盐碱水环境对脊尾白虾基因组DNA甲基化的影响*
秦 桢 李吉涛 李明栋 王佳佳 葛倩倩 刘 萍 李 健
盐碱水环境对脊尾白虾基因组DNA甲基化的影响*
秦 桢1,2李吉涛2①李明栋2王佳佳2葛倩倩2刘 萍2李 健2
(1. 上海海洋大学 水产科学国家级实验教学示范中心 上海 201306;2. 中国水产科学研究院黄海水产研究所 农业农村部海洋渔业可持续发展重点实验室 青岛海洋科学与技术试点国家实验室海洋渔业科学与食物产出过程功能实验室 山东 青岛 266071)
为探讨盐碱水环境对脊尾白虾()基因组DNA甲基化的影响,本研究利用MethylRAD-Seq技术探究了长期盐碱水养殖组(SAS)和正常海水养殖对照组(SW)脊尾白虾鳃组织中的DNA甲基化水平,并对关键通路和基因进行了差异表达分析。结果显示,脊尾白虾鳃组织基因组中CG和CWG位点(W=A或T)分别检测到2347003和416176处甲基化,甲基化普遍存在于基因组的基因间区和内含子区域,共筛选到8805个(8189个CG-DMSs和616个CWG-DMSs)差异甲基化位点,盐碱水环境下DNA甲基化水平略有增强。通过KEGG富集分析发现,DMS所在差异表达基因显著富集在HIF-1信号通路和剪接体通路,通路中、和等关键基因在脊尾白虾盐碱水环境适应中可能发挥着重要作用;对SW和SAS组差异甲基化基因(DMG)进行筛选,得到158个CG-DMGs和94个CWG-DMGs,其中,富集到脂质代谢和囊泡介导的转运通路中的DMG最多;此外,有一些DNA甲基化位点与基因表达呈负相关,表明DNA甲基化与基因调控之间存在复杂的联系,大部分基因组DNA甲基化对基因表达有正调控效应。本研究结果首次分析了在盐碱水环境下脊尾白虾鳃组织的DNA甲基化水平特征,为解析甲壳类盐碱水环境适应机制提供了基础信息。
脊尾白虾;盐碱水环境;DNA甲基化;差异表达基因
受气候、地形等自然和人为因素的影响,全球土壤和水的盐渍化程度正在增加(常玉梅等, 2021)。据研究推测,这些地区在未来将日趋扩大,严重威胁水产养殖的发展空间(张建峰, 2008)。目前,已有大量研究集中在盐碱地管理和盐碱水对水生动物(包括甲壳类)的毒性影响(Arooj, 2021; Chen, 2020; Conrado, 2017; 杨富亿等, 2004)。为促进盐碱地水产养殖的发展,对一些适应性较强的品种开展了盐碱地池塘养殖,例如耐盐碱鱼类、广盐性鱼类和虾蟹类等,目前应用于盐碱水规模化养殖的品种包括罗非鱼()、梭鲈()和凡纳滨对虾()等(徐文龙等, 2021; 来琦芳等, 2021)。对鱼类耐盐碱机制的研究主要集中在生理和分子调控机制(渗透压调节、氨氮代谢、激素调节等)方面,而对甲壳动物耐盐碱的研究则主要集中在对碱度胁迫的耐受能力及其生理变化(常玉梅等, 2021)。然而,目前对甲壳类动物耐盐碱的分子机制尚不清楚。
越来越多的证据表明,表观遗传学调控机制在生物对环境胁迫后的适应性调控过程中占据着至关重要的地位(Xu, 2020; Han, 2021)。研究发现,生物体可以通过DNA甲基化调节当代和后代的环境适应能力,比如对低氧、低盐、盐碱等环境胁迫的适应(Wang, 2021; 王会等, 2017; 环朋朋等, 2019; Su, 2020)。在甲壳动物中,有学者开展关于DNA甲基化对其生长发育、防御生物和非生物胁迫(包括热应激、盐度、重金属等)的影响。例如,薛蓓等(2017)研究了脊尾白虾() 4个生长发育阶段线粒体基因组的甲基化水平,发现脊尾白虾通过甲基化调节能量代谢影响机体的生长发育进程。热应激下,卤虫()和海鞘()的胞嘧啶甲基化导致基因的差异表达(Norouzitallab, 2014; Hawes, 2018)。Lovett等(2001)研究发现,甲基法尼酯(methyl farnesoate)能够调节岸蟹()机体渗透压,使其适应盐度变化很大的河口环境。年龄、取样地点、水温、禁食和镉暴露均能显著影响淡水钩虾()基因组胞嘧啶的甲基化水平(Cribiu, 2018)。
脊尾白虾俗称海水小白虾、白米虾,是我国东部沿海广温广盐的重要中小型经济虾类,以黄、渤海产量最高,年产量数千吨(Ge, 2015; Fan, 2020)。近年来,脊尾白虾养殖业迅速扩大,成为促进我国沿海渔业经济发展的重要品种(柳飞等, 2016; 李明栋等, 2021)。在滨海盐碱水试养脊尾白虾过程中,发现脊尾白虾虽然能够正常生存,但其生长及繁殖能力在高碳酸盐碱度下明显降低(Ge, 2019; 柳飞等, 2016)。开展脊尾白虾盐碱适应机制解析,对耐盐碱良种培育和盐碱水养殖具有重要意义。本研究利用河北沧州养殖基地脊尾白虾耐盐碱品系和山东日照试验基地脊尾白虾“黄育1号”作为研究对象,利用高效、低成本全基因组DNA甲基化检测技术(MethylRAD-Seq)对脊尾白虾鳃组织DNA甲基化水平进行分析,筛选与盐碱胁迫相关的关键基因和重要甲基化位点,旨在为解析脊尾白虾适应盐碱水环境的分子机制提供基础数据,为甲壳类盐碱适应机制研究提供参考。
1 材料与方法
1.1 实验材料
本研究用脊尾白虾“黄育1号”养殖于山东省日照海辰水产有限公司正常海水中(盐度25、pH 8.2、碳酸盐碱度2.0~3.0 mmol/L),脊尾白虾耐盐碱品系养殖于河北沧州滨海型盐碱水中(盐度15~20、pH 8.3~9.2、碳酸盐碱度3.5~8.0 mmol/L)。根据实验目的,正常海水养殖对照组(SW)取自日照养殖的“黄育1号”健康成虾,长期盐碱水养殖组(SAS)为2019年“黄育1号”引入河北沧州滨海型盐碱水选育3代的成虾个体。实验用脊尾白虾体长为(5.65±0.50) cm,体重为(1.52±0.38) g,随机选取体质健康、活力良好的个体。进行实验前3 d停止喂食,待虾状态稳定后解剖其鳃组织于–80℃下保存。SW和SAS组分别取6个生物平行,每个平行3份鳃组织,各组织样品分别标记为SW1~3和SAS1~3,用于后期DNA和总RNA提取。
1.2 DNA提取和文库构建
