TYMS/KRAS/BRAF molecular profiling predicts survival following adjuvant chemotherapy in colorectal cancer
Anastasios Ntavatzikos,Aris Spathis,Paul Patapis,Nikolaos Machairas,Georgia Vourli,George Peros,Iordanis Papadopoulos,Ioannis Panayiotides,Anna Koumarianou
Abstract
Key words: Colorectal neoplasms; Thymidylate synthase; Untranslated regions;Fluorouracil; KRAS; BRAF; Prognosis
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer in the United States of America while worldwide it is expected to increase by 60% to more than 2.2 million new cases and 1.1 million deaths by 2030[1,2].In 2014,almost 153000 patients died from CRC in the European Union,where it is the second leading cause of cancer death(Eurostat.Cancer statistics - specific cancers)[3].At diagnosis,74%-76% of patients have a localized or regional CRC.Fluoropyrimidines remain the backbone of adjuvant chemotherapy for early stage CRC patients after curative surgery[4,5].Fluoropyrimidines exert their action by different ways mainly by inhibiting the de novo formation of thymidylate (dTMP) from uridylate (dUMP)[6].Other mechanisms of action are more complex than simply inhibition of TS expression,as they involve inhibition of DNA synthesis and function through misincorporation of FdUTP into cellular DNA and inhibition of RNA processing and mRNA translation through the incorporation of FUTP into cellular RNA[7].The use of fluoropyrimidines is associated with reduction of recurrence in only 25% of patients with stage III CRC[8,9].Only 3%-7% of patients with stage II CRC will benefit from adjuvant chemotherapy[10].The variability of observed survival outcomes has been largely attributed to molecular heterogeneity andKRAS,BRAFand thymidylate synthase (TYMS)are being investigated to this end[11].KRASbelongs to the RAS subfamily of genes that encodes a 21-kDa small-GTPase[12].Activating mutations in RAS result in activation of major signaling pathways downstream of epidermal growth factor receptor (EGFR)stimulating cell proliferation and inhibiting apoptosis[13].In the metastatic disease setting,KRASmutations (mKRAS) is a predictor of resistance to EGFR inhibitors and is directly linked to poor patient survival,while its role in the adjuvant setting is under investigation[14-16].
BRAFis an essential part of the RAS/RAF/MAP2K (MEK)-MAPK signaling cascade and its mutations have been likewise associated with inferior survival in CRC patients after curative resection and adjuvant chemotherapy[17,18].
TheTYMSgene (GeneID 7298) is located on the short arm of chromosome 18(18p11.32).There is conflicting evidence on the role ofTYMSpolymorphisms in predicting response to 5FU-based chemotherapy[19-25].The loss of heterozygosity(LOH) at theTYMSlocus on chromosome 18 has been implicated as a factor affecting theTYMS-related resistance to fluoropyrimidine-based therapy[26].
ATYMSpolymorphism of the 5' untranslated region (5'UTR) results by the insertion of a 28 base-pair (bp) sequence (rs34743033)[19].From the resulting alleles that may include two or three 28bp tandem repeats (2R or 3R respectively),the 3R allele was associated with increased TYMS protein expression and TYMS enzyme activity[27,28].G->C single nucleotide polymorphism (SNP) in the tandem repeat sequence [rs2853542] was found to reduce the translational efficiency of a 3R to a 2R[19,29].Based on the presence of SNP polymorphisms (G or C) 3R are characterized as 3RG and 3RC.In addition,the 3'UTR may contain a 6 bp polymorphism (rs34489327)affecting theTYMSmRNA stability,and resulting in increased intratumoralTYMSmRNA[19,30].Depending on the presence of this 6 bp polymorphism,the three resulting genotypes are ins/ins (homozygous for insertion of 6bp),del/del (homozygous for deletion) and ins/del (heterozygous).
Based on all the above,the identification of potential markers that could elucidate which patients' subgroups could benefit most from fluoropyrimidine-based therapy remains an unmet clinical need.
The present study aims to investigate the associations ofTYMSpolymorphisms,LOH,mKRASandBRAFmutations (mBRAF) with clinicopathologic characteristics and survival outcomes of patients with CRC treated with fluoropyrimidine-based adjuvant chemotherapy.
