Efficacy of Xuebijing injection for the treatment of coronavirus disease 2019 via network pharmacology
Yu-Liang Zhang, Qian Cui, Dou Zhang, Xin Ma, Guo-Wei Zhang*
Efficacy of Xuebijing injection for the treatment of coronavirus disease 2019 via network pharmacology
Yu-Liang Zhang1, Qian Cui1, Dou Zhang1, Xin Ma1, Guo-Wei Zhang1*
1College of Traditional Chinese Medicine, Hebei University, Baoding 071000, China.
: To evaluate the mechanism of Chinese patent drug Xuebijing (XBJ) injection in the treatment of a new coronavirus disease 2019 (COVID-19) based on network pharmacology and molecular docking technology.: The TCMSP database was employed to collect and screen the active ingredients of the Chinese herb contained in the XBJ injection. The GeneCards database and STRING database were applied to collect and expand the targets of COVID-19 and compare and screen the related targets of COVID-19 by XBJ injection. Cytoscape was employed to build a network connecting Chinese medicine, compounds, targets, disease, and topology analysis was performed via the Network Analyzer to screen the key ingredients and targets. The software of Schrödinger molecular docking was used to verify the binding activity of the key ingredients of XBJ injection and the key targets of COVID-19. Metascape platform and DAVID database were utilized to conduct Gene Ontology analysis and Kyoto Encyclopedia of Genes and Genomes analysis on the key targets of COVID-19 treated by XBJ injection.Eight key compounds and 15 key targets were screened and verified by molecular docking; these key compounds included luteolin, quercetin, baicalein, and kaempferol. The key targets included DPP4, AR, ESR1, CALM1, and protein kinase 1. Gene Ontology analysis involved an apoptosis and hypoxia reaction and the changes in blood vessel morphology. Kyoto Encyclopedia of Genes and Genomes analysis involved signaling pathways of hypoxia inducible factor-1, VEGF, and PI3K/AKT/NF-κB.: The mechanism of XBJ injection when used to treat COVID-19 should be further investigated as the key compounds in XBJ regulated the expression of key targets such as protein kinase 1, VEGF-A, B-cell lymphoma-2, and TNF, which affected the COVID-19 receptors such as angiotensin-converting enzyme 2 and signaling pathways like hypoxia inducible factor-1, PI3K-Akt, and NF-κB, which alleviated the inflammation, respiratory distress, and hypoxia caused by COVID-19 infection.
Network pharmacology, Molecular docking, COVID-19, Xuebijing injection, Luteolin, Quercetin
The Chinese drug formula Xuebijing (XBJ) injection ameliorated the inflammatory reaction, respiratory distress, and hypoxia caused by coronavirus disease 2019 infection. Its underlying mechanism is via affecting the angiotensin-converting enzyme 2 and signaling pathways of hypoxia inducible factor-1, PI3K-Akt, and NF-κB by regulating the expression of protein kinase 1, VEGF-A, B-cell lymphoma-2, TNF, and other targets.
The prescription of XBJ injection was first introduced in the Xuefu Zhuyu decoction in the medical book Yilin Gaicuo (), written by Wang Qingren, a famous Chinese physician in the 1830s of the Qing Dynasty of China. The decoction is composed of such main ingredients as Honghua (),Chishao (),Chuanxiong (), Danshen ()and Danggui () with glucose as an auxiliary material that can clear away heat and toxic material (equivalent to an anti-inflammatory effect in Western medicine). XBJ injection has obtained the production approval of the State Food and Drug Administration of China and a new drug certificate for the drug of second class (approval number: Z20040033) in 2004. It has been employed for the clinical treatment of pneumonia for more than ten years, primarily as a treatment for severe pneumonia and severe pneumonia with sepsis, respiratory distress, respiratory failure, and fever with profound effects. It was designated as a severe drug in the(trial version 7) and obtained an approval letter of supplementary application by the State Food and Drug Administration of China for use as an additional drug application for new indications including severe pneumonia, critical systemic inflammatory response syndrome, or multiple organ failure from the novel coronavirus.
Background
A new coronavirus disease 2019 (COVID-19) occurred in Wuhan, Hubei Province, China, in December of 2019, which quickly spread to other countries. The fast sprawl of this epidemic and the potentially deadly respiratory symptoms of this disease have created a global public health hazard of pandemic proportions [1, 2]. Coronaviruses are species of virus in the family Coronaviridae, which are associated with common colds and significantly more serious diseases such as Middle East Respiratory Syndrome and Severe Acute Respiratory Syndrome [3]. The new coronavirus belongs to a new strain that has never been found in the human body before. No targeted, effective drug, primarily aimed at the symptomatic treatment of COVID-19, is currently available. Thus, the formulation of a plan for the rapid and effective treatment of this disease is an overarching problem [4].
Traditional Chinese medicine (TCM) has proved to be useful for viral pneumonia treatment and has shown favorable efficacy in the treatment of the Severe Acute Respiratory Syndrome outbreak in 2003. The treatment of COVID-19 with TCM is one of the vital treatment protocols in theissued and continuously updated by the National Health Commission of China. The prescription of Xuebijing (XBJ) injection came from the Xuefu Zhuyu decoction in the medical book Yilin Gaicuo (), written by Wang Qingren, a famous Chinese physician in the 1830s of the Qing Dynasty of China. The decoction is composed of Honghua (),Chishao (),Chuanxiong (), Danshen ()and Danggui () with glucose as an auxiliary material and is associated with clearing away heat and toxic material (equivalent to an anti-inflammatory effect in Western medicine). XBJ injection has obtained production approval from the State Food and Drug Administration of China and a new drug certificate as a drug of second class (approval number: Z20040033) in 2004. It has been employed for the clinical treatment of pneumonia for more than ten years, primarily for severe pneumonia, severe pneumonia with sepsis, respiratory distress, respiratory failure, and fever with profound effects [5]. It was designated as a severe drug in the(trial version 7) and has obtained an approval letter for the supplementary application by the State Food and Drug Administration of China for an additional drug application for newer indications such as severe pneumonia, critical systemic inflammatory response syndrome, or multiple organ failure caused by the novel coronavirus [6].
Analysis of the multiple ingredients and targets can be conducted via network pharmacology, a holistic approach used to explore the potential effects of the drugs on diseases [7]. Furthermore, this research method coincided with the overall concept of the TCM treatment of diseases, placing a bedrock for research into the mechanism of action of TCM. Molecular docking is a method of designing drug molecules through computer-aided means. By simulating the geometric structure of the molecules and the intermolecular forces, it can be used to predict the interaction between drugs and targets and can screen the effects of the active ingredients of TCM quickly and accurately, which has played a paramount role in the study of the mechanism of the effects of drugs [8].