采用TIANamp Marine Animals DNA Kit (TIANGEN)提取DNA,利用核酸定量仪(Thermo, NanoDrop 2000)和1%琼脂糖检测DNA质量、浓度和完整性。利用修饰依赖性内切酶EI 5U于37℃酶切4 h消化基因组DNA,按照10 µL酶切产物与0.80 µL特异性接头、1 µL 10×T4ligase buffer、1 µL 10 mmol/L ATP和2 µL T4DNA ligase (400 U/µL)的比例混合,加超纯水使总体积为20 µL。4℃连接6~8 h,连接产物经特异性引物反应扩增,反应条件:98℃初始变性30 s,98℃ 20个循环变性5 s,60℃退火20 s,72℃片段延伸10 s。每个样品平行扩增3管,用于后续回收目的片段。取3 µL PCR产物用8%聚丙烯酰胺凝胶进行非变性电泳检查。然后按照制造商说明将PCR产物用SteadyPure Agarose Gel DNA Purification Kit (Accurate Biotechnology Co., Ltd)进行切胶回收。利用QIAquick PCR Purification Kit和Qubit对PCR产物进行纯化和定量,基于MethylRAD技术构建标签文库,由上海欧易生物医学科技有限公司采用Illumina SE sequencing (50 bp)测序平台进行文库测序。
1.3 数据处理与统计分析
1.3.1 原始数据的质控 利用Illumina测序平台得到的原始图像数据文件经碱基识别转化为原始测序序列(raw reads)。在组装之前,通过预处理(随机截取103%的数据,去除距5′或3′端13~17 bp的片段、有待检测酶切位点的片段和低质量片段)对原始数据进行过滤与质控,得到各样品的高质量的有效数据(clean reads),进行后续的数据分析。
1.3.2 甲基化位点水平的定量 以本课题组前期组装的脊尾白虾全基因组序列为参考基因组(数据尚未发表),使用Bowtie 2软件(V2.3.4.1)对clean reads进行比对,参数设置为:--no-unal。比对完成后,去除能够同时比对到参考序列的多个位置的reads,采用RPM (reads per million)为单位对甲基化位点水平进行定量,计算公式为位点甲基化水平定量值=位点覆盖reads数/文库高质量reads×1000000。根据比对结果,统计CG位点(CmCGG、GmCGG、CmCGA、AmCGG、CmCGC、CmCGT、TmCGG和GmCGC)和CWG位点(mCAG和mCTG)的数目、电子酶切位点数目及reads深度,从而描绘整条染色体上2种位点的分布情况。
通过修饰依赖型核酸内切酶(EI酶和PI酶)进行电子酶切,识别C5-甲基化胞嘧啶(5-mC)和C5-羟甲基化胞嘧啶(5-hmC),得到一个30 bp (28~ 37 bp)左右的酶切片段。图1为EI酶和PI酶的酶切位点示意图。对于mCG位点,只选择EI酶的CmCGG位点和PI酶的8种比较稳定的CG位点进行下游数据分析(Boers, 2018);对于mCHG (H代表简并碱基A、C、T)位点,EI酶和PI酶都采用mCAG和mCTG结果,用CWG表示(W代表简并碱基A、T)。
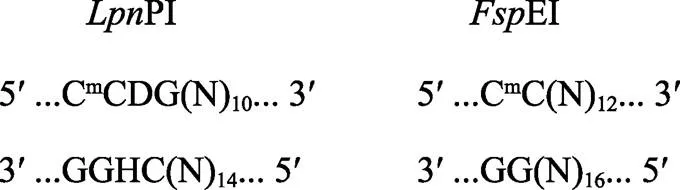
图1 LpnPI酶和FspEI酶的酶切位点
1.3.3 甲基化位点注释及分布 根据甲基化位点的位置信息,使用SnpEff软件(V 4.1g)位点进行注释,给出每个位点所在的基因元件以及位点注释的详细信息,利用bedtools软件(V 2.25.0)统计各个样品中不同基因元件中甲基化位点的分布情况,进一步选取基因转录起始位置(TSS)上下游各2 kb区段,转录终止位置(TTS)上下游各2 kb区段及基因体(genebody),将分布在不同区域的每个基因的序列均分为多个窗口,统计每个窗口的RPM值,将所有基因相同窗口的RPM值取平均,作为该窗口的RPM值,描述测序reads在上述区段的分布趋势。
1.3.4 甲基化位点的差异分析 将各个样本的base mean值作为测序深度的表达量估算值,利用DESeq包(V 1.36.0)对测序深度进行标准化处理,计算差异倍数,并采用负二项分布检验对reads数进行差异显著性检验,以此来筛选差异甲基化位点(DMS)。默认<0.05且差异倍数(fold change)大于2为筛选差异的条件。使用bedtools软件对各个比较组中不同基因元件中甲基化位点的分布情况进行统计。记录DMS的分布,并统计DMS在不同基因功能元件上的分布。统计DMS的甲基化水平,用以描述DMS所在基因的甲基化水平,获得DMS所在差异表达基因(DEG)。根据GO (Gene Ontology, http://www. geneontology.org/)和KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/)数据库进行GO和KEGG pathway功能和富集分析。< 0.05的GO term被认为是显著富集,Benjamini- Hochberg多重检验校正所有值以获得FDR。
1.3.5 基因水平的甲基化差异分析 以某一基因内的所有甲基化位点水平之和代表该基因的甲基化水平,对SW和SAS两组样品进行组间比较。同样计算差异倍数,利用负二项分布检验的方式对基因进行差异显著性检验,最终根据差异倍数及差异显著性检验结果来筛选差异甲基化基因(DMG)。由DMG组成基因集,根据GO和KEGG数据库进行GO和KEGG pathway功能和富集分析。
1.3.6 通过qRT-PCR验证差异甲基化基因和差异表达基因 利用TRIzol试剂(Invitrogen)提取鳃组织样本的总RNA,经核酸定量仪(Thermo, NanoDrop 2000)和1%琼脂糖凝胶电泳检测RNA的浓度和质量。按照HiScriptⅡ Q Select RT SuperMix for qPCR (Vazyme)说明书进行RNA反转录,合成cDNA。以cDNA为模板进行引物的验证和后续2组脊尾白虾鳃组织的甲基化特征分析。为了验证由MethylRAD-seq确定的脊尾白虾鳃组织中DMS相关的DEG,选取显著富集的KEGG通路相关的12个DEGs进行了qRT-PCR分析。用于qRT-PCR分析的mRNAs引物见表1。以18S为内参,引物序列如下:5′-TATACGCTA GTGGAGCTGGAA-3′和3′-GGGGAGGTAGTGACGA AAAAT-5′ (李美玉等, 2012; Wang, 2015)。在7500 fast Real-Time PCR系统(Applied Biosystems)中,使用SYBR Green PCR Master Mix (Life Technologies)进行qRT-PCR。取每个时间点的3个转录组样本进行qRT-PCR,每个生物重复进行3个技术重复。使用CT方法(2−ΔΔCt)计算目的基因的相对表达量(Tapia, 2017)。