MATERIALS AND METHODS
Patients and clinical data
This was a retrospective study carried out by a single institution (University General Hospital “ATTIKON”).Formalin-fixed paraffin-embedded tissues (FFPE) and clinical data of consecutive patients with CRC referred for adjuvant chemotherapy from January 2005 to January 2007 were retrieved.Of these,only patients with histologies reporting R0 surgical margins and treated with fluoropyrimidine-based adjuvant chemotherapy (and therefore with no redisual disease) were included in the analysis.In these cases,the integrity of mesocolon/mesorectum was preserved.
DNA extraction protocol
DNA was extracted from 5 μm thick FFPE sections,containing at least 30% malignant cells,using a commercially available kit (Purelink Genomic DNA kit,Thermo Fisher Scientific,Germany).DNA was quantified by qPCR (Quant-iT™ PicoGreen®dsDNA Assay Kit,Thermo Fisher Scientific,Germany) and was diluted accordingly to achieve a concentration of 10 ng/μL forTYMSpolymorphisms and 4 ng/μL for mKRASdetection.
TYMS polymorphisms
Analysis was carried-out as previously described[31,32].PCR was performed using 1U of Platinum®Taq DNA Polymerase (Thermo Fisher Scientific,Germany),1.5 mmol/L of Mg and 200 nmol/L of dNTPs and primers.Althoug the same primers were used,5'-UTR amplification was performed using a GC rich amplification kit (PCRX Enhancer System,Thermo Fisher Scientific,Germany) adding 1× of PRCx Enhancer.Genotyping for the 2R/3R polymorphism was performed by running 10 μL of the PCR product on a 1.5% agarose gel and staining with Ethidium Bromide as previously described (Ntavatzikoset al[31]).Similarly,for the 12G>C substitution,10 μL of PCR product was digested with 1U of HaeIII restriction enzyme (Takara,Japan) at 37oC for 1 h and run on an 8% 19:1 polyacrylamide gel.Polyacrylamide gels were used for the analysis of the 3'UTR.LOH analysis was achieved by analyzing the intensity of the 5'UTR and 3'UTR bands of the pictures acquired using the GeneTools software(Syngene,United Kingdom).The sample was categorized as having LOH if one of the bands had an intensity score of < 50% of the other.Samples showing LOH were defined as 2R/3RGLOH,2RLOH/3RG,2R/3RCLOH and 2RLOH/3RC indicating the allele that was partially lost.For quality control,selected products were sequenced to verify the sequence amplified.The amplified product was 242 bp for 3R and 214 bp for 2R polymorphisms,as revealed by the blast of the sequenced products and the alignment with the latest human assemblies.
Mutational analysis
Detection of mKRAS incodons 12 and 13 andBRAFactivating mutation V600E were performed as previously described with a commercially available Real-Time PCR kit(Therascreen KRAS,DxS Diagnostics,United Kingdom) detecting 6 mutations of codon 12 (G12D,G12A,G12V,G12S,G12R,G12C) and 1 mutation of codon 13(G13D)[31,33].A positive reaction mix for all mutations was included.To avoid false negative results caused by PCR inhibitors,a second exogenous reaction was simultaneously taking place.If the sample's ΔCt (Ct of control reaction-Ct mutation reaction) was lower than the value set by the manufacturer,then it was characterized as bearing a mutation.BRAF activating mutation V600E was identified using molecular beacons as previously described[33].One beacon for the wild type and one for the mutant allele were added at a final concentration of 100 nmol/L in a 25 μL PCR reaction containing 1× PCR Buffer,6 mmol/L MgCl2,200 nmol/L dNTPs,300 nmol/L of each primer and 1U of Platinum®Taq.PCR profile applied was 95oC 2 min,followed by 40 cycles of 95oC for 10 sec,62oC for 60 sec and 72oC for 20 sec.DNA extracts from the series of melanoma cell lines SKMEL2 and SKMEL20 were used as positive controls for both the wild type and mutant allele (CLS,Germany).The ABI 7500 Fast (Thermo Fisher Scientific,Germany) was used to perform all Real-Time PCR experiments.