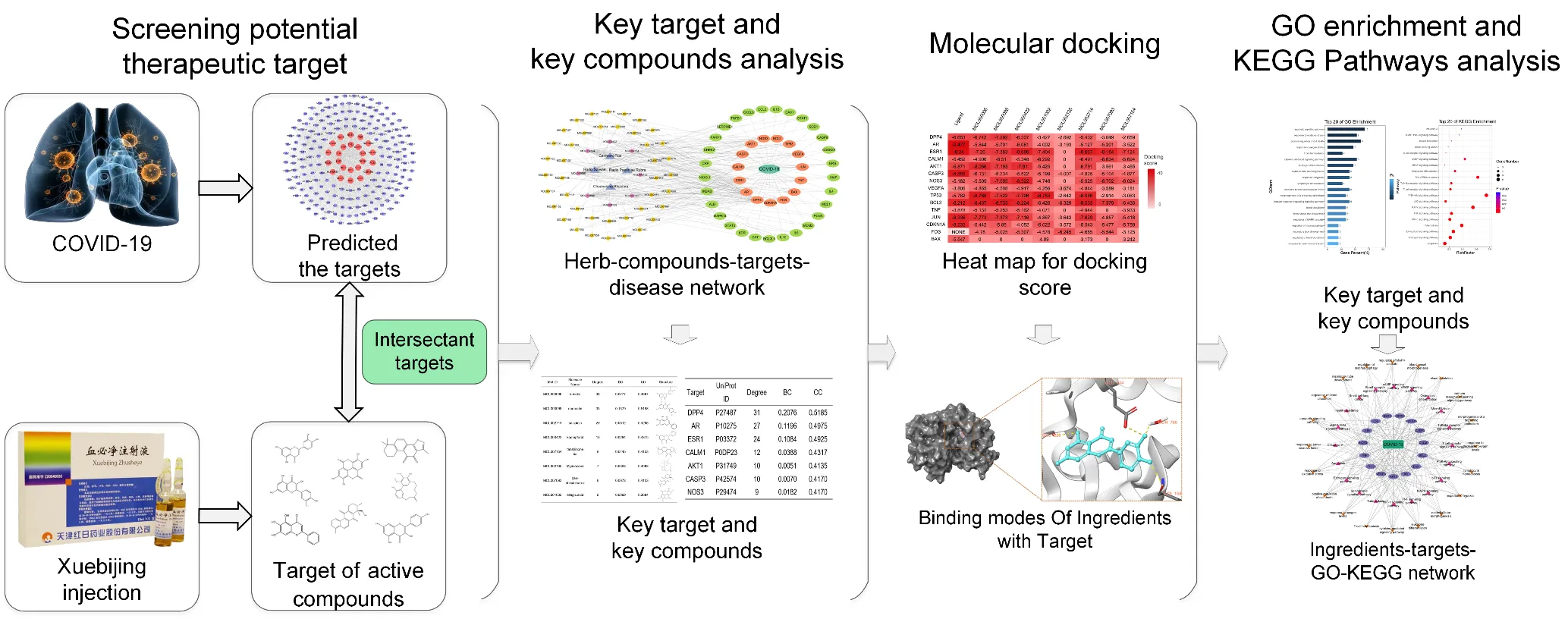
This study analyzed the potential mechanism of XBJ’s treatment in COVID-19 through multiple targets, channels, and pathways by network pharmacology and molecular docking. The research flow chart is shown in Figure 1.

Figure 1 Flow chart of this study
Material and methods
Screening of effective compound ingredients and the targets of XBJ injection
Candidate active compounds and targets were ultimately acquired via screening conditions by using TCMSP (http://tcmspw.com/tcmsp.php) [9] database to search the XBJ related active ingredients with oral bioavailability ≥ 30% according to the parameters of the human body absorption distribution metabolism and drug-likeness ≥ 0.18.
Prediction of the potential therapeutic targets of COVID-19
Based on the GeneCards [10] database with “coronavirus disease 2019” as a keyword, the related disease targets were collected and screened with a relevance score > 20 and then the target proteins related to COVID-19 were expanded by the STRING (https://string-db.org/) database [11]. The protein-protein interactions included in the STRING database could adequately reflect which proteins conducted the interaction. With the medium confidence set to 0.400 and the limitation of species as “Homo sapiens”, the targets expanded thereafter were the ones that interacted with the known target of COVID-19, which was capable of being used as a potential therapeutic target of COVID-19.
Screening of the related targets for XBJ injection in the treatment of COVID-19
The collected targets of XBJ were compared with the potential therapeutic targets of COVID-19 through the graphics option in TB tools to find the intersection of tow targets as the potential target, and then the Venn diagram of the collection of the injection targets of XBJ and the COVID-19-related targets were drawn to collect the intersection of the target genes, which were the targets in the XBJ treatment of COVID-19. Finally, the gene symbols of the related targets were mapped to UniProt ID through the UniProt (https://www.uniprot.org/) database.
Screening of the key targets and key compounds involved in XBJ treatment of COVID-19
In order to further screen the key compounds and key targets, the related targets of COVID-19 were treated with XBJ injection and were mapped to the corresponding chemical ingredients, and Cytoscape 3.7.2 [12] was applied to construct a herb-compound-targets-disease network to visualize this relationship. Subsequently, the key ingredients and key targets were screened via a topology analysis of the network performed by Network Analyzer.
Verification of the binding activity of the key ingredients of XBJ injection and the key targets of COVID-19 by molecular docking
The 3D structure of the compound in the mol2 format was obtained in the Pubchem (https://pubchem.ncbi.nlm.nih.gov/). Then, searched and screened for the target protein in the Protein Data Bank (PDB) database (https://www.rcsb.org/). The Ligand Docking module of Schrödinger was used for molecular docking. The PDB database stores structural data such as molecular crystallography, nuclear paramagnetic resonance spectroscopy, and 3D electron microscopy [13]. Screening conditions of this protein were as follows: (1) the analysis method was single-crystal X-ray diffraction; (2) resolution < 2A; (3) time of discovery was as late as possible; (4) preference was given to those containing small molecule ligands, of which angiotensin-converting enzyme 2 (ACE2) was currently selected due to the smaller database structure, and the conditions did not include resolution. Then, pretreatment of the compounds and proteins before docking involved the following the small molecule ligand was docked based on the active site of the small molecule ligand, and those without were docked with the active site calculated by using the binding site detect function of the Schrödinger software. The docking precision was set to standard precision, and the reference was the original ligand and ACE2, a known target of COVID-19. The results of the molecular docking and scoring were drawn as a heat map, and the binding mode was analyzed to verify the degree of binding of the key ingredients of XBJ injection to the key targets of COVID-19.
Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of key targets
The GO annotated gene targets the following three aspects: cellular components, molecular function, and biological processes. KEGG pathway analysis was used to comprehend the advanced functions and applications of the biological systems from the perspective of the molecular level. In order to explore the mechanism of XBJ treatment of COVID-19, GO annotation analysis was carried out via Metascape (https://metascape.org/) [14] platform using the UniProt ID of the key target. The threshold was< 0.01 and minimum count3, enrichment factor > 1.5; KEGG pathway analysis was created by the UniProt ID of key targets in the DAVID (https://david.ncifcrf.gov/) [15] database. Cytoscape 3.7.2 was used to construct the ingredients-targets-GO-KEGG (I-T-G-K) diagram for the results of the analysis.
Results
Active ingredients and targets of XBJ injection
The TCMSP database was applied, removing the sterol vitamins and sugars shared in the plants and vitamins, 115 effective compounds of XBJ were obtained, including 19 of Honghua (), 25 of Chishao (), 6 of Chuanxiong (), 65 of Danshen () and none of Danggui (). Among them, there are 411 predicted targets of Honghua (), 275 of Chishao (), 72 of Chuanxiong (), 197 of Danshen ()Five hundred thirty predicted targets of XBJ were obtained after removing the duplicates.
Prediction of the potential therapeutic targets of COVID-19
The COVID-19-related disease targets collected using the GeneCards database are shown in Figure 2 after the STRING database is expanded. The red nodes in the central area collected 24 targets for the GeneCards database, and the purple nodes in the outer circle expanded the STRING database. A total of 218 related disease targets were collected following expansion.
Screening of the related targets during XBJ treatment of COVID-19
The Venn diagram is a comparison between the COVID-19 potential therapeutic targets expanded, and the XBJ target and is depicted in Figure 3. A total of 44 intersection targets were visible, which are shown in Table 1. It has been suggested that these targets might be related to the targets for XBJ treatment of COVID-19. The intersection targets of the drug and the target of the disease do not include the currently found targets related to coronavirus infection, such as ACE2, which indicates that XBJ injection may not directly act on the ACE2 protein, and maybe an indirect relationship exists between them.