表1 用于qRT-PCR分析的mRNA引物

Tab.1 Primers of mRNAs used for the qRT-PCR analysis
2 结果与分析
2.1 DNA甲基化测序结果统计
对SAS和SW的6个样本进行MethylRAD测序和数据统计,测序结果显示,平均每个样本获得184 868 782条原始数据。对完成过滤质控的CG和CWG位点进行酶切,共获得55 642 471条有效数据,占总reads的35.04%~37.49%。统计每条scaffold上各种酶切位点的数目,每组有效数据平均88.71%可比对到脊尾白虾的参考基因组,共有22 633 946条有效数据在参考基因组上有唯一比对位置,具体测序信息见表2。
2.2 组间甲基化位点的分布统计
根据参考基因组比对结果,统计6个样品筛选到的甲基化位点数目及平均测序深度,在SW组的3个样品中平均发现377 441和64 655个CG和CWG型DNA甲基化位点,平均甲基化位点覆盖深度分别为7.990和5.203 (表3)。在SAS中共发现404 893个和74 071个CG和CWG型DNA甲基化位点,平均甲基化覆盖率分别为7.986和5.183,每个样本DNA甲基化位点(CG位点和CWG位点)的测序覆盖深度见表3。此外,与SW相比,SAS中CG处的DNA甲基化水平增加了0.05%,CWG处的DNA甲基化也增加了0.38%。这些结果表明,对于脊尾白虾鳃组织来说,mCG二核苷酸的胞嘧啶上发生突变是甲基化的主要表现形式,同时盐碱胁迫下,脊尾白虾鳃组织基因组的甲基化水平有所升高。
表2 MethylRAD文库测序数据统计

Tab.2 Statistics of sequencing data of MethylRAD library
根据甲基化位点的位置信息,对位点进行注释,发现这些DNA甲基化位点主要分布在5′端的核酸区、外显子、基因区、基因间区、内含子、剪接位点、3′端的核酸区域,CG型甲基化位点在不同功能元件上的分布数量均显著多于CWG型,但CG和CWG两种类型位点的分布趋势基本一致,基因间区中分布的甲基化位点比例最高,其次是内含子区。总之,盐碱胁迫导致基因组功能组成元件中CG和CWG位点分布具有重叠变化(图2)。统计reads在转录起始位置(TSS)上下游各2 kb区段,转录终止位置(TTS)上下游各2 kb区段和基因体的分布趋势,结果发现各个样品之间的DNA甲基化水平趋势是相似的,DNA甲基化位点多分布在基因体,DNA甲基化位点分布曲线在TSS的下游序列和TTS的上游序列甲基化标签频率明显高于其他序列(图3)。
2.3 差异甲基化位点表达分析
测序深度信息进行标准化处理后,分别筛选到8189个和616个CG型差异甲基化位点(CG-DMSs)和CWG型差异甲基化位点(CWG-DMSs) (图4)。CG-DMS和CWG-DMS大多数分布在基因间区和内含子区,DMS在不同功能元件上的分布详情见图5A和B。可以看出CG-DMS的数目显著多于CWG-DMS,基因间区和内含子区筛选到的DMS占较大比例。
2.4 位点水平的甲基化差异分析
对DMS所在的差异表达基因(DEG)进行GO功能富集分析,分析了CG和CWG甲基化水平中DEG最显著丰富的前30条GO terms。分析基于筛选3个类别中具有2个以上不同表达位点相关基因的GO条目,根据每个条目对应的–lg10从大到小排序10个,分为生物过程、细胞成分和分子功能。在CG-DEG中,最显著富集的GO term包括RNA剪接(RNA splicing)、剪接体复合体(spliceosomal complex)和ATP依赖的5′-3′DNA解旋酶活性(ATP-dependent 5′-3′ DNA helicase activity) (图6A)。CWG-DEG的GO富集分析结果表明,生物过程、细胞成分和分子功能中最显著富集的GO term分别是上皮细胞迁移,开放性气管系统(epithelial cell migration, open tracheal system)、中心体(centrosome)和蛋白质丝氨酸/苏氨酸激酶活性(protein serine/threonine kinase activity)(图6B)。
表3 甲基化位点数据及其深度统计

Tab.3 Statistics of methylation site data and depth
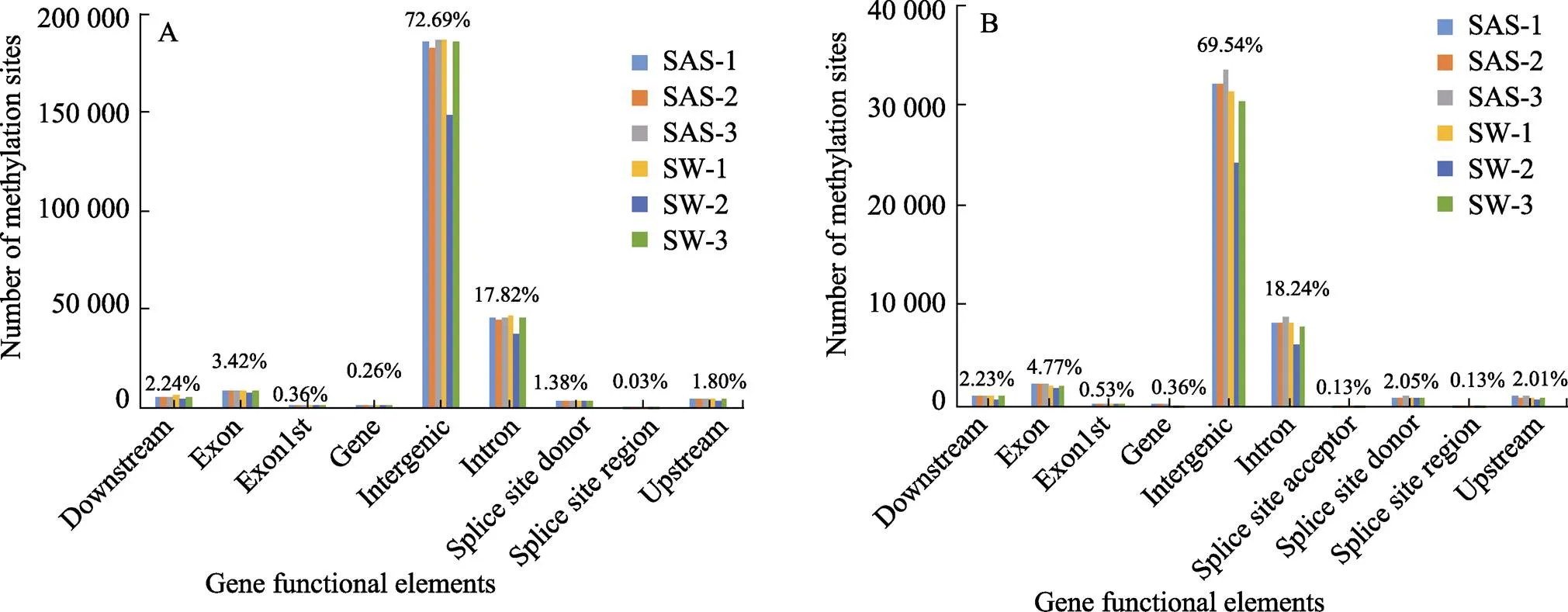
图2 甲基化位点在不同基因功能元件上的分布
A:CG位点的分布;B:CWG位点的分布
A: Distribution of CG methylation site; B: Distribution of CWG methylation site

图3 甲基化位点在TSS、TTS和Genebody的分布
A:CG位点在TSS的分布;B:CG位点在TTS的分布;C:CG位点在TSS、TTS和Genebody的分布;D:CWG位点在TSS的分布;E:CWG位点在TTS的分布;F:CWG位点在TSS、TTS和Genebody的分布
A: Distribution of CG sites in TSS; B: Distribution of CG sites in TTS; C: Distribution of CG sites in TSS, TTS, and Genebody; D: Distribution of CWG sites in TSS; E: Distribution of CWG sites in TTS; F: Distribution of CG sites in TSS, TTS, and Genebody

图4 差异甲基化位点统计
