Highly efficient CRISPR-SaKKH tools for plant multiplex cytosine base editing
ABSTRACT Base editing, as an expanded clustered regularly interspaced short palindromic repeats(CRISPR)-Cas genome editing strategy, permits precise and irreversible nucleotide conversion. SaKKH, an efficient variant of a Cas9 ortholog from Staphylococcus aureus(SaCas9),is important in genome editing because it can edit sites with HHHAAT protospacer adjacent motif (PAM) that the canonical Streptococcus pyogenes Cas9 (SpCas9) or its variants(e.g. xCas9, Cas9-NG) cannot. However, several technical parameters of SaKKH involved base editors have not been well defined and this uncertainty limits their application. We developed an effective multiplex cytosine base editor (SaKKHn-pBE) and showed that it recognized NNARRT,NNCRRT,NNGRGT,and NNTRGT PAMs.Based on 27 targets tested,we defined technical parameters of SaKKHn-pBE including the editing window, the preferred sequence context, and the mutation type. The editing efficiency was further improved by modification of the SaKKH sgRNA. These advances can be applied in future research and molecular breeding in rice and other plants.
1. Introduction
Genetic diversity is a key resource for genetic research and trait improvement in plants. To create new cultivars, scientists and breeders have used multiple methods, including cross breeding,mutation breeding and transgenic breeding to introduce heritable mutations into plant genomes. But to date, generation of precise nucleotide mutations by these approaches is still time-consuming and inefficient [1-4].Base editing,as a newly developed genome editing approach based on the CRISPR/Cas9 system, creates precise and irreversible base mutations in a programmable manner with high efficiency, reduced time cost, and simplicity [5]. The first reported cytosine base editors(CBEs)and subsequent adenine base editors (ABEs) have both been rapidly applied to the efficient generation of point mutations for basic research and molecular breeding[6-9].
The canonical SpCas9 with NGG PAM recognition has been widely used to generate CBEs[6,10,11].For better and effective application, the technical parameters of these base editors[6,11]and of the recently reported SpCas9-based adenine base editors catalyzing cytosine conversions [12] have been well defined, including the editing window, preferred sequence context around the target cytosine (C), and the target C that will show the maximum editing efficiency.
Another Cas9 ortholog from Staphylococcus aureus (SaCas9)recognizes NNGRRT PAM, and its variant SaKKH shows relaxed NNNRRT PAM recognition[13,14].Despite recognizing a long PAM sequence,SaKKH can edit sites including HHHAAT(H = A, T, or C) PAMs that are not recognized by SpCas9 or its variants (e.g. xCas9 and Cas9-NG) [15-18]. For this reason, it has also been used to develop CBEs in animals and plants[19-21]. However, in rice, no base mutation was detected at the single target site tested [20] and only one target with NNNAAT PAM and two targets with NNNGGT PAM showed base mutations [21]. Thus, based on these few edited target sites, the technical parameters of the reported SaKKH involved base editors have not been well defined. These parameters include their editing capability at sites with any other NNNRRT PAMs excluding NNNAAT and NNNGGT, the editing window, the preferred sequence context, and the mutation type.This uncertainty limits their utility.
The objectives of this study were to define these parameters for a new SaKKH-based multiplex cytosine base-editing system in rice and to further improve its editing efficiency by modifying the scaffold of its single guide RNA(Sa-sgRNA).
2. Materials and methods
2.1. Vector construction
The SaKKH [13] was codon-optimized for rice and then synthesized by GenScript Corp. (Nanjing, China). Its D10A nickase (SaKKHn) was point-mutated by PCR. SaKKHn was used to replace the SpCas9n(D10A)in the SpCas9n-pBE-basic vector [22], generating the vector SaKKHn-pBE-basic. Following Xie et al. [23], target sequences were cloned before the scaffold of SaCas9 sgRNA (Sa-sgRNA) using BsaI to generate SaKKHn-pBE constructions. The scaffold-modified Sa-sgRNA generated by extending the duplex by 3 or 5 bp was used to replace the original Sa-sgRNA in the SaKKHn-pBE constructions to obtain the corresponding SaKKHn-pBE constructions with the modified Sa-sgRNA. Target sequences and targets addressed with one construct are shown in Tables S1 and S2 respectively.The primers used in this study are listed in Table S3.
