Identification and characterization of resistance and pathogenicity of Enterococcus spp.in samples of donor breast milk
Luana Andrade Mendes Santana, Nívea Nara Novais Andrade, Lucas Santana Coelho da Silva, Caline Novais Teixeira Oliveira, Breno Bittencourt de Brito, Fabrício Freire de Melo, Cláudio Lima Souza, Lucas Miranda Marques, Márcio Vasconcelos Oliveira
Abstract
Key Words:Enterococcus spp.;Breast milk;Virulence;Human milk;Pasteurization;Antimicrobial resistance
INTRODUCTION
There is no ideal substitute for human milk (HM) during early life despite current technological advances in baby food production, processing, preservation, and preparation[1].Unquestionably, HM supplies newborns (NBs) with all the nutrients that adequately meet the physiological and metabolic needs in this age group[2].
Premature births and hospitalizations in neonatal intensive care units as well as maternal illness, death, and low milk production can hinder the introduction and maintenance of breastfeeding.In this context, the use of donated HM has become an efficient alternative to breastfeeding[2]and allows NBs to be fed with milk obtained from Human Milk Banks (HMBs)[3].This strategy involves the collection, processing,and quality control of colostrum, transition milk, and mature milk for later distribution to NBs under a doctor or nutritionist prescription[4].
Enterococcusspecies are associated with important infections in NBs and are potential contaminants of milked human milk (MHM) samples.Enterococcus spp.are part of the gastrointestinal tract microbiota in healthy humans[5]and may be present in significant numbers on the hands and utensils of milk donors.The presence of these bacteria in human milk indicates that they are due to direct or indirect contact with fecal material, compromising the quality of the MHM[6].SomeEnterococcus spp.,especiallyEnterococcus faecalis(E.faecalis)andEnterococcus faecium(E.faecium), are highly pathogenic in NBs and must be effectively inactivated through pasteurization at HMBs.
To date, there are no published data on the isolation of enteric bacteria with hygienic-sanitary conditions of household collections performed by MHM donors.Considering the propensity of donated milk to be contaminated by these microorganisms, the clinical importance of such pathogens, and their ability to be dispersed through vectors such as health professionals and insects[7], it is important to investigate their virulence and dispersion capacity.Moreover, it is crucial to evaluate the quality of the collection, preservation, and processing of the HM offered to NBs.These procedures are essential for a better understanding of the pathogenicity ofEnterococcus spp.present in raw and pasteurized human milk samples and in samples taken from the hands and nipple-areolar region of donors linked to HMBs.
MATERIALS AND METHODS
Study design, period, region, and population
This is a cross-sectional study performed using samples from the nipple-areolar region, hands, and aliquots of raw and pasteurized MHM from healthy donors registered at the HMB of the Municipal Hospital Esaú Matos in the city of Vitória da Conquista, State of Bahia, Brazil.
The present study recruited donors registered at the above-mentioned HMB who had a donation history with a mean weekly volume of donated milk greater than or equal to 400 mL.Donated samples of raw milk with a volume less than 400 mL were pooled with milk samples from other donors for pasteurization, thus losing the traceability and specificity of the sample from a specific donor.The exclusion criteria were:donors with wounds on the hand or nipple-areolar region and donation of raw milk samples with a milk volume lower than that required for individual pasteurization.
The microbial isolates analyzed in the present study were obtained by the single collection of samples from the nipple-areolar region and hands as well as aliquots of MHM before and after pasteurization from 30 donors between March and August 2018.The collections were carried out at the donors’ residences during scheduled visits.In addition to microbiological evaluation of the above-mentioned biological samples, questionnaires on hygienic-sanitary issues regarding the collection,packaging, and preservation of the donated milk were completed by the participants.
Ethical considerations
The study was approved by the Ethics Committee on Research with Human Beings of the Multidisciplinary Institute of Health of the Universidade Federal da Bahia, under the Certificate of Presentation for Ethics Apprehension number 80800717.8.0000.5556 and committee opinion number 2.475.023.The study followed all the conditions established by the Resolution 466/2012 of the Brazilian National Health Council.
Isolation, identification, evaluation of antimicrobial activity, and detection of constitutive, virulence, and resistance genes in Enterococcus spp.