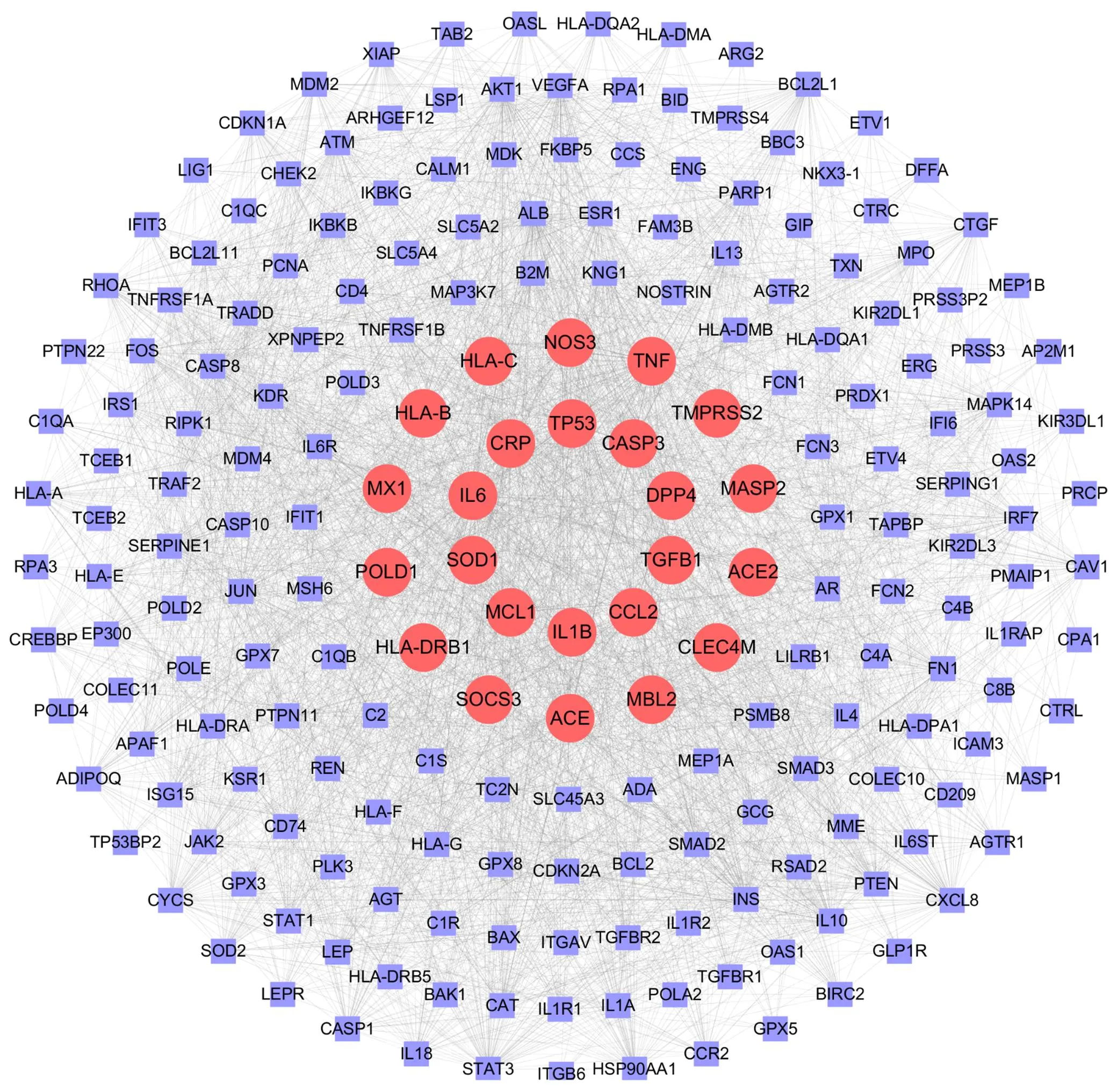
Figure 2 Network of expanding targets. The red nodes collect 24 targets for the GeneCards database, and the purple nodes expand the STRING database.
Figure 3 Venn diagram of coincidence targets
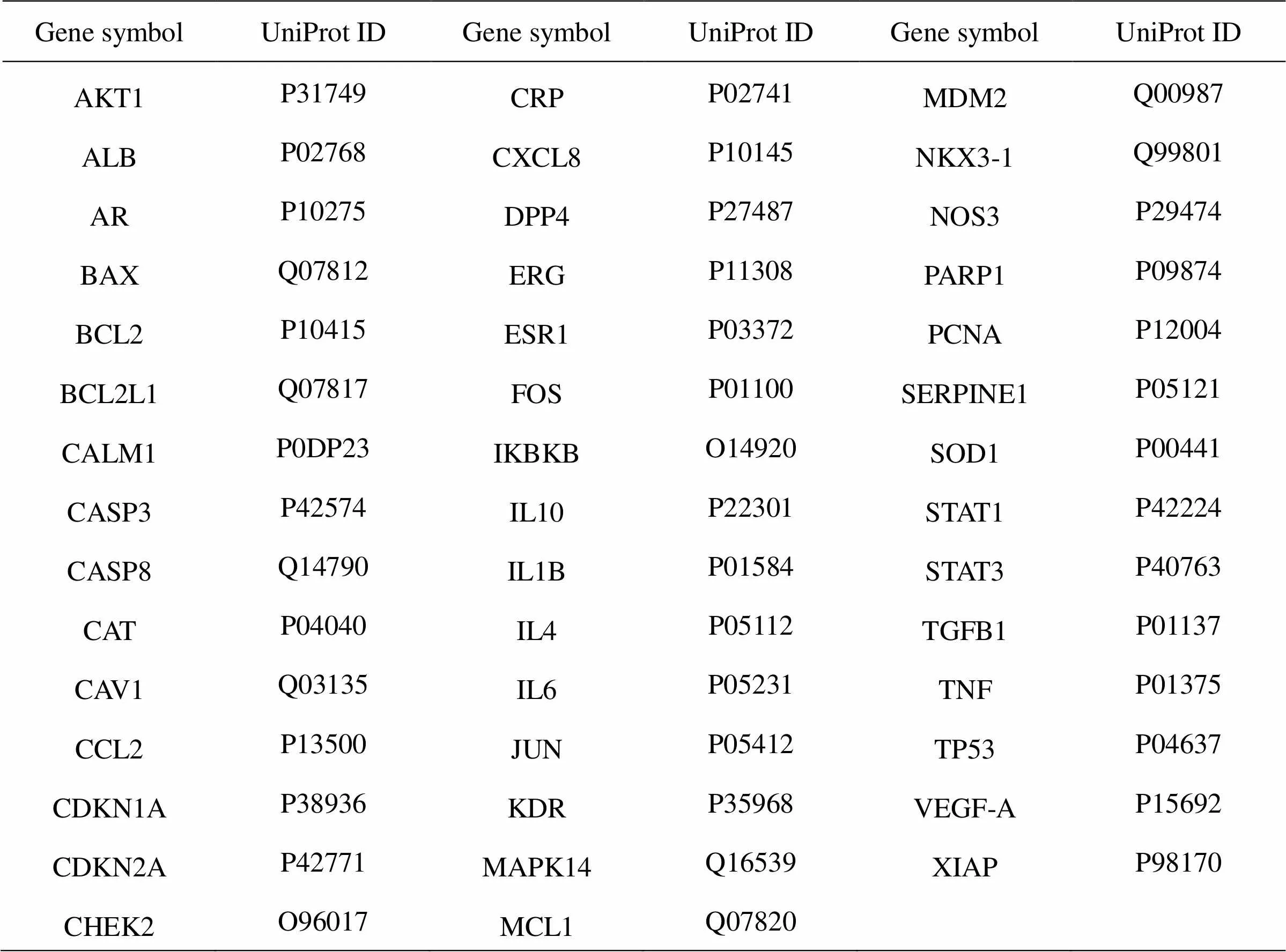
Table 1 Targets of COVID-19
Key targets and compounds of the XBJ treatment for COVID-19
The related targets of XBJ injection treatment of COVID-19 were mapped to the corresponding chemical ingredients, and Cytoscape was applied to construct a network diagram of the herb-compound-target-disease, as depicted in Figure 4. In order to further screen the key compounds and targets, Network Analyzer was employed to perform a topological analysis on the network in Figure 4 to obtain three network topology parameters such as degree centrality, betweenness centrality (BC), and closeness centrality (CC). The screening was based on parameters twice than the median [16], and eight key compounds were selected, as shown in Table 2. 15 key targets were selected, and the related characteristics are shown in Table 3. It has been suggested that XBJ treatment of COVID-19 might involve such compounds as luteolin, quercetin, baicalein, kaempferol, and others acting on the key target proteins like dipeptidyl peptidase-4 (DPP4), AR, ESR1, CALM1, protein kinase 1 (AKT1), of which DPP4, TP53, TNF, CASP3, and NOS3 were the targets without expanding protein-protein interactions, which played a significant role in the treatment.
