B, Wei H L, Cao Q. 2009. Analysis and evaluation of polysaccharides and triterpenes contents of fruitbody of Ganoderma lucidum strains. Edible Fungi of China,4, 38–40. (in Chinese)
Gao Y, Chen G, Daiy X, Gao Y, Chen G, Daiy X, Yey J, Zhou S.2004. A Phase I/II study of Ling Zhi mushroom Ganoderma lucidum (W.Curt.:Fr.) lloyd (Aphyllophoromycetideae)extract in patients with coronary heart disease. International Journal of Medicinal Mushrooms,6, 33–40.
Gardes M, Bruns T D. 1993. ITS primers with enhanced speci ficity for basidiomycetes-application to the identi fication of mycorrhizae and rusts. Molecular Ecology,2, 112–118.
Hoogendoorn B, Norton N, Kirov G, Williams N, Hamshere M,Spurlock G, Austin J, Stephens M K, Buckland P R, Owen M J, O’Donovan M C. 2000. Cheap, accurate and rapid allele frequency estimation of single nucleotide polymorphisms by primer extension and DHPLC in DNA pools. Human Genetics,107, 488–493.
Hseu R S, Wang H H, Wang H F, Moncalvo J M. 1996.Differentiation and grouping of isolates of the Ganoderma lucidum complex by random ampli fied polymorphic DNAPCR compared with grouping on the basis of internal transcribed spacer sequences. Applied and Environmental Microbiology,62, 1354–1363.
Kulkarni R K. 1991. DNA polymorphisms in Lentinula edodes,the shiitake mushroom. Applied and Environmental Microbiology,57, 1735–1739.
Li H B, Wu X Q, Peng H Z, Fu L Z, Wei H L, Wu Q Q, Jin Q Y,Li N. 2008. New available SCAR markers: Potentially useful in distinguishing a commercial strain of the superior type from other strains of Lentinula edodes. Applied Microbiology Biotechnology,81, 303–309.
Li X Z, Yuan X J, Jiang S, Du H, Pan J S, Wu A Z, Cai R. 2007.Development of sequence characterized ampli fied region(SCAR) markers in cucumber. Molecular Plant Breeding,3, 393–402.
Lin X S, Li K B, Chen T Q, He X J, Lin Y Q, Zeng X S, Yu Y R.2001. A study on the cultivation properties of eleven strains of Ganoderma lucidum and their esterase isozymes. Acta Agriculturae University Jiangxiensis,1, 81–84. (in Chinese)
Lindroos K, Sigurdsson S, Johansson K, Rönnblom L, Syvänen A C. 2002. Multiplex SNP genotyping in pooled DNA samples by a four-colour microarray system. Nucleic Acids Research,30, e70.
Merlera G G, Muñoz S, Coelho I, Cavaglieri L R, Torres A M, Reynoso M M. 2015. Diversity of black Aspergilli isolated from raisins in Argentina: Polyphasic approach to species identi fication and development of SCAR markers for Aspergillus ibericus. International Journal of Food Microbiology,210, 92–101.
Moncalvo J M, Wang H F, Hseu R S. 1995. Phylogeny of the Ganoderma lucidum complex based on ribosomal DNA sequences. Comparison with traditional taxonomic characters. Mycological Research,12, 1489–1499.
Murata H, Babasaki K, Yamada A. 2005. Highly polymorphic DNA markers to specify strains of the ectomycorrhizal basidiomycete Tricholoma matsutake based on σmarY1, the long terminal repeat of gypsy-type retroelement marY1.Mycorrhiza,15, 179–186.
Noguchi M, Kakuma T, Tomiyasu K, Yamada A, Itoh K, Konishi F, Kumamoto S, Shimizu K, Kondo R, Matsuoka K. 2008.Randomized clinical trial of an ethanol extract of ganoderma lucidum, in men with lower urinary tract symptoms. Asian Journal of Andrology,10, 777–785.
Nusaibah S A, Rajinder S, Idris A S. 2010. Somatic incompatibility and AFLP analysis of four species of Ganoderma strain from oil palm. Journal of Oil Palm Research,22, 814–821.
Qin L H, Tang Q, Chen M J, Pan Y J. 2006. Use of intersimple sequence repeats markers to develop strain-speci fic SCAR markers for Lentinula edodes. FEMS Microbiology Letters,257, 112–116.
Piergiorge R M, Pontes M N, Duarte A V B, Gusmão J. 2014.Haplotype-speci fic single-locus multiplex PCR assay for molecular identi fication of sea-bob shrimp, Xiphopenaeus kroyeri (Heller, 1862), cryptic species from the Southwest Atlantic using a DNA pooling strategy for simultaneous identi fication of multiple samples. Biochemical Systematics& Ecology,54, 348–353.
Sham P, Bader J S, Craig I, O’Donovan M, Owen M. 2002. DNA Pooling: A tool for large-scale association studies. Nature Reviews Genetics,3, 862–871.
Sliva D. 2006. Ganoderma lucidum in cancer research.Leukemia Research,30, 767–768.
Silva S N, Guerreiro D, Gomes M, Azevedo A P, Bezerra D C G, Rueff J, Gaspar J F. 2012. SNPs/Pools: A methodology for the identi fication of relevant SNPs in breast cancer epidemiology. Oncology Reports,27, 511–516.