2.2. Rice transformation
All the constructed binary vectors were introduced into Agrobacterium tumefaciens strain EHA105 using a freeze/thaw method.Embryogenic calli induced from mature seeds of the rice japonica cultivar Nipponbare were infected with the above Agrobacterium for rice transformation as described previously[24]. After incubation with Agrobacterium for 10 min, the calli were recovered for three days and then were cultured on selection medium containing 50 μg mL−1hygromycin. After four weeks, hygromycin-resistant calli were transferred to regeneration medium (not containing hygromycin) for shoot induction for one month.The 4-5-cm shoots were transferred to rooting medium for root induction. After two weeks, T0plants were harvested for further study.
2.3. Identification of transgenic T0 plants
Genomic DNA of each harvested T0plant was extracted using a DNA-quick Plant System kit (TianGen Biotech, Beijing,China). The target locus was amplified by PCR using Cas9-specific primers (Table S3) and samples showing an 898-bp nucleic acid fragment on agarose gel electrophoresis were identified as transgenic T0plants.
2.4. Mutant identification
Several transgenic T0plants were used to identify C-to-T conversions.Fragments covering the target site were amplified with specific primers(Table S3).The PCR products were purified using an EasyPure PCR Purification Kit (TransGen Biotech,Beijing, China) and then sent for Sanger sequencing (Tsingke Biological Technology, Beijing, China) to identify mutations.Base-editing efficiency was defined as the percentage of T0plants with any target C-to-T conversion among all the transgenic samples. Editing efficiency of single C-to-T conversion was defined as the percentage of mutants with C-to-T substitution at a specific single position among all the transgenic samples. A homozygous T0plant was defined as one in which all C-to-T mutations in the target were homozygous.
3. Results and discussion
3.1. SaKKHn-pBE enables efficient multiplex base editing
The tRNA-sgRNA system has been identified as an effective platform for multiplex genome editing and also improves sgRNA expression level [23], so we choose to develop a tRNA-sgRNA-based SaKKH-CBE. Because SaKKH requires NNNRRT PAMs [13], including NNNAAT, NNNGGT,NNNAGT and NNNGAT, and only two PAMs (NNCAAT and NNTGGT)were reported in previous study[21],we first tested the base-editing capability at sites with all four PAMs.Considering the four bases(G, A,C, or T) at the third position of the PAM, we chose 16 targets (T1-T16) with 16 different PAMs from the OsPDS, OsWaxy, OsGRF4, OsALS, OsMPK5, OsNRT1.1B, and OsMPK2 genes (Fig. 1, Table S1). We reoptimized the codons of SaKKH [13] for rice and obtained its D10A nickase (SaKKHn) by point mutation. SaKKHn fused with the commonly used Petromyzon marinus cytidine deaminase 1 (PmCDA1) and uracil DNA glycosylase inhibitor (UGI)was then placed under control of the Oryza sativa ubiquitin(OsUbq) promoter in a tRNA-sgRNA system, generating a PmCDA1-based CBE designated as SaKKHn-pBE (Fig. S1).Following Xie et al. [23], each target sequence was cloned before the Sa-sgRNA scaffold to construct one base-editing plasmid for Agrobacterium-mediated transformation (Table S2).
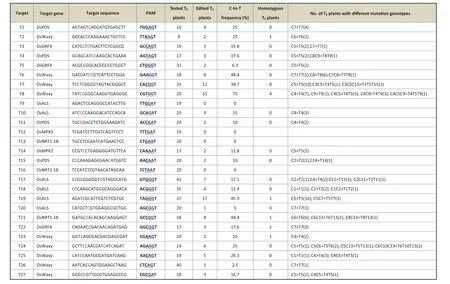
Fig.1- Base-editing efficiencies of SaKKHn-pBE in rice transgenic T0 plants.The Cs targeted for mutation are shown in pink.NRR at protospacer adjacent motif(PAM)position 3,4,and 5 is underlined and RR is shown in blue.
In stable transgenic T0plants, of 16 target sites, 12 were base-edited by SaKKHn-pBE with frequencies ranging from 6.3% to 75.0%, including four NNARRT, four NNCRRT, two NNGRGT, and two NNTRGT PAMs (Fig. 1). No C-to-T base mutation was detected at two NNGRAT sites(T9 and T13)and two NNTRAT PAM sites (T12 and T16), possibly because they were hardly edited (Fig. 1). These results suggested that SaKKHn-pBE could efficiently edit the rice genome at NNNRRT PAM sites. To further investigate the multiplex base-editing capability of SaKKHn-pBE, 15 target sites in OsALS, OsNRT1.1B, OsGRF4, and OsWaxy, T17-T31, were designed, including nine NNNGGT, five NNNAGT, and one NNNGAT PAM site (Table S1). There were three target sequences in each plasmid. Finally, C-to-T base conversions were detected at 11 target sites(T17-T27),with mutation rates ranging from 2.5% to 45.9% (Fig. 1). This result indicates that SaKKHn-pBE permits efficient multiplex base editing.