The verification of the binding activity of the XBJ key compounds and the COVID-19 key targets via molecular docking
Molecular docking using Schrödinger software verified whether the strong binding activity is between the smaller molecular compounds and the target. Protein Data Bank-identification (ID), Ligand-ID and resolution can all be seen in Table 4. A component-target docking score of less than 0 indicated a successful docking, the higher the absolute value of the score, the tighter the binding [17]. The fact that the score of the docking compound is close to the original ligand proves that the compound plays a significant role in determining the targets during treatment. The heat map of the molecular docking results is depicted in Figure 5. The depth of the color represents the docking score. The comparison of the scores of the key targets and the positive control ACE2 to the original ligand demonstrated that each key compound exhibited effective binding activity with at least 8 COVID-19 key targets. This result proved that the XBJ key compound performed binding activity to the COVID-19 key targets.
Through a detailed analysis of the image of the molecular docking result, the small molecule could be tightly bound to the active site of the target protein. In order to show a link between the target protein and the small molecules through a variety of chemical bonds, this is the typical binding model involved in the relevant chemical bond in the selection. In this paper, the combination mode of ellagic acid (MOL001002) docking ACE2, quercetin (MOL000098) docking DPP4, luteolin (MOL000006) docking ATK1, and baicalein (MOL002714) docking B-cell lymphoma-2 (BCL2) are depicted in Figure 6. From the overall diagram on the left, the small molecule compounds could bind tightly to the active sites of the receptor proteins. Small molecules were interlocked through their hydrogen bonds, salt bridge, and pi-pi interactions (Figure 6). The salt bridge exhibited by the purple dotted line is the most common force between the proteins, and the effect of binding on the two atomic groups together is due to a strong electrostatic interaction between the positive and negative charges. This force is weaker than the hydrogen bonds; however, it has a significant influence on the overall stability of the combination as it is found in large amounts [18]. The pi-pi stacking showed by the red dotted line tends to occur in the weak interactions between the electron-rich aromatic ring and the electron-deficient aromatic ring. To a large extent, it affects the reliability of the combination [19]. The hydrogen bond of the yellow dotted line is a kind of intermolecular force between the permanent dipoles. Hydrogen bonds occurred between a hydrogen atom bound to other atoms by covalent bonds, which result in another atom afforded a more substantial binding capacity second only to the covalent bond. Hydrogen bonds are the primary determinant as to whether the small molecules could bind to the target protein or not [20].

Figure 4 Herb-compounds-targets-disease network. The blue bold text is the target before expansion.
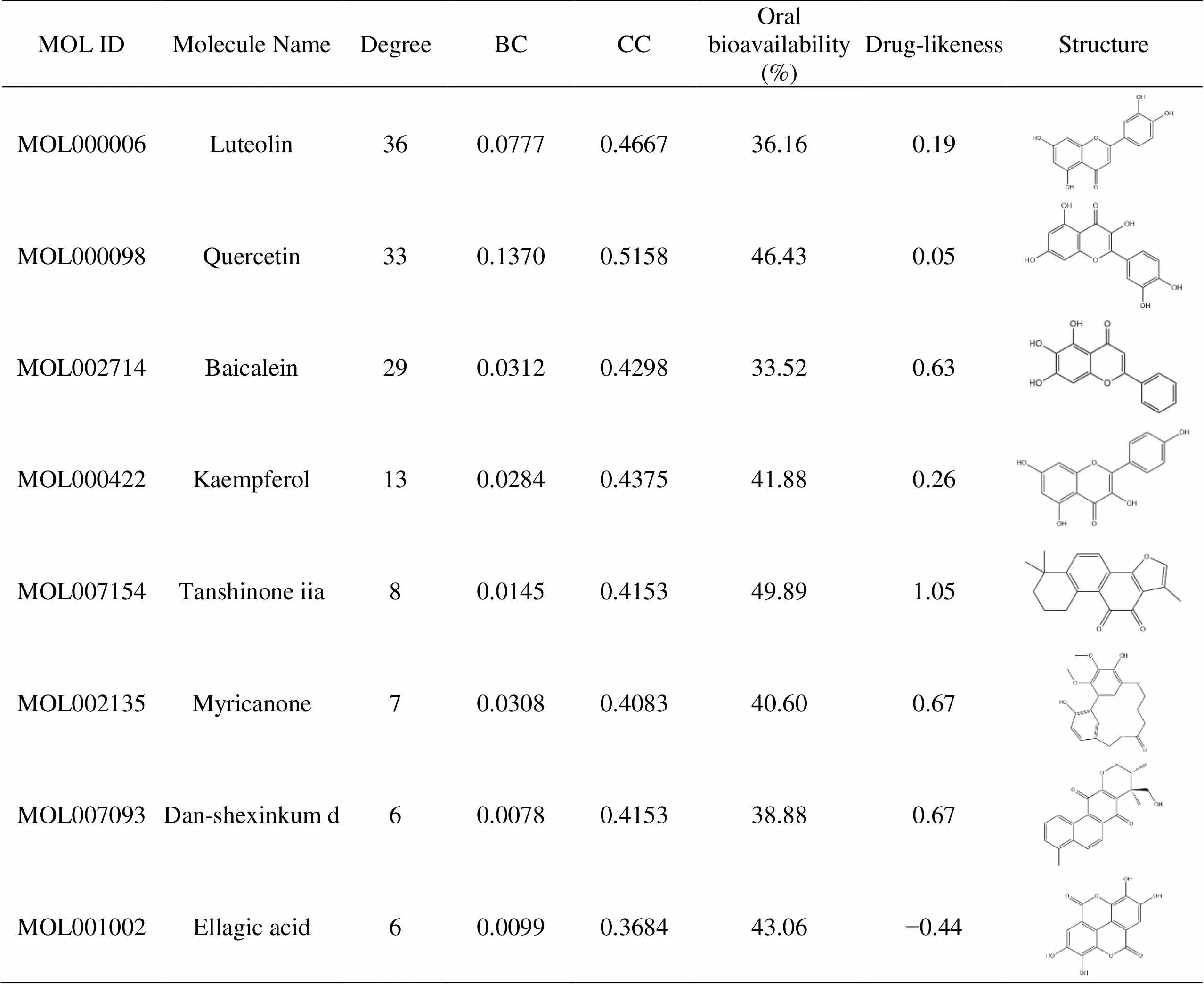
Table 2 Related characteristics of the key compounds
BC, betweenness centrality; CC, closeness centrality.

Table 3 Related characteristics of key targets
ID, identification; BC, betweenness centrality; CC, closeness centrality.