KEGG通路分析用于筛查DEG在CG和CWG甲基化水平上的生物学通路和信号转导。图7显示了CG-DMG富集到的前20个KEGG通路。结果显示,CG-DMG在HIF-1信号通路(HIF-1 signaling pathway)、剪接体(spliceosome)、孕酮介导的卵母细胞成熟(progesterone-mediated oocyte maturation)、卵母细胞减数分裂(oocyte meiosis)和细胞周期(cell cycle)通路显著富集(图7A)。下调DEG在富集剪接体通路,而上调DEG在HIF-1信号通路、孕酮介导的卵母细胞成熟、卵母细胞减数分裂、细胞周期和cAMP信号通路(cAMP signaling pathway)显著富集(图7B, C)。HIF脯氨酰羟化酶()、己糖激酶()、丝氨酸/苏氨酸蛋白激酶mTOR样()和溶质载体家族2 (促进葡萄糖转运蛋白)成员1 ()在HIF-1信号通路中显著富集上调表达(表4)。细胞分裂周期5样蛋白()、U1小核核糖核蛋白()、前mRNA处理因子()、剪接因子()、WW结构域结合蛋白()基因参与剪接体途径且显著下调表达(表4)。
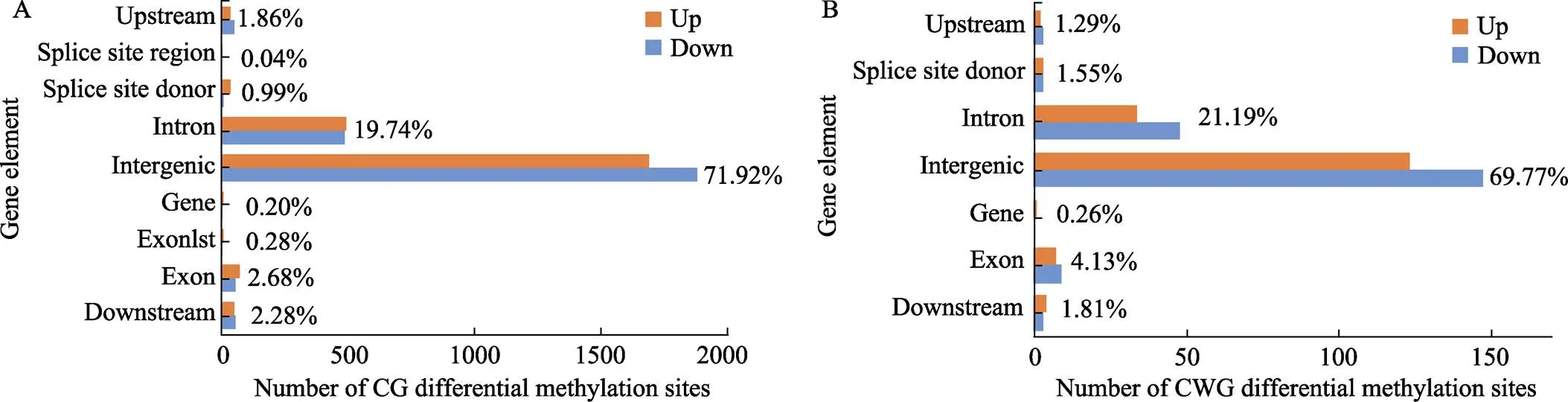
图5 差异甲基化位点在不同基因功能元件上的分布
A:CG位点的分布;B:CWG位点的分布
A: Distribution of CG methylation site; B: Distribution of CWG methylation site
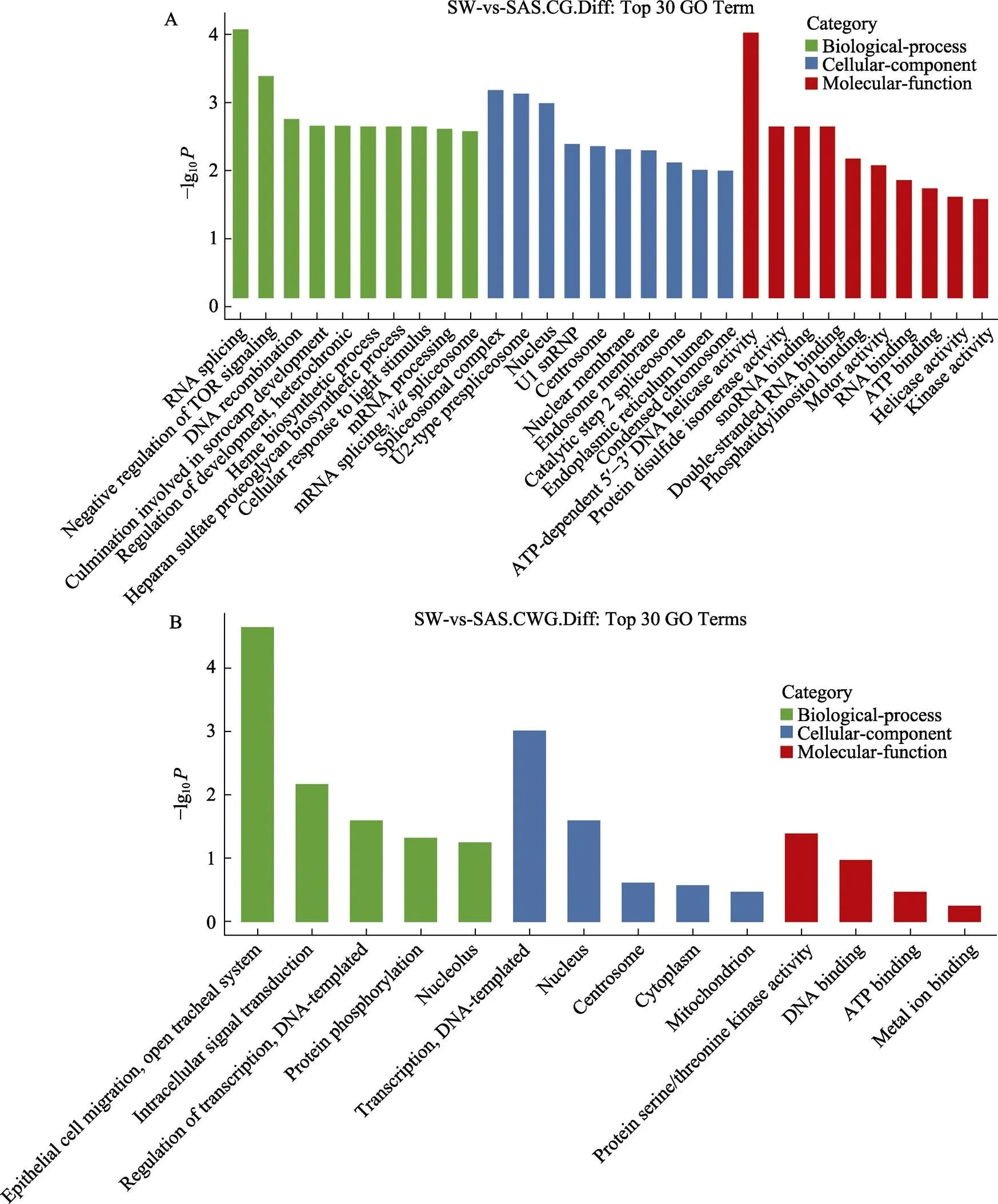
图6 CG和CWG以及上调和下调差异甲基化位点所在基因的前30个GO功能条形图
A:CG位点DEG的GO分类前30;B:CWG位点DEG的GO分类前30
A: The top 30 GO classification of the DEGs at the CG site; B: The top 30 GO classification of the DEGs at the CWG site

图7 CG差异甲基化位点所在基因的前20条KEGG富集分析
A:CG差异甲基化位点的KEGG富集分析top 20;B:下调表达CG差异甲基化位点的KEGG富集分析top 20;C:上调表达CG差异甲基化位点的KEGG富集分析top 20
A: The top 20 KEGG enrichment analyses of the CG-DEG; B: The top 20 KEGG enrichment analyses of down-regulated expression of CG-DEG; C: The top 20 KEGG enrichment analyses of up-regulated expression of CG-DEG
表4 HIF-1信号通路和剪切体通路中差异表达基因
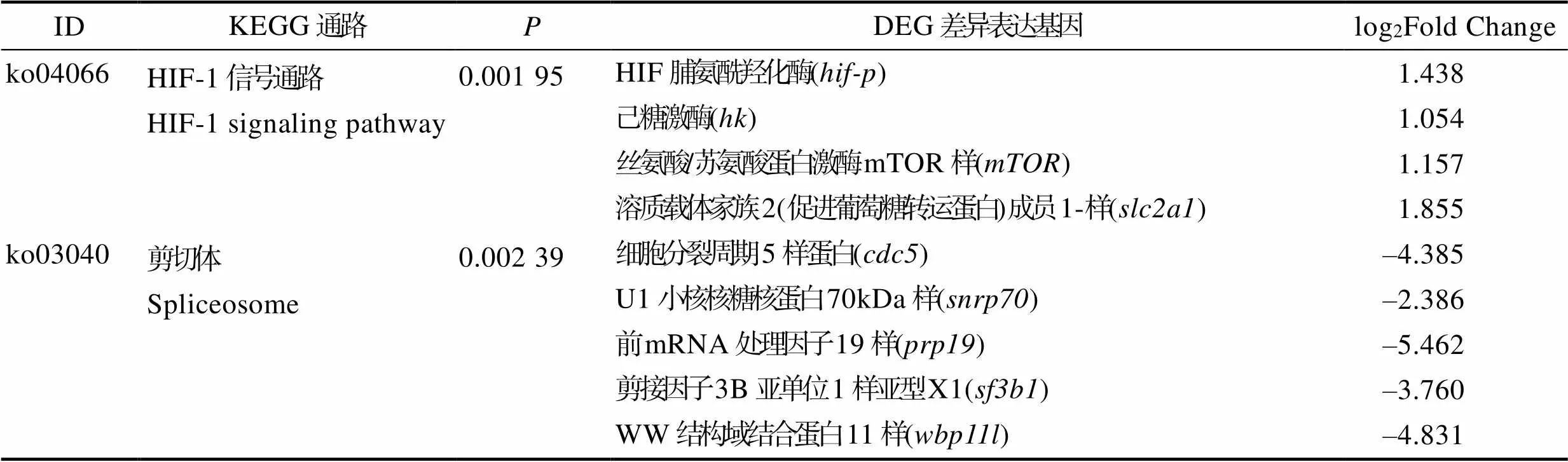
Tab.4 Differentially expressed genes of HIF-1 signaling pathway and spliceosome
2.5 基因水平的甲基化差异分析
对不同比较组间的差异表达甲基化位点所在基因进行统计,差异甲基化基因数目见图8。CG位点共注释到12892个表达甲基化基因,其中筛选到158个CG型差异甲基化基因(CG-DMGs),包括77个(48.73%)上调表达和81个(51.27%)下调表达;CWG位点注释到8666个甲基化基因,其中94个显著差异表达,上调基因33个(35.11%),下调基因61个(64.89%)。与DMS表达相同,下调表达基因多于上调表达。
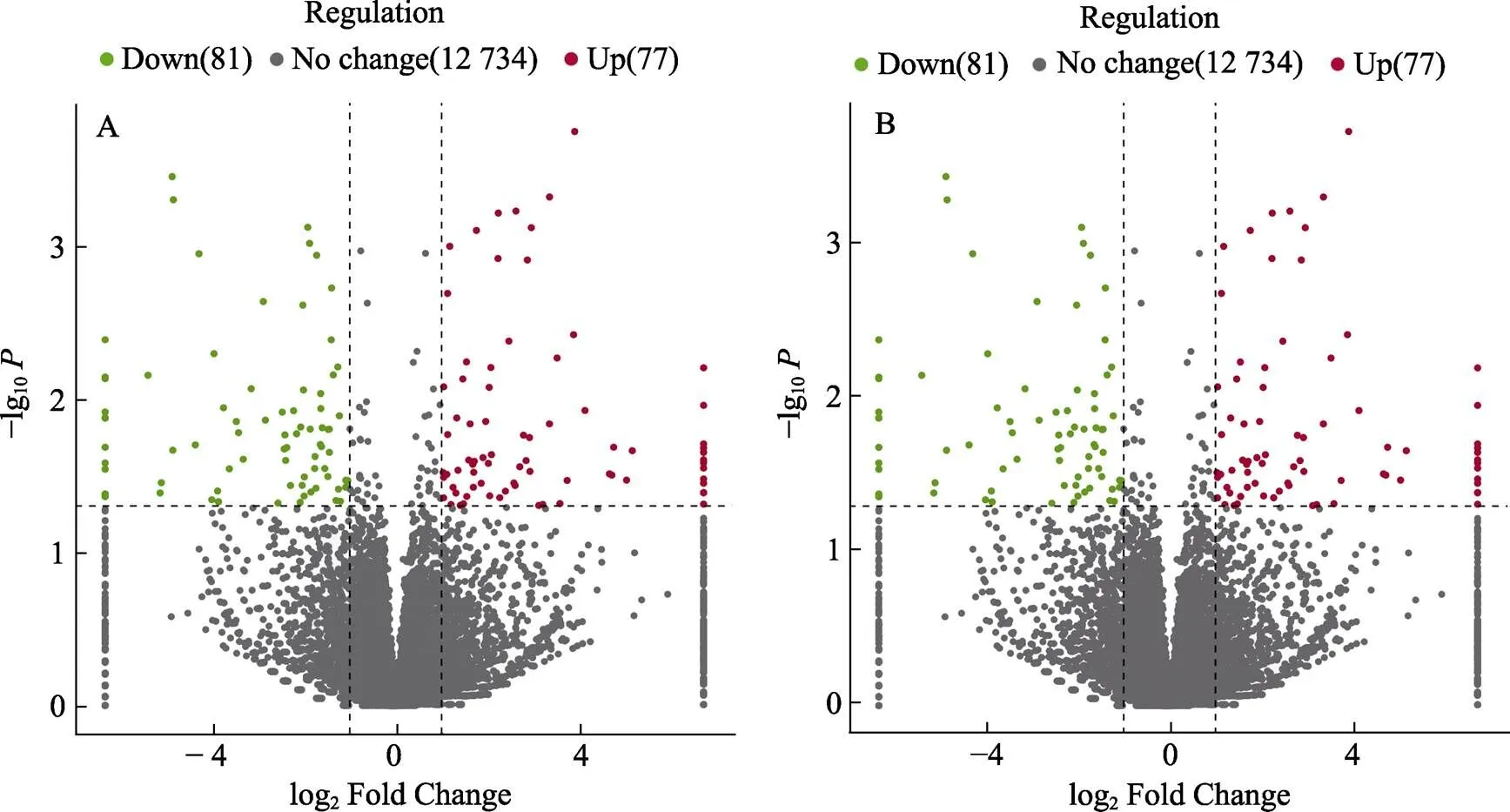
图8 组间差异甲基化基因火山图
A:CG型差异甲基化基因分布;B:CWG型差异甲基化基因分布
A: Distribution of CG-DMG; B: Distribution of CWG-DMG
通过对差异甲基化基因进行GO功能富集,对基因的功能进行描述。GO富集分析前30如图9所示:CG-DMG在肌肉组织发育(muscle organ development)过程显著富集(图9A);CWG-DMG在囊泡介导的转运(vesicle-mediated transport)、细胞膜(membrane)和铁离子结合(zinc ion binding)等离子转运过程中显著富集(图9B)。
CG-DMG共富集到39条显著富集KEGG通路(0.05),可分为5类,包括cellular processes (4条)、environmental information processing (6条)、genetic information processing (2条)、metabolism (16条)和organismal systems (11条)(图9C)。发现41%的相关基因与代谢过程(16条)相关,其中脂质代谢通路(6个)中富集到的基因数目最多(图9C)。CWG-DMG富集到21条KEGG通路,与CG型不同的是,CWG型缺少genetic information processing条目,大部分基因与生物过程(6条)相关,但信号转导通路(4个)中富集到的基因数目最多(图9D)。
2.6 组间差异甲基化位点在差异甲基化基因上的分布
对2组DMS和DMG进行比对分析,196个CG- DMSs对应到125个CG-DMGs上,36个CWG-DMSs比对到34个CWG-DMGs (表5)。基因和甲基化位点表达大部分呈正相关,但有一少部分位点与基因呈负相关。共13个CG-DMSs与对应的9个CG-DMGs呈负相关,其中,有8个CG-DMSs高甲基化而基因表达下调,5个CG-DMSs低甲基化而基因表达上调,只有2个CWG-DMSs与对应的2个CWG-DMGs呈负相关,均是高甲基化而基因下调表达(表6)。对所筛选到的负相关的甲基化位点所在功能元件进行统计,发现CG-DMS和CWG-DMS绝大比例分布在基因间区,仅有1个CG-DMS分布在内含子区,具体详情见表6。
2.7 qRT-PCR验证
为了验证MethylRAD-seq检测到的DMS所在DEG的甲基化水平变化及其在盐碱环境下基因表达水平之间的相关性,利用qRT-PCR分析评估了与HIF-1信号通路、剪接体、孕酮介导的卵母细胞成熟、卵母细胞减数分裂和细胞周期通路相关的12个DEGs。结果显示,这些基因存在显著差异表达:、、、、和上调表达;、、、、和下调表达。长期盐碱水养殖环境使脊尾白虾鳃组织中这些基因的DNA甲基化状态发生显著变化 (图10)。
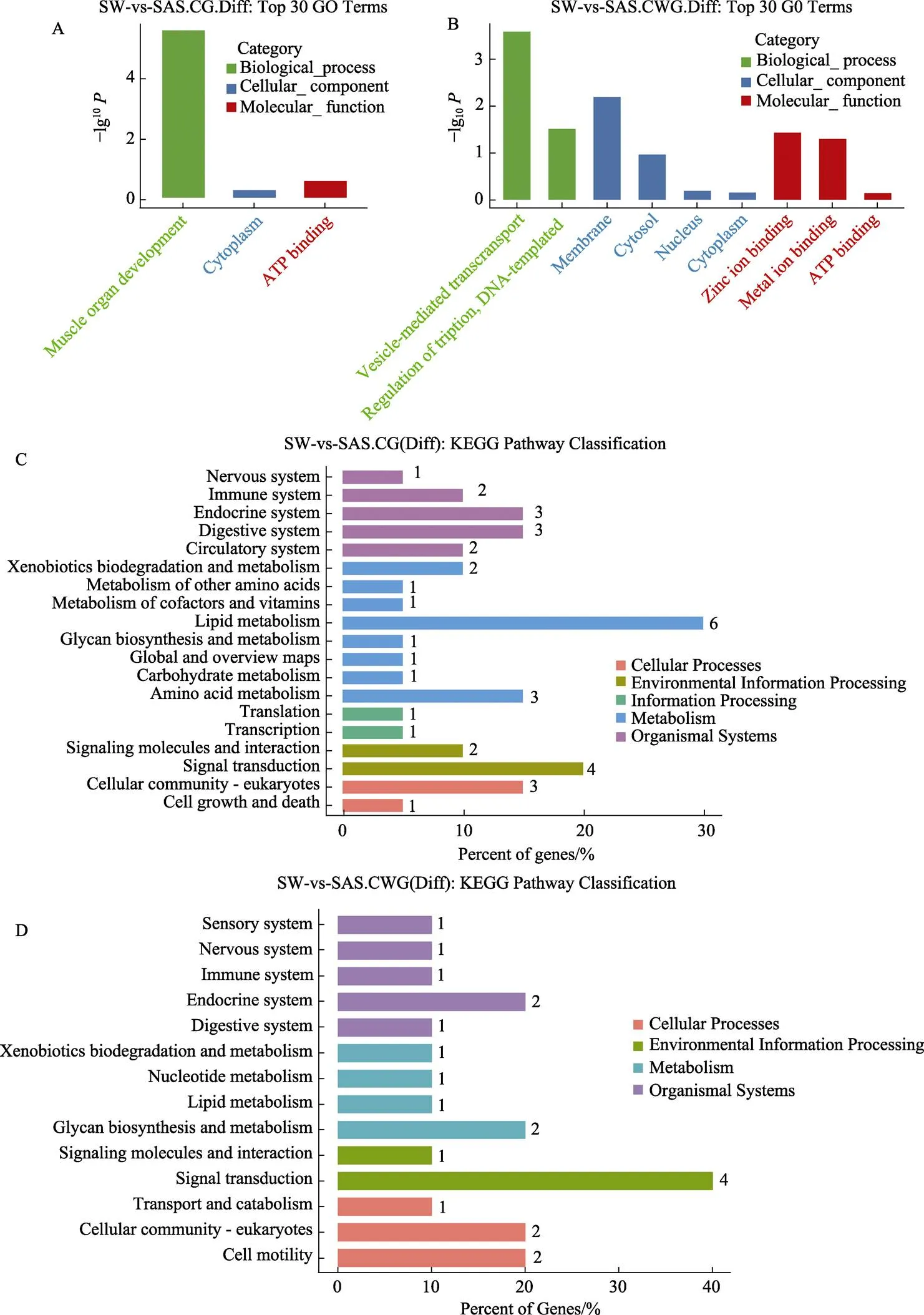