TYMS-gene polymorphisms stratification model
Based on the predictedTYMSprotein expression,5'UTR polymorphisms were assigned into low (2RG/2RG,2RG/3RC,3RC/3RC),medium (2RG/3RG,2RG/3RCLOH,2RG/3RGLOH,2RGLOH/3RC) and high TYMS protein expression group (3RG/3RG,3RG/3RC,2RGLOH/3RG)[31].The effect of each 3'UTR polymorphism was examined against all the others by applying univariate analysis and it was found that only the ins/LOH polymorphism had a statistically significant effect.Based on this finding,3'UTR polymorphisms were allocated into two groups depending on the presence or not of ins/LOH.This classification is depicted in Table 1.
Statistical analysis
Association ofTYMSpolymorphisms with selected clinicopathological characteristics was performed using theχ2test with a 2-sided significance of 0.05.Time-to-event distributions were estimated using the Kaplan-Meier method.For all associations,the level of statistical significance was set at a = 0.05.Overall survival (OS) was defined as the interval between initiation of adjuvant chemotherapy and death of any cause.Disease-free survival (DFS) was defined as the time from adjuvant chemotherapy initiation to the first recurrence or death by any cause.
Surviving patients were censored at the date of last contact.Cox proportional hazards model was used to estimate the relationship of clinicopathological parameters andTYMSpolymorphisms with OS and DFS.The relationship ofTYMSpolymorphisms and the groups to which classified with OS and DFS was assessed by univariate Cox regression analysis.The final multivariate model was selected using a backward selection procedure,starting from an initial model that included all potential risk factors andTYMSpolymorphisms.Model selection was based on likelihood ratio test,while the removal criterion was set at 0.10.All statistical analyses were performed using the SPSS software version 24.0 (SPSS Inc,Chicago,IL,United States).The statistical methods of this study were reviewed by Georgia Vourli from the Department of Hygiene,Epidemiology and Medical Statistics,Medical School University of Athens.

Table 1 TYMS polymorphisms' groups according to risk group and level of expression respectively
RESULTS
Patient characteristics
Medical records of 130 consecutive patients and their FFPE were retrieved for analysis.Patients' clinicopathologic data including age,gender,primary tumor site,histological grade,treatment and survival are shown in Table 2.With a median follow-up of 71.2 mo (range 0.5-157),51 patients (39.2%) experienced disease recurrence while 45 patients (34.6%) died.The 5-year OS and DFS rate was 73.9% and 61.6% respectively.
The frequency ofTYMSpolymorphisms involving G>C SNP and LOH are presented in Table 3.Significant associations were found among patients' tumor characteristics and polymorphisms as shown in Table 4.
Univariate survival analysis
Univariate Cox regression analysis ofTYMSpolymorphisms,mKRASand mBRAF,LOH and selected clinicopathological patients' characteristics are shown in Table 5.Univariate analysis indicated a trend for a better DFS and OS in the group of 5'UTR polymorphisms with medium expression profile (group B),while ins/LOH polymorphism of the 3'UTR were associated with a trend for worse DFS and OS.The analysis of mKRASshowed no significant effect on survival whereasBRAFV600E mutation was associated with increased risk of death.Clinical variables,close to statistical significance,were age (< 65years oldvs≥ 65years old),primary site (rectalvscolon),histological grade (III-IVvsI-II) and stage (IIIvsI and II).
Multivariate survival analysis
Results of the multivariate analysis includingTYMSpolymorphisms,mBRAFand selected clinicopathological characteristics are shown in Table 6.From the 5'UTR polymorphisms,the group A (2RG/2RG,2RG/3RC,3RC/3RC) and group C(3RG/3RG,3RG/LOH,3RG/3RC) were associated with higher risk for disease recurrence and death as compared to group B (2RG/3RG,2RG/LOH and 3RC/LOH).Similarly,group B of 3'UTR polymorphism (ins/LOH) was associated with increased risk of relapse and death as compared to group A.
Kaplan-Meier curves for DFS and OS according toTYMS3'UTR and 5'UTR polymorphisms groups are shown in Figure 1.Stage III increased independently the risk for relapse while theBRAFmutation increased independently the risk for death.Kaplan-Meier curves for OS according to mBRAFare shown in Figure 2.