3.2. Technical parameters of SaKKHn-pBE
All 23 edited target sites were taken together to investigate the technical parameters of SaKKHn-pBE. Analysis of the editing window showed that it spanned positions 1 to 15 within the protospacer from the 5′ end of the target (Fig. 1,Fig.2A),which was wider than that in the reported SpCas9-related CBE (3 to 9) [11]. Given that editing of C4 to C9 showed high preference over Cs from other positions (Fig.2A), the editing window spanned mainly positions 4 to 9 in the target.
We next investigated the preferred sequence context around the target C. First, we found that among all NCs being edited within the 23 target sites, CC and TC were predominant in the order CC > TC ≫GC > AC (Fig. 2B). This finding indicated that C positioned just 3′ to a C or T was preferred for editing by SaKKHn-pBE. Second, we calculated the proportion of each NC being edited in the total corresponding NC in the editing window,which further confirmed that CC and TC were far more likely to be edited than GC and AC (Fig. 2C). We further investigated it in a divided editing window and found that TC in the main editing window(4-9 bp)showed the highest editing preference,while CC was second in 4-9 bp and third in 1-3 bp (Fig.2C). By comparison,the editing preferences for CC and TC in 10-15 bp were relatively low(Fig.2C).Additionally,in 1-3 bp,GC and AC were rarely edited(Fig.2C).In conclusion, GC and AC are relatively difficult to edit, especially AC, in contrast to the poor editing only of GC by the reported SpCas9 CBEs with rat cytidine deaminase rAPOBEC1 [6]. Just as SpCas9-CBEs were further evolved with improved deaminases to overcome target sequence with poor GC [25], SaKKHn-pBE could be improved to overcome AC and GC in future. Lastly, we also examined the effect of the nucleotide just 3′ of the target C and found that NCG was most preferred for editing, while NCT was the least common when the upstream nucleotide was fixed as T or C (Fig.2D).
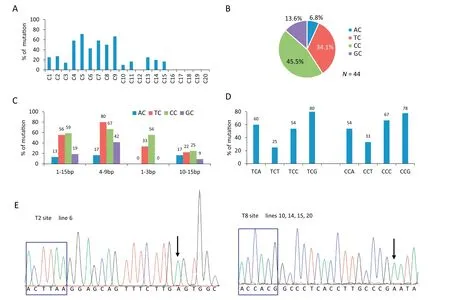
Fig.2- Cytosine base mutation by SaKKHn-pBE in transgenic T0 rice plants.(A)Proportion of target sites with mutation at a given position among all edited target sites containing C at the position.X-axis represents the position of C from the 5′end of the target.(B) Distribution of each NC among all 44 NC being edited.(C)The proportion of each NC being edited in the total corresponding NC in the editing window.(D)The proportion of each TCN or CCN being edited in the total corresponding NCN in the editing window.(E)Sequencing chromatograms at T2 and T8 sites of all homozygous T0 lines produced by SaKKHn-pBE.Gto-A conversions in the opposite strand are shown.Arrows indicate edited bases.PAM sequences are shown in blue boxes.
For mutation type, single C-to-T mutants were most common, and Cs with the highest editing efficiency at each target almost all corresponded to single mutants in T0plants(Fig.1).Homozygous mutations were seldom generated(Fig.1,Fig.2E).