图9 差异甲基化基因的GO功能和KEGG通路富集
A:CG型差异甲基化基因的GO分类;B:CWG型差异甲基化基因的GO分类;C:CG型差异甲基化基因的KEGG富集通路;D:CWG型差异甲基化基因的KEGG富集通路
A: GO functional classification of CG-DMG; B: GO functional classification of CWG-DMG; C: KEGG pathway enrichment of CG-DMG; D: KEGG pathway enrichment of CWG-DMG
表5 差异甲基化位点映射到差异甲基化基因的数量统计

Tab.5 Quantitative statistics of differential methylation sites mapped to differential methylation genes
表6 差异甲基化位点与差异甲基化基因呈负相关的基因
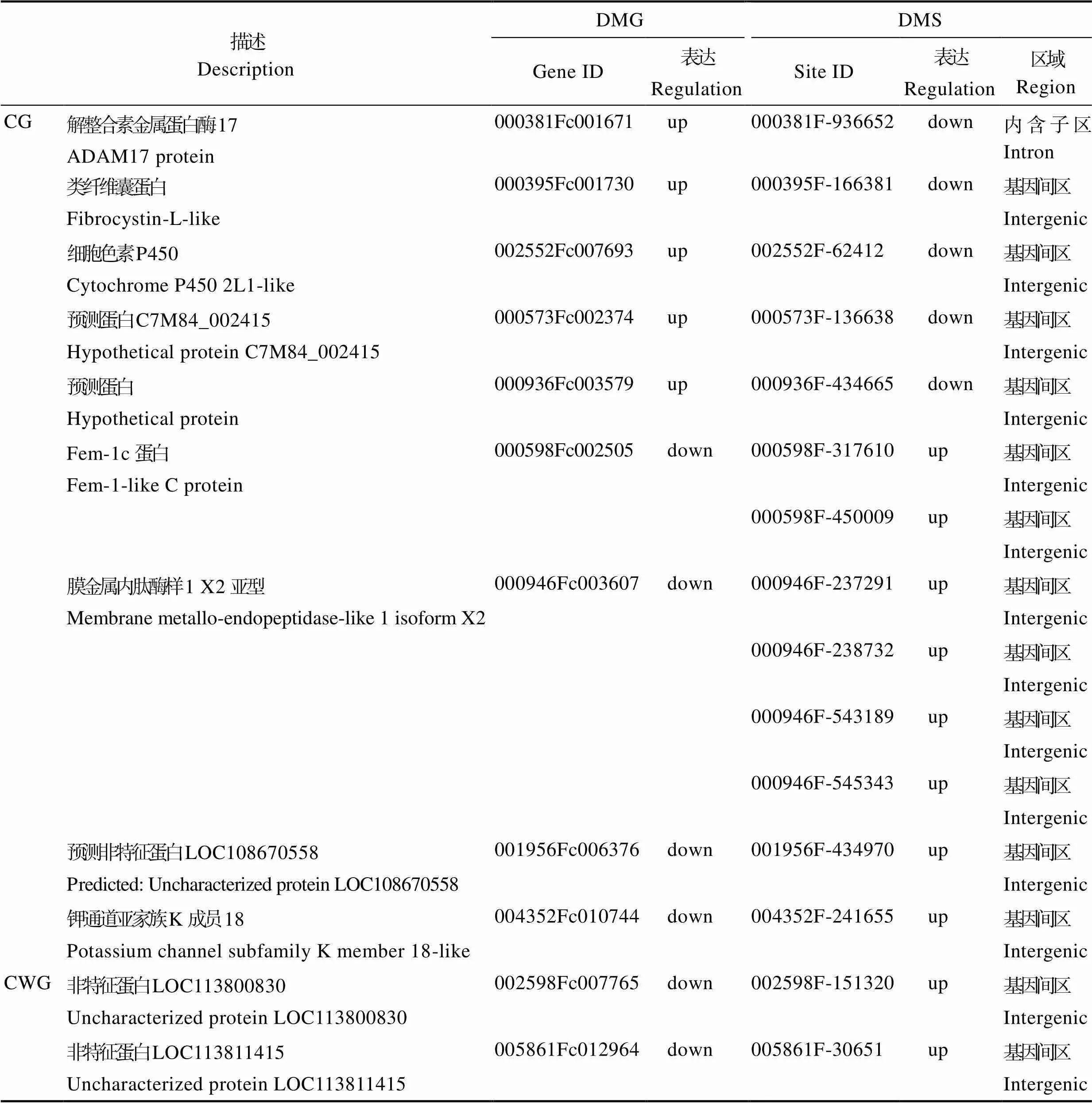
Tab.6 Genes with negative correlation between differential methylation sites and differential methylation genes

图10 KEGG富集途径中差异表达基因的qRT-PCR结果
3 讨论
DNA甲基化对真核生物生长发育至关重要,包括调节配子的形成、早期胚胎发育、细胞分化、衰老和致癌等关键过程(Varriale, 2006; 蔡影等, 2018)。无脊椎动物的整体基因组胞嘧啶甲基化水平较低,甲壳类动物和昆虫相比其他无脊椎动物如软体动物的胞嘧啶甲基化水平更低(Gavery, 2013)。本研究中,通过对正常海水养殖和长期盐碱水养殖的脊尾白虾鳃组织构建DNA甲基化文库,利用MethylRAD测序,发现了一些可能与脊尾白虾鳃组织响应盐碱胁迫的差异甲基化位点及其相关基因和差异甲基化基因。结果显示,SAS组鳃组织基因组中CG和CWG位点略有增加,表明盐碱水环境诱导脊尾白虾鳃组织基因组中DNA甲基化升高,通过激活或抑制某些通路的表达来适应盐碱环境,在脊尾白虾适应外界环境的变化中发挥着非常重要的作用。环境变化总能引起水生生物DNA甲基化的变化,研究发现,水环境中化合物和温度等外界环境变化能够引起斑马鱼()和仿刺参() DNA甲基化水平升高(陈宏姗等, 2016; Yang, 2020)。随着环境条件的变化,水生生物DNA甲基化的水平可能出现趋势性变化,例如,随着干露胁迫时间的增长,太平洋牡蛎()基因组甲基化水平呈现先增高后降低的趋势;斑马鱼ZF4细胞在短期低温培养时基因组DNA甲基化水平明显增高,但长期低温培养后DNA甲基化水平反而下降(张鑫等, 2017; 侯艳雯等, 2019)。也有研究表明,高温胁迫使得仿刺参、虾夷扇贝()和近江牡蛎() CG平均甲基化水平显著降低(温争争等, 2021; 吴彪等, 2016; 王翠丽等, 2019)。之前的研究发现,CG位点在各种物种中显示出较高水平,范围从虾夷扇贝约89.5%到仿刺参约91% (吕佳, 2013; 李玉强等, 2018)。除CG外,CWG甲基化在基因内并不常见,但在基因组的基因间和内含子区域中更为丰富(Lister, 2009)。本研究中,甲基化位点水平相对定量结果显示,脊尾白虾CG型甲基化与其他海洋无脊椎动物类似,是甲基化主要的表现形式。
DNA甲基化水平根据基因组功能元件的不同而变化,且在基因组的基因间区和内含子区域中更为丰富。研究表明,DNA甲基化的分布主要分布在基因间、外显子、内含子、下游和上游区域(Saha, 2020; Li, 2021)。此外,大量研究发现,脊椎动物甲基化位点主要在启动子区(promoter)分布,启动子区CG型甲基化水平决定转录水平结果(Xu, 1999; Yoder, 1997; Jiang, 2019; 齐云峰等, 2019)。但海洋无脊椎动物香港巨牡蛎()、仿刺参等甲基化位点在基因间区和内含子区中更丰富(Rajan, 2021; 李玉强等, 2018;李欣容, 2022)。这与本研究中CG和CWG位点在不同功能元件中的分布结果相一致。同样,本研究中,基因体的甲基化水平高于TSS和TTS。DNA甲基化会形成不易去除的局部异染色质化状态,表明基因体具有相对稳定的染色体结构,抑制转录水平的进行。基因启动子区通常位于TSS的上游,负责基因的表达调控(Jiang, 2019)。在人()体组织中的研究表明,TSS下游区域的甲基化是基因表达的高度信息。本研究中,甲基化水平高峰期明显处于TSS区下游,进一步推测脊尾白虾基因间区CG型甲基化水平的高低可能影响基因在转录水平的表达,并且对转录水平抑制表达有着重要作用。在位点水平上对DEG进行GO分析发现,这些基因主要分为3类GO terms,包括生物过程、细胞成分和分子功能。这些功能包括RNA剪接、DNA重组转录调控和DNA模板,表明盐碱胁迫诱导的甲基化变异抗性基因(Ackah, 2022)。进一步对DEG进行KEGG途径富集分析表明,HIF-1信号通路、孕酮介导的卵母细胞成熟、卵母细胞减数分裂、细胞周期和剪接体途径在脊尾白虾鳃组织的盐碱胁迫响应中起关键作用。
鳃是水生动物氧气运输、渗透压调节、离子转运、酸碱调节等的主要组织,环境因子如盐度、pH、低氧等均可能造成鳃组织损伤,从而影响呼吸和离子转运等功能(Nikinm, 2014)。缺氧诱导因子1(HIF-1)是一种碱性螺旋–环–螺旋–PAS结构域转录因子,能够调控机体对环境适应性应答(杨梦思等, 2016)。另外,HIF-1可增强氧气供给并介导低氧适应性反应。以往的研究发现,脊尾白虾在低溶氧环境下易诱导HIF产生,从而刺激血液氧的供应能力(曹梅等, 2021)。高盐碱胁迫会改变机体的渗透压和酸碱平衡,机体不仅能通过离子调控、应激蛋白合成等维持,也会产生自我调节机制适应环境胁迫(王楠等, 2015)。在高盐胁迫下,水生生物同样易产生氧化应激及渗透压失衡,Bal等(2021)观察到高盐水环境中印度囊鳃鲶()血红蛋白含量显著下降,耗氧率升高。在高碱度下,水环境无法维持罗非鱼机体中足够的溶解氧水平,最终导致死亡,若能保持水中充足氧含量,在高碱度下生物死亡率不会增加(Colt, 2013)。本研究发现,、、和在HIF-1信号通路中显著富集上调表达。作为一种转移酶,能够催化葡萄糖的磷酸化,这是葡萄糖代谢的第一步,并能抑制ADP (Majewski, 2004)。mTOR属于磷脂酰肌醇3-激酶相关激酶蛋白家族,能够促进物质代谢,参与细胞凋亡、生长、增殖和运动,调节蛋白质合成和转录(Sarbassov, 2005)。同时,qRT-PCR结果显示,基因的表达水平同甲基化水平均显著上调,这意味着、、mTOR和可通过保持高甲基化水平促进其相应靶基因表达以响应盐碱胁迫。