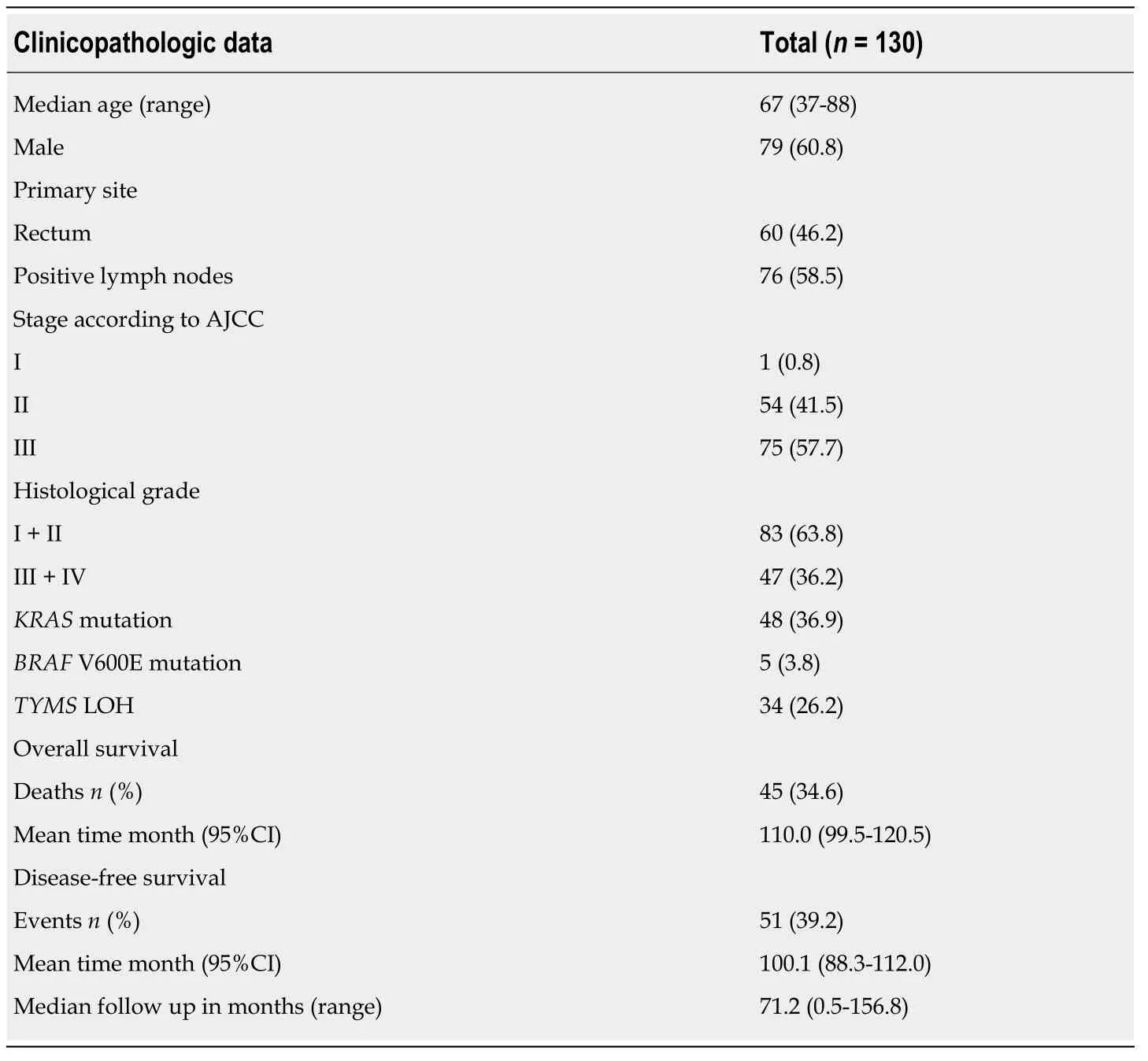
Table 2 Clinicopathologic data for colorectal cancer patients treated with adjuvant chemotherapy
DISCUSSION
This is a retrospective study of 130 patients with CRC treated with surgery and adjuvant chemotherapy,studying for the first time the correlation ofTYMSpolymorphisms,LOH,mKRASand mBRAFwith survival outcomes.We report that the 3'UTR and 5'UTRTYMSpolymorphisms were independent factors associated with risk of disease relapse and death.In particular,ins/LOH increased risk of disease relapse and death,while the group of 5'UTR polymorphisms containing 2RG/3RG,2RG/LOH and 3RC/LOH decreased the risk of disease relapse and death.The study of mKRASpointed out that it did not associate with disease relapse or related death,while the mBRAFincreased independently the risk of death.