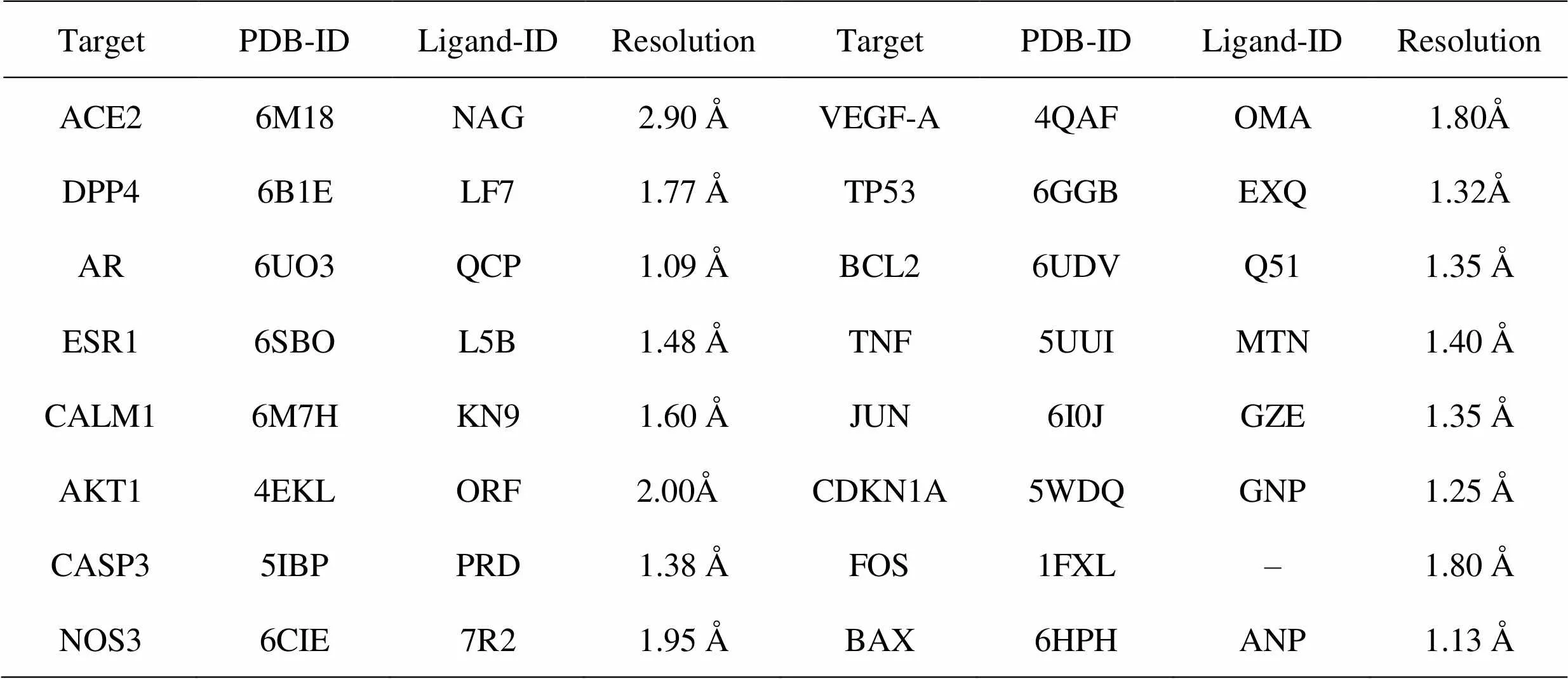
Table 4 Information on 15 proteins involved in molecular docking
PDB, Protein Data Bank; ID, identification;–, not mentioned.
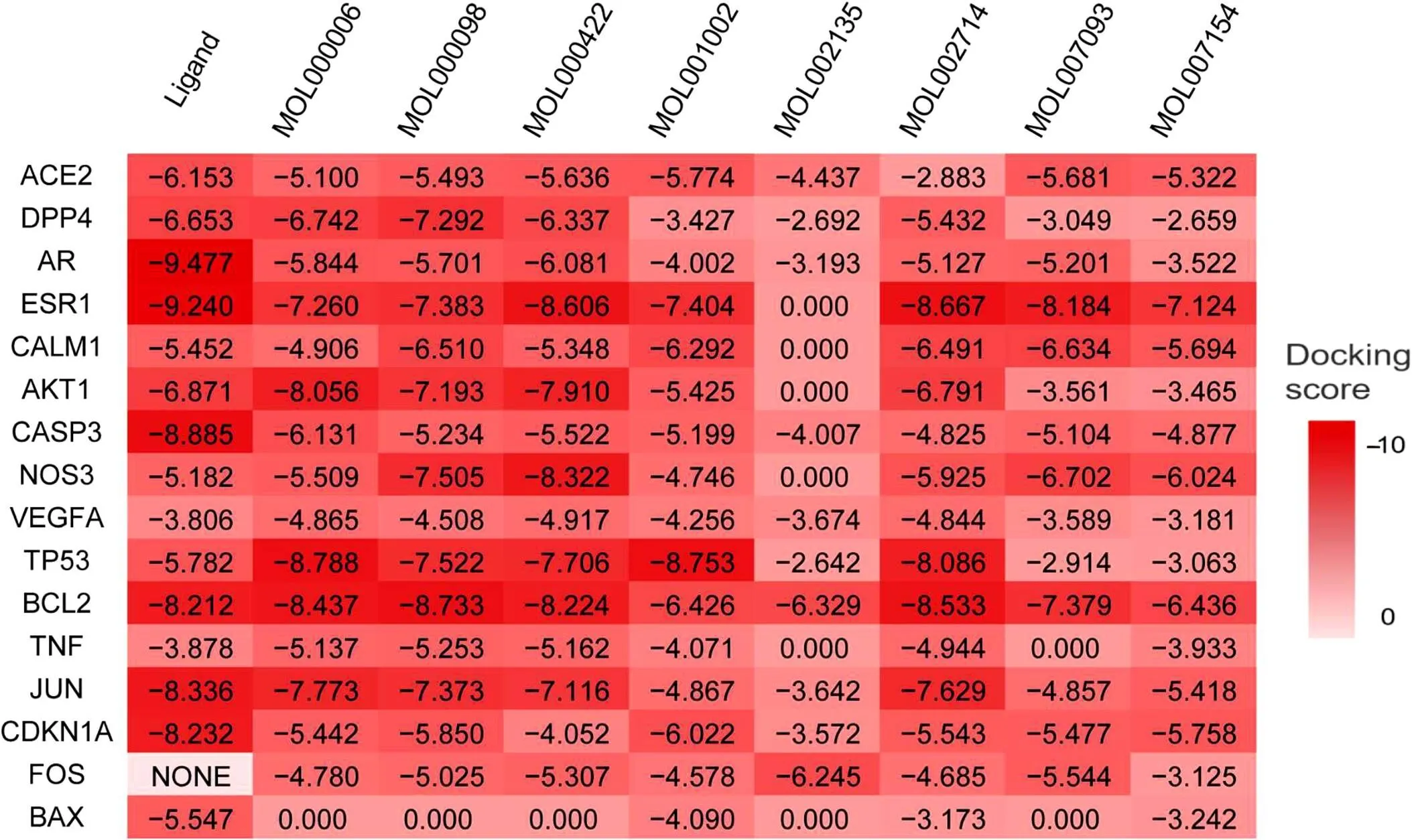
Figure 5 Heat map for docking score. The depth of color represents the docking score; a docking score of less than 0 indicates a successful docking, the higher the absolute value of the score, the tighter the binding.
Analysis of the GO biological process and the KEGG pathway for the key targets
GO biological process analysis. GO biological process analysis obtained a total of 172 enrichment pathways, including 33 pathways related to COVID-19. It selected the 20 GO terms with the smallest-value to display in a graphical table (Figure 7). In Figure 7, the ordinate is a GO term, and the abscissa is the percentage of the target gene count of the GO term to the total count of the target genes, the darker the color, the smaller the-value. The value of the column was the count of the genes in a GO term.

A. ACE2 and MOL001002B. DPP4 and MOL000098 C. ATK1 and MOL000006D. BCL2 and MOL002714
Figure 6 Binding modes of targets and compounds. The purple dotted line is the salt bridge, the red dotted line is pi-pi stacking, and the yellow dotted line is the hydrogen bond. ACE2, angiotensin-converting enzyme 2; DPP4, dipeptidyl peptidase-4; AKT1, protein kinase 1; BCL2, B-cell lymphoma-2.
Figure 7 Enrichment analysis of the GO biological process. The ordinate is a GO term and the abscissa is the percentage of the target gene count of the GO term to the count of all of the target genes. The color represents the-value, the darker the color, the smaller the-value. The value of the column is the count of genes in a GO term.
The biological processes indicated that the treatment of COVID-19 by XBJ involved apoptosis, response to hypoxia, blood vessel morphogenesis, regulation of macroautophagy, and response to TNF. This result shows that XBJ can affect these biological processes in the treatment of diseases.