After collection, the biological samples were immediately inoculated into enrichment brain heart infusion broth (HIMEDIA®) and incubated at 35-37ºC for 24 h.After that,they were placed in Petri dishes with 5% Blood Agar Medium (Base Agar, HIMEDIA®)and Chromagar Orientation (BD™ CHROMagar™ Orientation) in a bacteriological oven at 35-37ºC for 24 h.Both methods were important for increasing the isolation and identification of microorganisms of interest in the present study.
The colonies with presumptive characteristics ofEnterococcus spp.selected in both media underwent Gram staining to evaluate the morphological characteristics as well as to perform the catalase test and subsequent tests to identify the genusEnterococcus,namely:hydrolysis of esculin in the presence of bile and growth in sodium chloride(NaCl) 6.5% media.
AllEnterococcus spp.isolates underwent evaluation of antimicrobial activity using the agar diffusion method[8-10], according to the recommendations of the Clinical and Laboratory Standards Institute (2017)[11].
With the aim of detecting the constitutive[12], virulence[13-16], and resistance[17,18]genes inEnterococcus spp., all samples that were positive in the identification step underwent polymerase chain reaction (PCR) after reactivation in brain heart infusion medium.
Statistical analysis
A descriptive analysis of the study variables was carried out from a database constituted in Microsoft Excel through the Statistical Program EPI INFO (version 7.2.2.6).For the frequency comparison, the chi-square test was used, with values ofP<0.05 (95% confidence interval) considered statistically significant.
RESULTS
Milk samples were collected from 30 donors registered at the HMB of the Municipal Hospital Esaú Matos, in Vitória da Conquista, Bahia State, Brazil.A total of 81 samples were collected:30 samples from swabs containing skin samples from the nippleareolar region and donors’ hands, 30 samples of raw MHM, and 21 samples of pasteurized MHM.Following the methodological criteria, it was not possible to obtain an appropriate aliquot of pasteurized milk from 9 (30%) participants as they did not donate enough milk volume to perform pasteurization in a single bottle.
Enterococcus spp.strains were isolated from 9 donors (30%), and a total of 11 strains were identified:7 strains (63.6%) from skin samples and 4 strains (36.4%) from raw MHM samples.AlthoughEnterococcus spp.were not detected in the pasteurized MHM samples, in 38% (n=8) of these samples other microorganisms, mainlyStaphylococcus aureus, were isolated.
The study participants were questioned about socioeconomic aspects and hygienicsanitary conditions in order to link selected variables to the route of human milk contamination byEnterococcus spp.In this context, most participants were aged 30 years or less, were married or living with a partner, had a family income above two minimum wages, had one child, and were educated to high school level.
With regard to the hygienic-sanitary procedures performed by the donors for milk collection, most participants reported that they always performed the following procedures:hand washing before the collection of milk (96%);held back hair before milking (80%);breast washing before milking (86.7%);used a mask while milking(86.7%);avoided talking during milking (60%);discarded the first jet of milk (53.3%);milk obtained using a manual/electric pump (80%).However, only 12 (80%)participants declared that they checked the size of their nails and trimmed them when necessary before milking (Table 1).
Socioeconomic variables of HM donors such as age (above or below 30 years),marital status (single or living as a couple), family income (higher or lower than 2 minimum wages), number of children (one or more than one), and educational level(high school or graduate) as well as variables related to hygienic-sanitary procedures including hand washing before milking, talking during milking, and discarding the first milk jet at the beginning of collection were not significantly associated with the detection ofEnterococcus spp.in samples (P<0.05) (Table 1).
Among the 11 isolates ofEnterococcus spp., antimicrobial resistance was found in 10 isolates (91%).Resistance to tetracycline was most common (63% of isolates,n=7),and intermediate sensitivity to ciprofloxacin was found in 27% of the isolates (n=3).Resistance to quinolones (ciprofloxacin, norfloxacin, and levofloxacin) was observed in 9% of the isolates (n=1) (Table 2).All isolates were sensitive to vancomycin.All specimens were analyzed by PCR to detect quinolone resistance genes (gyrA andparC genes), but no positive results were obtained.
TheEnterococcus spp.isolates were analyzed by PCR for detection of the constitutiveddlgene encoding the D-alanine ligases, which are species-specific forE.faecalisandE.faecium.This analysis showed that among the 11 enterococcal strains from 9 participants, 6 isolates were identified asEnterococcus spp.and 5 isolates were identified asE.faecalis.However, threeE.faecalisisolates were found to be simultaneously colonized byE.faeciumstrains.Thus, three donors hadE.faecalisandE.faeciumstrains.Such detection was possible because, after the initial identification of theEnterococcusgenus, the stored colonies were collected from the CHROMagar Orientation medium (BD™ CHROMagar™ Orientation), which identified the genus and not the species ofEnterococcus;thus, colonies of both species were collected.