Since the early studies of adjuvant chemotherapy treatment with 5FU,23 years ago,there have been two landmark advances in the field[34].The first one involved the incorporation of oral capecitabine as an alternative to intravenously administered 5FU[35].The second was the addition of oxaliplatin to 5FU that lead to a 4.2% absolute improvement in OS of patients with T4 and N1 disease (stage III disease; MOSAIC trial) whereas stage II patients did not benefit[36,37].As clinicopathologic parameters are important but not sufficiently useful in deciding which patients with stage II-III will benefit from adjuvant chemotherapy,molecular markers are essential[38].Several studies reported the association ofTYMSpolymorphisms,TYMSmRNA andTYMSprotein expression with survival in patients with CRC but with inconsistent findings[20-22,24,39-43].A meta-analysis indicated that patients with advanced CRC tumors expressing high levels of TYMS had a poorer OS compared to tumors expressing low levels[44].On the contrary,a subsequent prospective,blinded analysis of TYMS expression in the adjuvant treatment of CRC concluded that TYMS expression did not show a significant prognostic value[45].None of the studies included in their multivariate analysis themBRAFstatus nor the differentTYMSpolymorphisms.
5'UTR polymorphisms
In this studyTYMSpolymorphisms emerged as prognostic factors for survival outcomes in patients treated with surgery and adjuvant chemotherapy.More specifically,the group B (2RG/3RG,2RG/3RCLOH,2RG/3RGLOH,2RGLOH/3RC)was shown to have the lowest risk of recurrence and a trend for lower risk of death when compared to the other two groups A (2RG/2RG,2RG/3RC,3RC/3RC) and C(3RG/3RG,3RG/3RC,2RGLOH/3RG).Similarly,a previous study showed that 5'UTR polymorphisms associated with survival.In particular,they reported that ‘low risk' polymorphisms (2RG/2RG,2RG/3RC,3RC/3RC) were associated with improved DFS regardless chemotherapy treatment[40].On the contrary,a previous study indicated thatTYMS5'UTR polymorphisms do not predict clinical outcome of CRC patients treated with 5-FU based chemotherapy[39].Nevertheless,neither of these two studies took into consideration a combined analysis of 3'UTR polymorphisms,LOH or mBRAFstatus.In addition,the categorization of theTYMS5'UTR polymorphisms into only two groups (high expression group:2RG/3RG,3RC/3RG,3RG/3RG and low expression group:2RG/2RG,2RG/3RC,3RC/3RC),albeit it facilitates statistical processing it also entails the risk of classification error.Indeed,in this way both studies placed the 2RG/3RG with the high expression 3RG/3RG,although 2RG/3RG is a member of the group of heterozygous 5'UTR polymorphisms group that are generally considered to have an intermediate expression profile[27,46].Our study identified heterozygotes such as 2RG/3RG,2RG/LOH and 3RC/LOH,as independent good prognostic factors for recurrence and death in CRC patients treated with surgery and adjuvant chemotherapy.

Table 3 Frequency of TYMS 5'UTR,3'UTR genotypes
3'UTR polymorphisms
In our study,3'UTR polymorphism ins/LOH was found to independently increase the risk for both relapse and death.Comparably,two other studies outlined the negative effect of the ins allele in the therapeutic outcome of CRC patients treated with adjuvant chemotherapy and neoadjuvant setting in rectal cancer patients[41,47].On the contrary,another study found that ins/ins with 2R/3R and any 3'UTR polymorphism with 3R/3R predict longer DFS and OS in CRC patients treated with adjuvant 5FU-based chemotherapy[22].However,in the later study the SNP G>C and LOH status were not taken into consideration.