KEGG pathway analysis. KEGG pathway enrichment obtained a total of 57 enriched pathways, of which 27 pathways were related to COVID-19. The 20 KEGG pathways with the smallest-value are depicted in the graphical display (Figure 8). The Y-axis represents the pathway name, the X-axis represents the enrichment degree, and the air bubble size indicates the number of targets contained in the pathway, the color of the significance of the pathway, is correlated to the data of the-value.
The KEGG pathway analysis results showed that the XBJ treatment of COVID-19 involved the signaling pathways of hypoxia inducible factor-1 (HIF-1), TNF, NF-κB, VEGF, PI3K-Akt, and p53. It illustrated that the active ingredients of XBJ might be able to treat diseases via influencing these signaling pathways.
The construction of the ingredients-targets-diseases-pathway relationship. In order to organize the I-T-G-K relationship, Cytoscape 3.7.2 was used to construct the I-T-G-K relationship diagram. The nodes with different colors and shapes represent the corresponding diseases, ingredients, and targets (Figure 9). The 15 key targets of XBJ were distributed in distinct metabolic pathways and biological processes which jointly regulated the organismal level. The targets reflected the mechanism of action involved in this multi-component TCM, which suggested potential as a multi-targeted therapy to treat COVID-19.
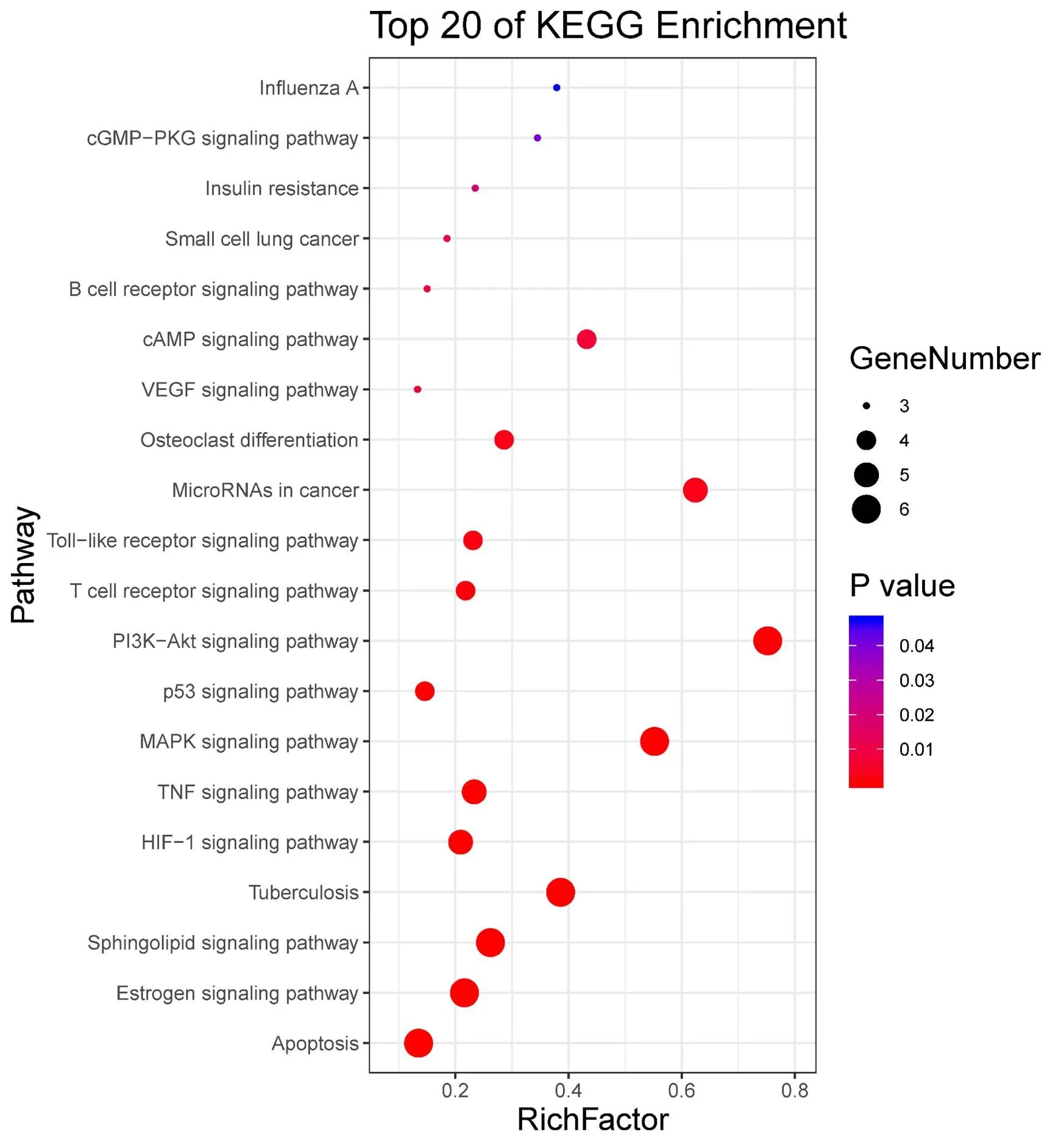
Figure 8 Enrichment analysis of the KEGG pathway. The Y-axis represents the pathway name, and the X-axis represents the enrichment degree; the air bubble size indicates the number of targets contained in the pathway and the color shows the data of the-value of the pathway.
Figure 9 I-T-G-K network. I-T-G-K, ingredients-targets-GO-KEGG; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Discussion
XBJ has been used as a specific, clinical, and evidence-based treatment of pneumonia for more than a decade. It is mostly used in severe pneumonia and severe pneumonia complicated with sepsis, respiratory distress, and respiratory failure, and fever [21]. Therefore, it has become a Chinese patent medicine as a joint recommendation for use as a therapy for both Chinese medicine and Western medicine and has been proven to be effective in improving community-acquired pneumonia outcomes [22]. Severe COVID-19 patients will experience symptoms such as acute respiratory distress syndrome, fibrosis of the lungs, septic shock, metabolic acidosis, and blood coagulation dysfunction. Therefore, the optimal treatment for severe patients should improve pulmonary function and hypoxia, reduce acute lung injury, pulmonary fibrosis, and enhance the antiviral immune response to reduce inflammation and other aspects [23, 24]. Therefore, the current study was based on research to determine a multi-component and multi-targeted therapy for COVID-19. Through network pharmacology technology and molecular docking technology, this study explored the potential targets of XBJ treatment for COVID-19 and stated the mechanism of action found to provide a scientific basis for clinical application. Our specific findings are as follows.
The analysis results of the XBJ treatment for the COVID-19 key targets and key compounds showed that eight key compounds regulated 15 key targets. Among the key compounds found in XBJ, the flavonoids accounted for the majority including luteolin (MOL000006), quercetin (MOL000098), baicalein (MOL002714), tanshinone iia (MOL007154), myricanone (MOL002135); then the phenols such as ellagic acid (MOL001002), kaempferol (MOL000422); and the quinone dan-shexinkum d (MOL007093). Flavonoids such as baicalein inhibit the inflammatory factors such as TNF and interleukin (IL) by interfering with the NF-κB pathway, which exert antipyretic and anti-inflammatory effects, inhibit inflammation and apoptosis, and reduce acute lung injury [25]. Quercetin reduces the production of NF-κB and ICAM-1 by inhibiting the NF-κB signaling pathway and achieves anti-inflammatory effects [26]; reportedly, it could improve lung fibrosis in murine models by inhibiting the SphK1/S1P signal transduction [27]. Phenolic compounds such as ellagic acid has anti-inflammatory, anti-allergic, antiviral, and anti-cancer properties. It is reported that ellagic acid inhibits the Akt, ERK, p38, MAPK, and PI3K/Akt pathways, and the NF-κB anti-inflammatory signaling pathways to trigger anti-inflammatory effects [28].