All isolates ofEnterococcus spp.underwent PCR for detection of the virulence genesace,efaA,gelE,as,cylA,hyl,esp,as,cylA,hyl,andesp(Table 3).Among the 11 isolates, 7 isolates (63%) had theefaA gene that encodes the surface antigen A, which is commonly associated with biofilm formation and endocarditis, and 3 isolates (27%)had theacegene, which encodes the protein that adheres to collagen and laminin,favoring bacterial adhesion commonly associated with endocarditis.The following enterocin genes were also genotypically investigated by PCR inEnterococcus spp.:Enterocin A, Enterocin B, Enterocin P, and Enterocin 31, also known as Bacteriocin.However, none of the isolates were positive for these genes.
DISCUSSION
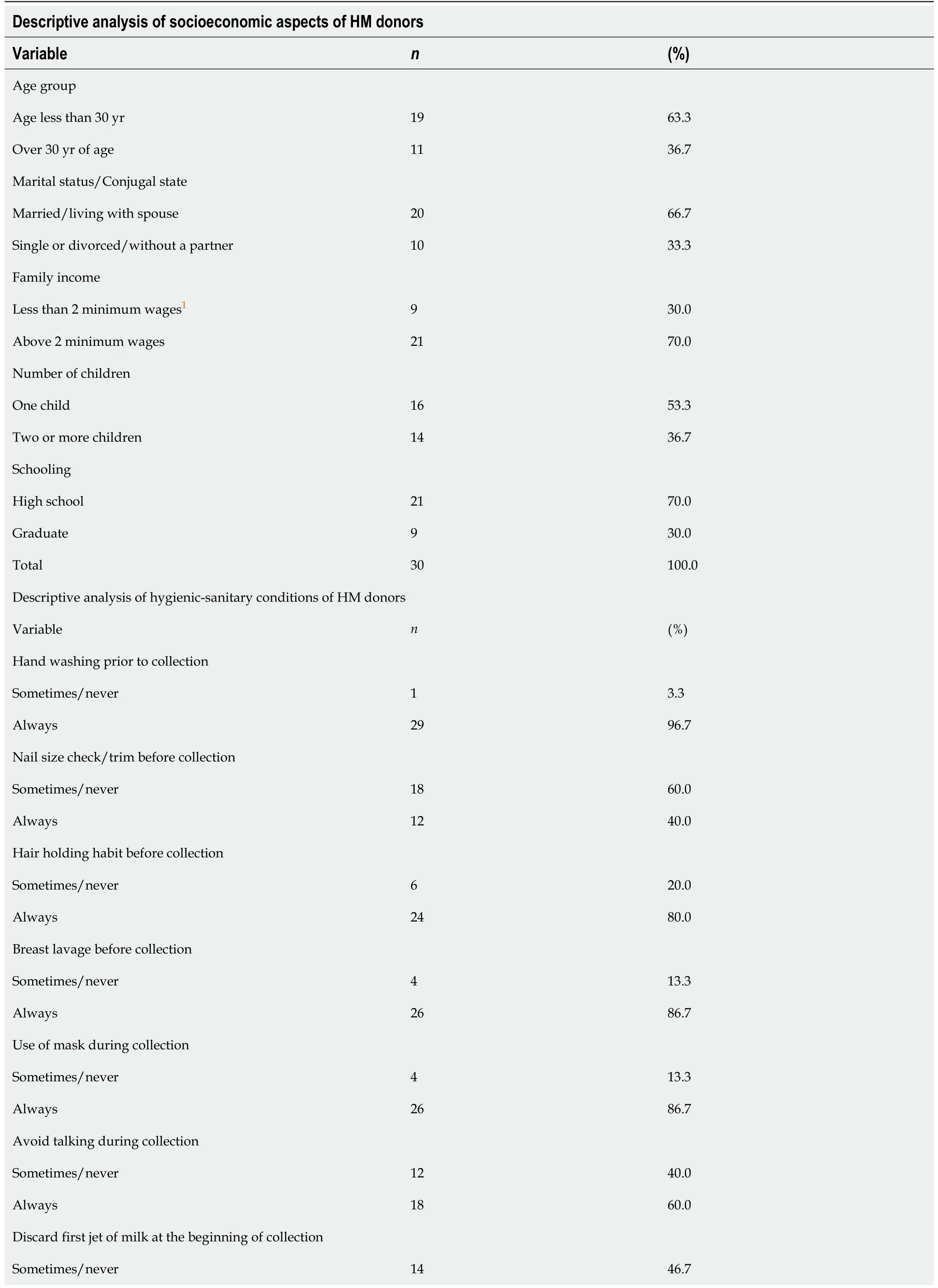
Table 1 Descriptive analysis of the socioeconomic aspects and hygienic-sanitary conditions related to the collection of human milk and to human milk donors (n=30) registered in the Human Milk Bank of the Municipal Hospital Esaú Matos in Vitória da Conquista, Bahia State, Brazil

1Minimum wage in the period:BRL954,00.2Question applied only to donors who reported performing milking by hand and using an electric pump.