KRAS and BRAF
The present study showed that the rate of mBRAFidentified in our population (3.8%)was lower than expected,as previously reported rates in the adjuvant setting ranged from 7.9% to 17%[17,36,48].Albeit mBRAFwas not associated with the risk for relapse,mBRAFindependently increased the risk of death.In agreement with our study,three previous studies linked mBRAFto poor survival in relation with MSI status[17,48,49].A fourth study reported that mBRAFwas an adverse prognostic factor for both DFS and OS,independently of MSI status[50].Contrary to these studies,another study indicated that BRAF mutations did not confer a worse prognosis[36].Differently to our study,none of the above studies took into considerationTYMSpolymorphisms.
In this study mutatedKRASdid not emerge as a predictive factor for survival in the univariate analysis.Similar to ours,two previous studies indicated that mKRASwas not associated with survival in stage II/III CRC patients[48,51].On the contrary,a more recent study reported that the risk of recurrence was higher for mKRAScompared to wild typeKRAStumors[52].More recently,another study reported that mKRAShad prognostic impact on DFS and OS independently of microsatellite instability status[50].None of the above studies took into considerationTYMSpolymorphisms.

Table 4 Associations between patient characteristics and TYMS polymorphisms
Other findings of the analysis
We found that patients born from 1943 onwards had more frequently the polymorphism 3RG/3RG and high-grade malignancy tumors (RR 1.730,95%CI:1.088-2.747;P= 0.030).Two previous studies have also linked age toTYMSpolymorphisms and protein expression in CRC[53,54].As more data gather,the differences in the frequency of polymorphisms among generations are of great interest.These differences could derive from epigenetic modifications induced by environmental changes during the course of human life[55].Another important open question is whether in younger generationsTYMSpolymorphisms associate with higher risk of developing aggressive cancer due to changes in the genetic substrate.
We report for the first time that mKRAShad a strong correlation with the polymorphism 3RG/3RC and with polymorphisms that contain only 3RC allele(3RC/3RC,3RC/LOH).Contrary to our findings,a previous study reported no significant relationship between any of theTYMSpolymorphisms with tumor characteristics[56].However,in the understudy grouping ofTYMSpolymorphisms,LOH was not considered.
Limitations
Although the size of this study's patient cohort is one of the largest reported,still it makes it difficult to analyze the large sum of polymorphisms resulting from the combination of 3'UTR and 5'UTR polymorphisms,SNP G>C and LOH.Another limitation is that subsequent chemotherapy lines following disease relapse were not included in the survival analysis.An important limitation is that classification ofTYMSpolymorphisms into groups was based on our statistical analysis and previously published data but requires further validation in prospective trials.
Another important limitation is that the levels ofTYMSprotein expression and activity were not examined.Although immunohistochemical analysis ofTYMSprotein expression is considered important,several studies have shown thatTYMSprotein expression is affected by several factors,like p53 mutation and other genes which proved to affect the final level ofTYMSexpression,like astrocyte elevated
gene-1 (AEG-1) and enolase superfamily member 1 (ENOSF1) during the course of the disease[57-60].It has been reported that there is discordance inTYMSmRNA expression and TYMS protein levels between primary tumors and their metastasis[61-63].Furthermore,the binding of TYMS protein to its own mRNA,as well as the binding of TYMS top53 mRNAcauses translational repression,in an autoregulatory translational manner[64-66].Other significant prognostic and predictive markers such as NRAS,PIK3CA exon 20 and MMR/MSI were not included in this analysis[64-66].

Table 5 Univariate Cox regression analysis for clinicopathological features and genotypes
In conclusion,the group ofTYMSpolymorphisms 2RG/3RG,2RG/LOH and 3RC/LOH and the absence of ins/LOH was associated with better prognosis in CRC patients treated with adjuvant chemotherapy while mBRAFwas associated with increased risk of death.Proof of concept,prospective studies are required to validate our findings.