Key targets included DPP4, AR, ESR1, CALM1, AKT1, CASP3, NOS3, VEGF-A, TP53, BCL2, TNF, JUN, CDKN1A, FOS and BAX. The results of molecular docking suggest that the above targets and ACE2 could effectively bind to the compound. AKT1 is a direct target downstream of the PI3K pathway and regulated many processes, including proliferation, apoptosis, inflammation, and angiogenesis [29]. The literature shows that AKT1 could regulate apoptotic resistance and pulmonary fibrosis by mediating the mitochondria [30]. BCL2 is a protein with inhibitory effects on apoptosis, through the interaction between proteins, and regulats the permeability of the outer mitochondrial membrane, thereby it controlls mitochondrial autophagy and affects apoptosis [31]. Other literature reports that in fibroid lungs and activated fibroblasts, regulating the expression of BCL2 and autophagy activity could be adjusted to reduce autophagy, which plays a role in inhibiting pulmonary fibrosis [32]. TP53 is a transcription factor, and transcriptional activation functioned to maintain the stability of the genome and plays multiple roles in apoptosis, including differentiation, aging, DNA repair, metabolism, and the immune response [33]. Another study has demonstrated that promoting the expression of TP53 could induce the secretion of the WNT ligands, stimulate tumor-associated macrophages to produce IL-1β, and inhibit the subsequent neutrophil inflammation [34]. Therefore, it might play a similar role in the pathogenesis of COVID-19. VEGF-A, a vascular regulator, plays a vital role in promoting angiogenesis, endothelial cell migration, and proliferation [35, 36]. Furthermore, ACE2 reportedly antagonized the increase in pulmonary vascular permeability mediated by VEGF-A and thereby improved pulmonary function following an acute lung injury [39]. Since ACE2 is considered as a viral receptor of COVID-19 [38–40], XBJ might affect the expression of the ACE2 receptors by influencing the VEGF-A enough to treat this disease.
Based on the analysis of the Go biological process, this article found that the relevant biological processes involved in XBJ as a potential treatment of COVID-19 primarily included apoptosis, response to hypoxia, blood vessel morphogenesis, regulation of macroautophagy, and the response to TNF. These processes might have regulated the mechanism of generation and development of COVID-19. The generation process of COVID-19 includes pulmonary injury and lung fibrosis, inflammation, and hypoxia. XBJ restored the impaired blood vessels and ameliorated hypoxia by interfering with macroautophagy and relieving inflammation. The analysis of the I-T-G-K network suggested that XBJ could treat COVID-19 by acting on multiple pathways.
The HIF-1 signaling pathway is crucial for an organism during a low oxygen concentration and hypoxia response [41]. HIF-1 regulates hundreds of gene transcriptions in the expression of cell-specific, including VEGF-A, IL6, AKT1, and NF-κB [42]. Under hypoxia, the HIF-1 signaling pathway activates the VEGF signaling pathway, which could be protective by up-regulating the reverse transcription of VEGF, and inflammation could be relieved by regulating IL6, TNK, AKT1, and NF-κB [43]. Thus, XBJ might improve pulmonary injury, restore impaired vessels, and hypoxia by affecting HIF-1 and VEGF.
NF-κB signaling pathway is found to be crucial to the inflammation and immune response of cells [44]. NF-κB pathway reportedly inhibits replication of the influenza a virus and exacerbates viral pneumonia [45, 46]. TNF, AKT1, and NF-κB are closely related to the PI3K-Akt signaling pathway and NF-kappaB signaling pathway. PI3K-Akt activates NF-κB to regulate many aspects, such as anti-inflammation-related to multiple cellular gene expressions [46]. In the development of cancer, NF-κB plays a vital role in regulation. Reportedly, lower transcription activity by NF-κB decreases expression of the inflammatory cytokines such as IL-1β, TNF-α, and IL-6; thus, reducing inflammation and apoptosis as well as alleviating pulmonary injury [47]. In vitro, TNF-α-induced acute lung injury could be attenuated via inhibiting NF-κB and activating Nrf2 [48]. Moreover, in vivo animal experiments found the release of proinflammatory cytokines-was mediated by PI3K/AKT/GSK3β and NF-κB [49]. Therefore, XBJ might have an anti-inflammatory effect by affecting the activation of PI3K/AKT/NF-κB, which could reduce inflammation caused by COVID-19.
Conclusion
Currently, no golden standard exists for the treatment of COVID-19. TCM plays a significant role in treating new diseases because it features multiple ingredients, targets, and approaches. These features were used to explore the complicated mechanism of XBJ in the treatment of COVID-19 via network pharmacology and molecular docking. The mechanism of action showed potential for the key compounds in XBJ, as they regulated the expression of key targets such as AKT1, VEGF-A, BCL2, and TNF, which affected the COVID-19 viral receptors such as ACE2 and the signaling pathways like HIF-1, PI3K-Akt, and NF-κB. Thus, XBJ might take effect by (1) affecting the key targets, and flavonoids in XBJ that attenuated acute lung injury by alleviating the inflammation and apoptosis; (2) affecting VEGF-A and then the expression of the ACE2 receptor; (3) activating VEGF by affecting HIF-1, which improved pulmonary injury, restored the impaired vessels, and hypoxia; (4) alleviating the inflammation caused by COVID-19 by activating PI3K/AKT/NF-κB.
At present, the pathogenesis of COVID-19 has not yet been elucidated. Thus, the mechanism of XBJ involved in the treatment of COVID-19 requires further research. The current study reflects the holistic and systematic characteristics of TCM treatment by studying the interactions between these therapeutic targets and pathways. This article has provided a basis for exploring the mechanism of XBJ in the treatment of COVID-19 and offers insight into the potential of XBJ for future researchers.
1. Hui DS, Azhar EI, Madani TA, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health-the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 2020, 91: 264–266.
2. Heymann DL, Shindo N. COVID-19: what is next for public health? Lancet 2020, 395: 542–545.
3. Henry BM. COVID-19, ECMO, and lymphopenia: a word of caution. Lancet Respir Med 2020.
4. Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov, 2020, 19: 149–150.
5. Liu S, Yao C, Zhang J, et al. Efficacy of Xuebijing injection for sepsis (EXIT-SEP): protocol for a randomised controlled trial. BMJ Open 2019, 9: e28664.
6. Scheme for diagnosis and treatment of 2019 novel coronavirus pneumonia (The 7th Trial Edition). Jiangsu Journal of Traditional Chinese Medicine 2020, 52: 1–6. (Chinese)
7. Boezio B, Audouze K, Ducrot P, et al. Network-based approaches in pharmacology. Mol Inform 2017, 36.
8. Saikia S, Bordoloi M. Molecular docking: challenges, advances and its use in drug discovery perspective. Curr Drug Targets 2019, 20: 501–521.
9. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014, 6: 13.
10. Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards Suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 2016, 54: 1–30.
11. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019, 47(D1): D607–D613.
12. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003, 13: 2498–2504.
13. Burley SK, Berman HM, Bhikadiya C, et al. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res 2019, 47(D1): D464–D474.
14. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019, 10: 1523.
15. Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009, 37: 1–13.
16. Cui Q, Zhang YL, Ma YH, et al. A network pharmacology approach to investigate the mechanism of Shuxuening injection in the treatment of ischemic stroke. J Ethnopharmacol 2020: 112891.
17. Etti I, Abdullah R, Hashim NM, et al. Artonin E and structural analogs from Artocarpus species abrogate estrogen receptor signaling in breast cancer. Molecules 2016, 21.
18. Zhang N, Wang Y, An L, et al. Entropy drives the formation of salt bridges in the protein GB3. Angew Chem Int Ed Engl 2017, 56: 7601–7604.
19. Zhuang W, Wang Y, Cui P, et al. Applications of pi-pi stacking interactions in the design of drug-deliverysystems. J Control Release 2019, 294: 311–326.
20. Arunan E, Mani D. Dynamics of the chemical bond: inter- and intra-molecular hydrogen bond. Faraday Discuss 2015, 177: 51–64.
21. Li C, Wang P, Zhang L, et al. Efficacy and safety of Xuebijing injection (a Chinese patent) for sepsis: a meta-analysis of randomized controlled trials. J Ethnopharmacol 2018, 224: 512–521.