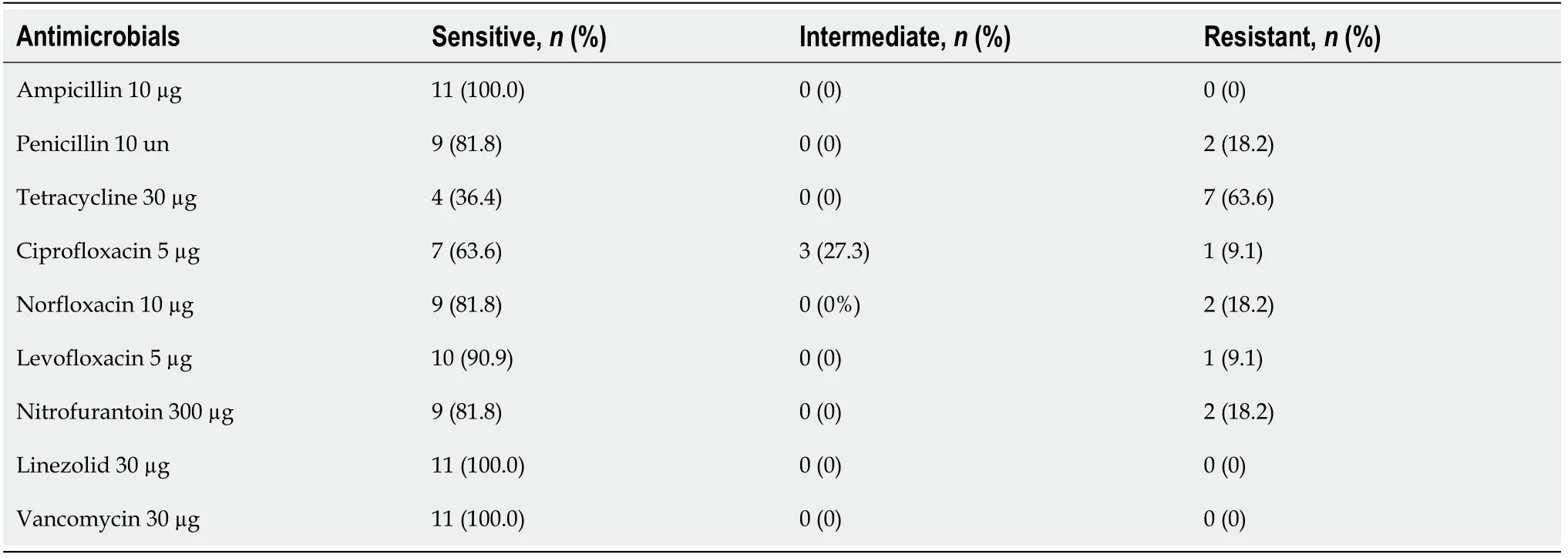
Table 2 Sensitivity profile of the isolates (n=11) of Enterococcus spp. found in relation to the list of antimicrobials evaluated following the recommendations of the Clinical and Laboratory Standards Institute (2017)

Table 3 Distribution of the virulence genes of Enterococcus faecium, Enterococcus faecalis, and Enterococcus spp.