ACKNOWLEDGEMENTS
We dedicate this manuscript to the late Petros Karakitsos,our mentor and colleague who founded the lab of cellular and molecular biology where this work was carried out.He will always be remembered with love.

Table 6 Multivariate Cox regression analysis for clinicopathological features and selected genotypes

Figure 1 Kaplan-Meier curves for disease free survival and overall survival according to thymidylate synthase polymorphisms:A:Disease free survival(DFS) according to 5' untranslated region (UTR); B:Overall survival (OS) according to 5'UTR; C:DFS according to 3'UTR; D:OS according to 3'UTR.

Figure 2 Kaplan-Meier survival curve for overall survival according to BRAF mutation status (V600E vs WT - wild type).aP < 0.05.
ARTICLE HIGHLIGHTS
Research background
A large proportion of patients with colorectal cancer (CRC) do not benefit from fluoropyrimidine-based adjuvant chemotherapy (FBAC).Fluoropyrimidines are thymidylate synthase(TYMS) inhibitors.Single nucleotide polymorphism (SNP) and various polymorphisms have been discovered in the 5' untranslated region (UTR) and in the 3'UTR of theTYMSgene and their association with the survival of CRC patients is under consideration but with conflicting results.Molecular profiling could help clinicians to identify patients with CRC who may benefit from adjuvant chemotherapy,as shown by the associations of BRAF mutations with inferior survival in CRC patients after adjuvant chemotherapy.Also,although KRAS mutations have been found to be associated with poor patient survival,their role in the adjuvant setting is under investigation
Research motivation
There is a need to study the association of the numerous combinations of TYMS polymorphisms(3'UTR,5'UTR and SNP) with CRC patient survival in a multivariate model including clinicopathological patients' features andKRAS/BRAFmutations.The loss of heterozygosity(LOH) affects polymorphisms and should be included in such a study.
Research objectives
This study aimed to investigate the association of all knownTYMSgene polymorphisms,LOH,KRASandBRAFmutations with the survival of CRC patients treated with adjuvant chemotherapy.
Research methods
Formalin-fixed paraffin-embedded tissues of 130 consecutive patients treated with FBAC were analysed for the detection of TYMS polymorphisms,mKRAS and mBRAF.Patients were classified according to 5'UTR TYMS polymorphisms and the predicted expression profile,into three groups (high,medium and low expression),utilizing the current literature.This categorization could reduce classification errors.Based on the presence or absence of the 3'UTR polymorphism ins/LOH patients were allocated into two groups (high and low risk of relapse),utilizing the results from univariate analysis of the 3'UTR TYMS polymorphisms.Cox regression models examined the associated 5-year survival outcomes
Research results
In this study,where BRAF,TYMS polymorphisms including SNP G>C and LOH were taken into consideration,both 3'UTR and 5'UTR polymorphisms emerged as independent prognostic factors of survival outcome after adjuvant chemotherapy for CRC.More specifically,the group of patients with tumors bearing 5'UTR polymorphisms 2RG/3RG,2RG/LOH and 3RC/LOH was associated with better survival.On the contrary,patients with ins/LOH polymorphism in the 3'UTR had worse survival outcome.Also,mBRAF was found to correlate independently with worse prognosis.
Research conclusions
Knowledge of TYMSgene polymorphisms andBRAFstatus indicates prognosis and could aid clinicians to distinguish the group of patients in need for adjuvant chemotherapy.
Research perspectives
The study of the effect on the survival of CRC patients of the numerous genotypes resulting from the combinations of the 3'UTR and 5'UTR polymorphisms,the SNP and LOH requires larger prospective studies.These studies could validate our findings.Also,they could facilitate the grouping of the TYMS polymorphisms in more than just two groups and thus reduce the classification errors.
 World Journal of Gastrointestinal Oncology2019年7期
World Journal of Gastrointestinal Oncology2019年7期
- World Journal of Gastrointestinal Oncology的其它文章
- Utilizing gastric cancer organoids to assess tumor biology and personalize medicine
- Sarcopenia in pancreatic cancer-effects on surgical outcomes and chemotherapy
- lntraoperative intraperitoneal chemotherapy increases the incidence of anastomotic leakage after anterior resection of rectal tumors