Su H, Wang L, Liu L, Chi X, Zhang Y. 2008. Use of inter-simple sequence repeat markers to develop strain-speci fic SCAR markers for Flammulina velutipes. Journal of Applied Genetics,49, 233–235.
Sun S J, Gao W, Lin S Q, Zhu J, Xie B G, Lin Z B. 2006.Analysis of genetic diversity in Ganoderma population with a novel molecular marker SRAP. Applied Microbiology &Biotechnology,72, 537–553.
Suprasert P, Apichartpiyakul C, Sakonwasun C, Nitisuwanraksa P, Phuackchantuck R. 2014. Clinical characteristics of gynecologic cancer patients who respond to salvage treatment with Lingzhi. Asian Paci fic Journal Cancer Prevention,15, 4193–4196.
Wang J B. 2002. ISSR markers and their applications in plant genetics. Hereditas,24, 613–616. (in Chinese)
Wang S H, Liang C J, Weng Y W, Chen Y H, Hsu H Y, Chien H F, Tsai J S, Tseng Y C, Li C Y, Chen Y L. 2012. Ganoderma lucidum polysaccharides prevent platelet-derived growth factor-stimulated smooth muscle cell proliferation in vitro and neointimal hyperplasia in the endothelial-denuded artery in vivo. Journal of Cellular Physiology,227, 3063–3071.
White T J, Bruns T, Lee S, Taylor J W. 1990. Ampli fication and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: A Guide to Methods and Applications. Academic Press, SanDiego. pp. 315–322.
Wu T S, Shi L S, Kuo S C. 2001. Cytotoxicity of Ganoderma lucidum triterpenes. Journal of Natural Products,64,1121–1122.
Wu X Q, Fu L Z, Cheng J W, He L, Li W Q, Li H B, Wei H L,Cao Q. 2009. Analysis and evaluation of polysaccharides and triterpenes contents of submerged culturing mycelium of strains of Ganoderma lucidum. Forest By-Product and Speciality in China,5, 1–3. (in Chinese)
Xiao Z T, Yan Y, He H Q, Tan Z Y, Guo J X, Li S W, Ye Y C. 2009. Comparative study of twenty-eight Ganoderma lucidum strains. Guangdong Agricultural Sciences,9,58–60. (in Chinese)
Xu J W, Xu Y N, Zhong J J. 2010a. Production of individual ganoderic acids and expression of biosynthetic genes in liquid static and shaking cultures of Ganoderma lucidum.Applied Microbiology & Biotechnology,85, 941–948.
Xu J W, Zhao W, Zhong J J. 2010b. Biotechnological production and application of ganoderic acids. Applied Microbiology &Biotechnology,87, 457–466.
Zhang R, Huang C, Zheng S, Zhang J, Ng T B, Jiang R, Zuo X, Wang H. 2007. Strain-typing of Lentinula edodes in China with inter simple sequence repeat markers. Applied Microbiology & Biotechnology,74, 140–145.
Zhang Y, Lin Z, Hu Y, Zhang Y, Lin Z, Hu Y, Wang F. 2008.Effect of ganoderma lucidum capsules on T-lymphocyte subsets in soccer players of living high-training low. British Journal of Sports Medicine,42, 819–822.
Zhao H, Zhang Q, Zhao L, Huang X, Wang J, Kang X. 2012.Spore powder of Ganoderma lucidum improves cancerrelated fatigue in breast cancer patients undergoing endocrine therapy: A pilot clinical trial. Evidence-Based Complementary and Alternative Medicine,2012, 809614.
Zhao J D. 1989. The Ganodermataceae in China. Science Press, Beijing. pp. 1-76. (in Chinese)
Zheng L Y, Jia D H, Fei X F, Luo X, Yang Z R. 2009. An assessment of the genetic diversity within Ganoderma strains with AFLP and ITS PCR-RFLP. Microbiological Research,164, 312–321.
Zietkiewicz E, Rafalski A, Labuda D. 1994. Genome fingerprinting by simple sequence repeats (SSR)-anchored polymerase chain reaction ampli fication. Genomics,20,176–183.
6 April, 2017 Accepted 12 June, 2017
Correspondence TANG Chuan-hong, E-mail: tangchuanhong123@163.com; ZHANG Jing-song, Tel: +86-21-62200746, Fax: +86-21-62201530, E-mail: zhangjs888@163.com
© 2018 CAAS. Publishing services by Elsevier B.V. All rights reserved.
10.1016/S2095-3119(17)61825-2
Section editor HUANG San-wen
Managing editor SHI Hong-liang
 Journal of Integrative Agriculture2018年1期
Journal of Integrative Agriculture2018年1期
- Journal of Integrative Agriculture的其它文章
- Climate change and agriculture: Impacts and adaptive responses in Iran
- Development of elite restoring lines by integrating blast resistance and low amylose content using MAS
- Evaluation of stability and yield potential of upland rice genotypes in North and Northeast Thailand
- ldenti fication of the resistance gene to powdery mildew in Chinese wheat landrace Baiyouyantiao
- New clues concerning pigment biosynthesis in green colored fiber provided by proteomics-based analysis
- Constitutive expression of feedback-insensitive cystathionine γ-synthase increases methionine levels in soybean leaves and seeds