Enterococcus spp.are present in the human microbiota and are associated with important infections in NBs, and are potential contaminants of HM samples[8].To date,there are no studies correlating NB infections and infant colonization withEnterococcus spp.in the city where this study was carried out.Moreover, there are no data associating socioeconomic aspects and hygienic-sanitary conditions regarding HM donors with the isolation of enteric bacteria.In the present study,Enterococcus spp.were isolated from 30% of the participants.Similarly,Enterococcus ssp.were isolated from raw MHM samples within four[19]and seven[20]days after delivery.In such studies, gram-positive bacteria, includingE.faecalis[21],E.faecium[21,22],Staphylococcus spp[23]and fecal coliforms[2]were identified in MHM[19], but they were not detected after milk pasteurization;however, other microorganisms continued to be detected after pasteurization.
Similar findings were observed in other HMBs in which the isolation rates were 2%[2], 6%[2], 7.5%[21], and even 70.4%[3]in non-pasteurized MHM samples and was 50.7%in pasteurized MHM[3]samples.These data reinforce the importance of strict hygienicsanitary procedures in HMBs as well as the microbiological control of HM samples.
Among the 11Enterococcus spp.isolated in this study, different types of resistance to antibiotics were found in 91% of the isolates (n=10).In the phenotypical analysis of antimicrobial resistance, most isolates were susceptible to ampicillin, linezolid, and vancomycin, but resistance to tetracycline (63%), ciprofloxacin (27%), norfloxacin,levofloxacin, nitrofurantoin (9%), and penicillin was detected.Enterococcus spp.resistance has been previously reported in samples from miscellaneous clinical materials[22], food, and clinical samples[23], with resistance rates of 5% to tetracycline[22]and of 3%[22]and 70.5%[23]to ciprofloxacin.
Despite the identification of quinolone resistance, the resistance genesgyrA andparC were not detected.This phenomenon may be associated with interferences,including differences in the pH of the Mueller Hinton media, in mean temperature,and in the density of the inoculum applied.Due toEnterococcus spp.resistance to antimicrobials such as penicillin, ampicillin, aminoglycosides, tetracycline, and vancomycin and its association with nosocomial infections[24], it is crucial to identify these bacteria in different types of biological samples, including HM.
In this study, the presence of virulence factors in the isolated strains was also evaluated, and the genesefaA andacewere found in 63% and 27% of the isolates,respectively.These genes are associated with enterococcal biofilm formation and endocarditis, making the treatment of infections caused by these microorganisms difficult.With regard to species identification, of the three isolates with theacegene,two wereEnteroccocus spp.and one hadE.faeciumandE.faecalisstrains.Among the seven isolates carrying theefaA gene, three wereEnterococcus spp., two wereE.faecalis,and two hadE.faecalisandE.faeciumstrains.
The genesaceandefaA have been previously detected inE.faecalisandE.faecium[22]hospital strains and theacegene has also been found inEnterococcus spp.of clinical origin[23].The pathogenicity mechanisms ofEnterococcus spp.are not yet fully understood.It is known that these microorganisms have virulence factors that are associated with the severity and duration of human infections as well as with the dissemination of these microorganisms in the hospital environment, where they mainly cause urinary infections, endocarditis, and bacteremia.The most frequently reported virulence factors are related to the adhesion to host tissues, invasion, abscess formation, modulation of the host inflammatory responses, and secretion of toxic products[25].
The high percentage ofEnterococcus spp.isolates in HM samples associated with the high antimicrobial resistance rate (mainly to tetracycline and ciprofloxacin) and the positivity of virulence factors (detection of the genesaceandefaA) found in this study confirm the importance of carrying out strict control from the collection to the distribution of MHM.In this context, it is necessary to take utmost care during milk processing in order to avoid the dispersion of microorganisms, as the entry of these microorganisms into the hospital environment can increase the risk of infection in hospitalized patients, mainly in those with a weakened immune system.
CONCLUSION
The present study demonstrates the necessity for microbiological control in HMBs and the requirement to develop alternatives to avoid the entry ofEnterococcus spp.species into the hospital environment, which can potentially lead to important NB infections that increase the length of hospitalizations and, consequently, health service costs.Considering that most premature NBs are transferred to intensive care units, thereby requiring food supplementation with MHM, and that the consumption of contaminated MHM can lead to mild-to-severe infections, the accurate identification of bacterial pathogens is an essential requirement for the detection of microorganism reservoirs, infection sources, HM distribution monitoring, and decision taking aiming to reduce health risks in NBs.Moreover, strict control from the collection to the distribution of donated HM may reduce the dispersion rates of virulent and multidrug-resistant microorganisms that can cause several problems in a hospital environment, leading to increases in morbidity, mortality, and costs related to health care.