剪接体是由大量蛋白质和小核RNA组成的大型核糖核蛋白复合物,主要存在于真核细胞的细胞核中,是一种在转录后的初级转录物(Marondedze, 2020)。本研究中,、、、和参与剪接体途径并且显著下调表达,这意味着盐碱胁迫减少了RNA过程和蛋白质合成通路。进一步表明,盐碱胁迫可能通过减少RNA的合成,降低鳃组织的代谢速率以及参与RNA剪接的基因的甲基化状态来抑制RNA剪接。总之,这些结果表明,DNA甲基化可能调节了以上基因以响应盐碱胁迫。我们推测脊尾白虾由于长期生活在盐碱环境,DNA甲基化水平变化在响应盐碱胁迫中发挥重要作用,盐碱胁迫致使HIF-1信号途径增强,同时抑制了剪接体的表达,从而进化出独特的生存机制。
基因水平的GO和KEGG富集结果表明,盐碱胁迫不仅影响细胞的生长发育,还与离子转运有关,表明脊尾白虾对盐碱有较强的适应性。同时,本研究发现一些差异甲基化基因在一些信号通路中富集。离子转运相关基因在许多生理功能和细胞过程中影响不同的渗透调节(Cao, 2019; Si, 2019; Zimmer, 2021)。另外,水生动物对外部非生物环境的适应不仅有内在的调节,而且取决于能量的支持(Song, 2021; Su, 2020; Root, 2021)。脊尾白虾因长期盐碱胁迫而导致脂质过氧化和生理代谢的变化,本研究中能量代谢通路和脂质代谢通路显著富集,推测脊尾白虾通过调节鳃组织DNA甲基化水平对能量代谢通路和脂质代谢通路的影响适应盐碱水环境。
解整合素金属蛋白酶17 (ADAM17 protein, ADAM17)是一种跨膜金属蛋白酶,能够参与多种蛋白质胞外域脱落,具有黏附和蛋白水解特性,参与细胞分子、生长因子受体及表皮生长因子受体过程,在膜结合型蛋白的翻译后修饰中发挥关键作用(王超男等, 2016; Li, 2015)。盐碱胁迫后ADAM17可能通过低甲基化促进膜结合型蛋白的翻译后修饰,促进质膜对胞内环境的维持。类纤维囊蛋白(fibrocystin- L-like, FPC-L)是一个单次跨膜的受体样蛋白,含有一个潜在蛋白激酶C磷酸化位点,胞外区存在被高度糖基化的可能(连培文等, 2011)。盐碱胁迫下脊尾白虾鳃组织中FPC-L可能发生糖基化,对翻译后蛋白进行了修饰,从而适应盐碱环境。细胞色素P450 (cytochrome P450 2L1-like, CYP450)主要分布在内质网和线粒体内膜上,能够参与环境化合物在内的外源性物质(在类固醇/甾醇)的代谢(Ventura, 2017)。盐碱水环境中,CYP450可能诱导脊尾白虾鳃组织细胞凋亡,内质网和线粒体相关的凋亡途径可能发挥了重要的作用。本研究中,我们发现ADAM17、FPC-L和CYP450呈现出低甲基化状态。然而,对应的差异甲基化基因却显示表达水平在盐碱环境下显著升高,这表明DNA甲基化可能通过激活蛋白激素和防御化合物来诱导脊尾白虾中ADAM17、FPC-L和CYP450的调节以适应盐碱水环境。
4 结论
本研究通过对正常海水养殖和长期盐碱水养殖脊尾白虾的甲基化水平比对分析发现,盐碱水环境下DNA甲基化水平略有增强,甲基化位点丰富呈现在基因间区和内含子区域并且在TSS区下游出现甲基化水平高峰期。分析了脊尾白虾鳃组织在2种养殖水环境中位点和基因水平的富集通路及组间DMS和DMG的表达情况,提示和基因共同参与的HIF-1信号通路和和基因共同参与的剪接体通路可能在脊尾白虾盐碱水环境适应中发挥重要作用。进一步研究参与HIF-1信号通路和剪接体通路的基因表达情况,为揭示脊尾白虾盐碱水环境适应机制提供新的见解。位点水平和基因表达呈负相关的ADAM17、FPC-L和CYP450可作为后期解析脊尾白虾盐碱水环境下表观调控的重点研究对象。因此,该研究将为揭示盐碱水环境对脊尾白虾的影响提供新的理论依据。
ACKAH M, GUO L, LI S,DNA methylation changes and its associated genes in mulberry (L.) Yu-711 response to drought stress using MethylRAD sequencing. Plants (Basel), 2022, 11(2): 190
AROOJ F, SADDAM H, SADAM H,Differential morphophysiological, biochemical, and molecular responses of maize hybrids to salinity and alkalinity stresses. Agronomy, 2021, 11(6): 1150
BAL A, PATI S G, PANDA F,. Low salinity induced challenges in the hardy fish: Future prospective of aquaculture in near coastal zones. Aquaculture, 2021, 543(8): 737007
BOERS R, BOERS J, HOON B D,Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme. Genome Research, 2018, 28(1): 88–99
CAI Y, MENG X L, LIU P,Molecular cloning of the DNA methyltransferase-2 gene and its expression during embryonic, larval, and gonadal development in the swimming crab. Journal of Fishery Sciences of China, 2018, 25(5): 928–935 [蔡影, 孟宪亮, 刘萍, 等. 三疣梭子蟹甲基转移酶2基因克隆及在胚胎、幼体和性腺发育过程中的表达分析. 中国水产科学, 2018, 25(5): 928–935]