22. Song Y, Yao C, Yao Y, et al. Xuebijing injection versus placebo for critically ill patients with severe community-acquired pneumonia: a randomized controlled trial. Crit Care Med 2019, 47: e735–e743.
23. Guan WJ, Zhong NS. Clinical characteristics of COVID-19 in China. N Engl J Med 2020, 382.
24. Jiang F, Deng L, Zhang L, et al. Review of the clinical characteristics of coronavirus disease 2019 (COVID-19). J Gen Intern Med 2020.
25. Chu L, Zhu F, Zhou W, et al. Baicalein attenuates acute lung injury induced by intestinal ischemia/reperfusion via inhibition of nuclear factor-kappaB pathway in mice. Chin Crit Care Med 2017, 29: 228–232. (Chinese)
26. Liu Y, Yu C, Ji K, et al. Quercetin reduces TNF-alpha-induced mesangial cell proliferation and inhibits PTX3 production: involvement of NF-kappaB signaling pathway. Phytother Res 2019, 33: 2401–2408.
27. Zhang X, Cai Y, Zhang W, et al. Quercetin ameliorates pulmonary fibrosis by inhibiting SphK1/S1P signaling. Biochem Cell Biol 2018, 96: 742–751.
28. Kowshik J, Giri H, Kishore TKK, et al. Ellagic acid inhibits VEGF/VEGFR2, PI3K/Akt and MAPK signaling cascades in the hamster cheek pouch carcinogenesis model. Anticancer Agents Med Chem 2014, 14: 1249–1260.
29. Rafalski VA, Brunet A. Energy metabolism in adult neural stem cell fate. Prog Neurobiol 2011, 93: 182–203.
30. Larson-Casey JL, Deshane JS, Ryan AJ, et al. Macrophage Akt1 kinase-mediated mitophagy modulates apoptosis resistance and pulmonary fibrosis. Immunity 2016, 44: 582–596.
31. Dai H, Meng W, Kaufmann S. BCL2 family, mitochondrial apoptosis, and beyond. Cancer Transl Med 2016, 2: 7.
32. Han R, Ji X, Rong R, et al. MiR-449a regulates autophagy to inhibit silica-induced pulmonary fibrosis through targeting Bcl2. J Mol Med 2016, 94: 1267–1279.
33. Kung C, Basu S, Murphy ME. A link between TP53 polymorphisms and metabolism. Mol Cell Oncol 2016, 3: e1173769.
34. Delbridge ARD, Kueh AJ, Ke F, et al. Loss of p53 causes stochastic aberrant X-chromosome inactivation and female-specific neural tube defects. Cell Rep 2019, 27: 442–454.
35. Li T, Jiang Y, Hu Y, et al. Interleukin-17-producing neutrophils link inflammatory stimuli to disease progression by promoting angiogenesis in gastric cancer. Clin Cancer Res 2017, 23: 1575–1585.
36. Asensio-Sanchez VM. VEGF-A and VEGF-Ax: bad protein and good protein. Arch Soc Esp Oftalmol 2015: 90, 149–150.
37. Yu X, Lin Q, Qin X, et al. ACE2 antagonizes VEGF-A to reduce vascular permeability during acute lung injury. Cell Physiol Biochem 2016, 38: 1055–1062.
38. Yang JM, Meng X, Xue F, et al. ACE2 in the context of 2019-nCoV infection: friend or foe? Zhonghua Xin Xue Guan Bing Za Zhi 2020, 48: E12. (Chinese)
39. Batlle D, Wysocki J, Satchell K. Soluble angiotensin-converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci 2020, 134: 543–545.
40. Diaz JH. Hypothesis: angiotensin-converting enzyme inhibitors and angiotensin receptor blockers may increase the risk of severe COVID-19. J Travel Med 2020.
41. Lu Y, Wang B, Shi Q, et al. Brusatol inhibits HIF-1 signaling pathway and suppresses glucose uptake under hypoxic conditions in HCT116 cells. Sci Rep 2016, 6: 39123.
42. Wilkins SE, Abboud MI, Hancock RL, et al. Targeting protein-protein interactions in the HIF system. ChemMedChem 2016, 11: 773–786.
43. Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med 2003, 9: 677–684.
44. Harikrishnan H, Jantan I, Haque MA, et al. Anti-inflammatory effects ofSchum. & Thonn. through inhibition of NF-kappaB, MAPK, and PI3K-Akt signaling pathways in LPS-induced human macrophages. BMC Complement Med Ther 2018, 18: 224.
45. Dai J, Gu L, Su Y, et al. Inhibition of curcumin on influenza a virus infection and influenzal pneumonia via oxidative stress, TLR2/4, p38/JNK MAPK and NF-kappaB pathways. Int Immunopharmacol 2018, 54: 177–187.
46. Xu Y, Liu L. Curcumin alleviates macrophage activation and lung inflammation induced by influenza virus infection through inhibiting the NF-kappaB signaling pathway. Influenza Other Respir Viruses 2017, 11: 457–463.
47. Ju M, Liu B, He H, et al. MicroRNA-27a alleviates LPS-induced acute lung injury in mice via inhibiting inflammation and apoptosis through modulating TLR4/MyD88/NF-kappaB pathway. Cell Cycle 2018, 17: 2001–2018.
48. Yang H, Zhuo J, Sun C, et al. Pogostone attenuates TNF-alpha-induced injury in A549 cells via inhibiting NF-kappaB and activating Nrf2 pathways. Int Immunopharmacol 2018, 62: 15–22.
49. Jiang K, Guo S, Yang C, et al. Barbaloin protects against lipopolysaccharide (LPS)-induced acute lung injury by inhibiting the ROS-mediated PI3K/AKT/NF-kappaB pathway. Int Immunopharmacol, 2018, 64: 140–150.
:
Yu-Liang Zhang analyzed most of the data from database, and wrote the initial draft of the paper; Guo-Wei Zhang developed the idea for the study; Qian Cui and Dou Zhang conducted the enrichment analyses; all authors analyzed the data and discussed the results and revised the manuscript.
:
This study was supported by the Foundation of Health Commission of Hebei Province (20190123) and the Natural Science Foundation of Hebei Province of China (H2018201179).
:
XBJ, Xuebijing; COVID-19, coronavirus disease 2019; TCM, traditional Chinese medcine; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; I-T-G-K, ingredients-targets-GO-KEGG; BC, betweenness centrality; CC, closeness centrality; IL, interleukin; AKT1, protein kinase 1; HIF-1, hypoxia inducible factor-1; BCL2, B-cell lymphoma-2; ACE2, angiotensin-converting enzyme 2; DPP4, dipeptidyl peptidase-4; PDB, Protein Data Bank.
:
The authors declare no conflicts of interest.
:
Yu-Liang Zhang, Qian Cui, Dou Zhang, et al. Efficacy of Xuebijing injection for the treatment of coronavirus disease 2019 via network pharmacology. Traditional Medicine Research 2020, 5 (4): 201–215.
:Rui-Wang Zhao.
:06 April 2020,
20 April 2020,
: 18 May 2020.
Guo-Wei Zhang. No.342, Yuhua Dong Road, College of Traditional Chinese Medicine, Hebei University, Baoding 071000, China. E-mail: xxzgw@126.com.
10.12032/TMR20200507178
 Traditional Medicine Research2020年4期
Traditional Medicine Research2020年4期
- Traditional Medicine Research的其它文章
- Treating COVID-19 by traditional Chinese medicine: a charming strategy?
- India’s indigenous idea of herd immunity: the solution for COVID-19?
- Does Liuzijue Qigong affect anxiety in patients with chronic obstructive pulmonary disease, even during the COVID-19 outbreak? a randomized, controlled trial
- Qingfei oral liquid downregulates TRPV1 expression to reduce airway inflammation and mucus hypersecretion injury caused by respiratory syncytial virus infection and asthma in mice
- The advances of traditional Chinese medicine in the treatment of liver diseases in 2019
- The selection rules of acupoints and meridians of traditional acupuncture for postoperative nausea and vomiting: a data mining-based literature study