对孩子而言,这样的标准似乎就更高了。但是专家提示,孩子的专注力不是天生的,需要家长耐心地培养。“专心”,不仅是孩子固定坐在位置上而已,而是应该眼到、手到、心到,在专注的过程中认真地观察、积极地思考。
ARTICLE HIGHLIGHTS
Research background
Human milk is the primary source of nutrition for newborns.Hospitalized babies often require nutritional support from Human Milk Banks.AsEnterococcus spp.are part of the microbiota of healthy donors, they are potential contaminants of pumped breast milk.
Research motivation
Donor Human Milk is an important alternative that allows the appropriate feeding of hospitalized newborns.A number of steps are needed to ensure the safe collection,processing, and quality control of colostrum, transition milk, and mature milk for subsequent distribution to newborns under a doctor or nutritionist prescription.However, such milk can be contaminated byEnterococcus spp.species that are highly pathogenic in newborns, and can increase morbidity, hospitalization time, and mortality among neonates.
Research objectives
The present study intended to identify and describe the bacterial virulence and resistance in samples obtained from the nipple-areolar region, hands, and breast milk aliquots of donors at the HMB of Municipal Hospital Esaú Matos in the city of Vitória da Conquista, Bahia State, Brazil.
Research methods
This cross-sectional study used samples from the nipple-areolar region, hands, and aliquots of raw and pasteurized milked human milk from healthy human milk donors.The study recruited donors who had a donation history of a mean weekly volume of donated milk equal to or higher than 400 mL.The microbial isolates from milk samples were analyzed before and after pasteurization.A total of 30 donors were assessed between March and August 2018.
The samples were immediately inoculated into enrichment brain heart infusion broth (HIMEDIA®) and incubated at 35-37ºC for 24 h after collection.Thereafter, they were placed in Petri dishes with 5% Blood Agar Medium (Base Agar, HIMEDIA®) and Chromagar Orientation (BD™ CHROMagar™ Orientation) in an incubator at 35-37ºC for 24 h.
Colonies in whichEnterococcus spp.were observed were selected from both media and underwent Gram staining to evaluate the morphological characteristics and to perform the catalase test and subsequent tests to identify the genus Enterococcus.
AllEnterococcus spp.isolates underwent evaluation of antimicrobial activity by the agar diffusion method, according to the recommendations of the Clinical and Laboratory Standards Institute (2017).
To detect the constitutive, virulence, and resistance genes inEnterococcus spp., all samples that were positive in the identification step underwent polymerase chain reaction after reactivation in brain heart infusion media.
A descriptive analysis of the study variables was carried out from a database constituted in Microsoft Excel through the Statistical Program EPI INFO (version 7.2.2.6).For the frequency comparison, the chi-square test was used, with values of P <0.05 (95% confidence interval) considered statistically significant.
Research results
Eighty-one samples were collected, with 30 samples from swabs containing nippleareolar region skin samples and donors’ hands, 30 samples of raw milked human milk,and 21 samples of pasteurized milked human milk.Enterococcus spp.strains were isolated from 30% of the donors and 11 strains were identified:7 strains (63.6%) from skin samples and 4 strains (36.4%) from raw MHM samples.
The majority of participants declared that they washed their hands before the collection of milk (96%), held back hair before milking (80%, washed the breasts before milking (86.7%), used a mask while milking (86.7%), avoided talking during milking(60%), discarded the first jet of milk (53.3%), and obtained milk using a manual/electric pump (80%).
No statistically significant associations between demographic and hygienic characteristics and the detection ofEnterococcus spp.were observed.Antimicrobial resistance was identified in 10 isolates (91%), mainly to tetracycline.
Research conclusions
Here we demonstrate the importance of appropriate microbiological control in Human Milk Banks in order to preventEnterococcus spp.in hospitals as these bacteria can lead to important newborn infections that increase morbidity, mortality, and hospitalizations among these patients, and increase health service costs.
Research perspectives
This study adds information to the microbiological control of donated milk, which, if well-performed from human milk collection to its distribution,may reduce the dispersion rates of virulent and multidrug-resistant microorganisms in a hospital environment, avoiding unwanted clinical outcomes among newborns.Putting such measures into practice may improve the health of children around the world.
 World Journal of Clinical Pediatrics2020年3期
World Journal of Clinical Pediatrics2020年3期
- World Journal of Clinical Pediatrics的其它文章
- Influenza B infections in children:A review