CAO J, SHI F. Comparative analysis of the aquaporin gene family in 12 fish species. Animals (Basel), 2019, 9(5): 233
CAO M, WANG X Q, QIN C X,Transcriptome analysis ofsubjected to hypoxic stress. Progress in Fishery Sciences, 2021, 42(2): 112–123 [曹梅, 王兴强, 秦传新, 等. 脊尾白虾对低氧响应的转录组学分析. 渔业科学进展, 2021, 42(2): 112–123]
CHANG Y M, LIANG L Q. Advances of research of physiological and molecular mechanisms related to alkali- saline adaptation for fish species inhabiting alkali-saline water. Journal of Fisheries of China, 2021, 45(5): 798–812 [常玉梅, 梁利群. 耐盐碱鱼类的生理和分子机制研究进展. 水产学报, 2021, 45(5): 798–812]
CHEN H S, SHENG L X, BIAN H F. Investigation of the effects of developmental toxicity and genomic DNA methylation upon naphthalene exposure in zebrafish (). Journal of Northeast Normal University (Natural Science), 2016, 48(3): 167–173 [陈宏姗, 盛连喜, 边红枫. 化合物萘对斑马鱼发育毒性及基因组DNA甲基化影响的研究. 东北师大学报(自然科学版), 2016, 48(3): 167–173]
CHEN M M, ZHANG S R, WU L P,Organic fertilization improves the availability and adsorptive capacity of phosphorus in saline-alkaline soils. Journal of Soil Science and Plant Nutrition, 2020, 21(1): 487–496
COLT J, KROEGER E. Impact of aeration and alkalinity on the water quality and product quality of transported tilapia: A simulation study. Aquacultural Engineering, 2013, 55(1): 46–58
CONRADO G V, JANET H BEffects of alkalinity and total hardness on growth and survival of postlarvae freshwater prawns,(De Man 1879). Aquaculture, 2017, 473: 521–527
CRIBIU P, CHAUMOT A, GEFFARD O,Natural variability and modulation by environmental stressors of global genomic cytosine methylation levels in a freshwater crustacean,. Aquatic Toxicology, 2018, 205: 11–18
FAN Z, PENG F, LI J,Effects of α-ketoglutarate on growth performance, antioxidant capacity and ammonia metabolization against chronic carbonate alkalinity stress in Songpu mirror carp (Songpu). Aquaculture Research, 2020, 51(5): 29–40
GAVERY M R, ROBERTS S B. Predominant intragenic methylation is associated with gene expression characteristics in a bivalve mollusc. PeerJ, 2013, 1: e215
GE Q Q, LI J T, WANG J J,Characterization, functional analysis, and expression levels of three carbonic anhydrases in response to pH and saline-alkaline stresses in the ridgetail white prawn. Cell Stress Chaperones, 2019, 24(3): 503–515
GE Q Q, LIANG J P, LI J T,Molecular cloning and expression analysis of Relish gene from the ridgetail white prawn. Fisheries Science, 2015, 81(4): 699–711
HAN J, HU Y, QI Y,High temperature induced masculinization of zebrafish by down-regulation ofandvia DNA methylation. Journal of Environmental Sciences, 2021, 107(9): 160–170