3.3. Increasing the editing efficiency of SaKKHn-pBE by SasgRNA optimization
Given that the scaffold of sgRNA from SpCas9(Sp-sgRNA)has been modified to increase editing efficiency[26]and shares a similar structure with Sa-sgRNA [14], we focused on SasgRNA optimization to improve the editing efficiency of SaKKHn-pBE. Because extending the duplex in Sp-sgRNA by 3 and 5 bp was reported to increase the knockout efficiency of SpCas9 [26], we speculated that similar optimizations in SasgRNA would also increase the editing efficiency of SaKKHnpBE.Three or five bp were introduced as O+3 bp or O+5 bp,respectively, for extension of the duplex (Fig. 3A). We first chose three NNCGGT (T7, T21, T22) and two NNCAGT (T25,T26) PAM sites. Both SaKKHn-pBE variants (O + 3 bp and O+5 bp)appeared to increase the editing frequency(Table 1).Another two NNGGGT sites (T5, T18) and one NNTGGT site(T8) were tested, further confirming the results (Table 1).Apart from an increase in the editing frequency overall,SaKKHn-pBE with O + 3 bp produced an especially marked increase in editing of more target Cs in the editing window(Fig. 3B). SaKKHn-pBE with O + 3 bp outperformed SaKKHnpBE with O + 5 bp at most targets (Table 1, Fig. 3B). These results indicated that, like Sp-sgRNA, Sa-sgRNA could be optimized for increased editing frequency.Other approaches,such as adopting high-efficiency nuclear localization signals and using surrogate systems, might further increase the frequency[27-31].
4. Conclusions
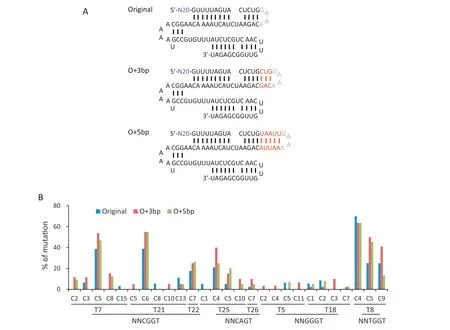
Fig.3-Sa-sgRNA optimization for improved efficiency in rice.(A)Schematic representation of Sa-sgRNA(original)and the two variants(O+3 bp and O+ 5 bp)with extending bp highlighted in red.(B) Summary of editing efficiencies of single C-to-T conversions for different base editors at eight target sites.
We developed SaKKHn-pBE, an effective SaKKH-based multiplex base-editing system with well-studied editing parameters. Among these parameters, the wide editing window and consistent production of single mutants of C with the highest editing efficiency in the target demonstrate its high potential. We improved the editing efficiency of SaKKHn-pBE by modifying the structure of Sa-sgRNA. Besides the further application of SaKKHn-pBE, these features also offer potential for future applications in adenine base editing [8] or even the recently developed prime editing technology [32] in rice and provide a reference for other plants and animals.
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2020.03.002.
Author contributions
J.Y. and C.Z. designed the experiments. C.Z., F.W., S.Z., G.K., J.S. and L.L. performed all the experiments. C.Z. analyzed the results. J.Y. supervised the project. J.Y. and C.Z. wrote the manuscript.
Declaration of competing interest
The authors have submitted patent applications based on the results reported in this paper.
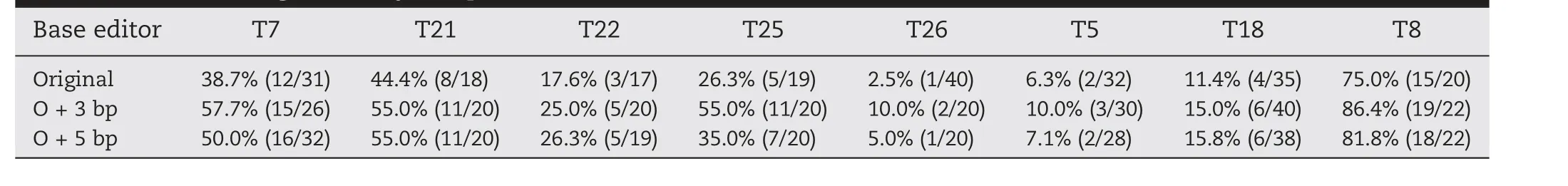
Table 1-Base-editing efficiency comparison of three base editors.
Acknowledgments
This work was supported by the Beijing Scholars Program[BSP041].
- The Crop Journal的其它文章
- Base editing in plants:Current status and challenges
- Increasing fidelity and efficiency by modifying cytidine base-editing systems in rice
- Improving the efficiency of the CRISPR-Cas12a system with tRNA-crRNA arrays
- Developing high-efficiency base editors by combining optimized synergistic core components with new types of nuclear localization signal peptide
- Intron-targeted gene insertion in rice using CRISPR/Cas9: A case study of the Pi-ta gene
- Generation of seed lipoxygenase-free soybean using CRISPR-Cas9