HAWES N A, TREMBLAY L A, POCHON X,Effects of temperature and salinity stress on DNA methylation in a highly invasive marine invertebrate, the colonial ascidian. PeerJ, 2018, 6: e5003
HOU Y W, LIU W, JIANG P L,. Effect of cold stress on genomic DNA methylation in zebrafish ZF4 cells. Journal of Fishery Sciences of China, 2019, 26(2): 271–279 [侯艳雯, 刘玮, 姜蓬垒, 等. 低温对斑马鱼ZF4细胞基因组DNA甲基化水平的影响. 中国水产科学, 2019, 26(2): 271–279]
HUAN P P, LÜ J J, SUN D F,The cloning of the PtDNMT1 gene ofand its expression analysis in low salinity adaptation. Progress in Fishery Sciences, 2019, 40(1): 92–100 [环朋朋, 吕建建, 孙东方, 等. 三疣梭子蟹PtDNMT1基因的克隆及其在低盐适应中的表达分析. 渔业科学进展, 2019, 40(1): 92– 100]
JIANG Y, LI J, REN F,PM2.5-induced extensive DNA methylation changes in the heart of zebrafish embryos and the protective effect of folic acid. Environmental Pollution, 2019, 255(3): 113331
LAI Q F, YAO Z L, GAO P C,. Saline-alkaline water environment and suitable culture species. Beijing: China Agriculture Press, 2021 [来琦芳, 么宗利, 高鹏程, 等. 盐碱水环境与增养殖种类. 北京: 中国农业出版社, 2021]
LI M D, LI J T, SHI K P,Estimation of heritability and genetic correlation of saline-alkali tolerance in. Progress in Fishery Sciences, 2021, 42(1): 117–123 [李明栋, 李吉涛, 史鲲鹏, 等. 脊尾白虾耐盐碱性状遗传力和遗传相关的估计. 渔业科学进展, 2021, 42(1): 117–123]
LI M Y, LI J, LIU P,Cloning and expression analysis of ferritin gene in. Oceanologia et Limnologia Sinica, 2012, 43(2): 306–312 [李美玉, 李健, 刘萍, 等. 脊尾白虾() ferritin基因克隆及表达分析. 海洋与湖沼, 2012, 43(2): 306–312]
LI Q, XIE J, HE L,Identification of ADAM10 and ADAM17 with potential roles in the spermatogenesis of the Chinese mitten crab,. Gene, 2015, 562(1): 117–127
LI X R, LIAO M J, LI B,Genomic DNA methylation levels and transcriptome differences ofin response toinfection and their association analysis. Progress in Fishery Sciences, 2022, 43(3): 176–185 [李欣容, 廖梅杰, 李彬, 等. 刺参响应灿烂弧菌侵染的基因组DNA甲基化水平和转录组差异及其关联分析. 渔业科学进展, 2022, 43(3): 176–185]
LI Y Q, WANG R J, LI Y L,Genome-wide profiling of DNA methylation inbased on MethylRAD-Seq. Periodical of Ocean University of China (Natural Science), 2018, 48(9): 41–50 [李玉强, 王睿甲, 李语丽, 等. 基于MethylRAD-Seq技术对仿刺参DNA甲基化图谱的研究. 中国海洋大学学报(自然科学版), 2018, 48(9): 41–50]
LI Z Q, TANG M Q, LUO D J,Integrated methylome and transcriptome analyses reveal the molecular mechanism by which DNA methylation regulates kenaf flowering. Frontiers in Plant Science, 2021, 12: 709030
LIAN P W, FU Y L, LI A,Preparation and characterization of a polyclonal antibody against human Fibrocystin-L. Chinese Journal of Cellular and Molecular Immunology, 2011, 27(1): 78–81 [连培文, 付玉龙, 李奥, 等. 新基因PKHDL71产物Fibrocystin-L的抗体制备及其亚细胞定位的初步研究. 细胞与分子免疫学杂志, 2011, 27(1): 78–81]
LISTER R, PELIZZOLA M, DOWEN R H,Human DNA methylomes at base resolution show widespread epigenomic differences. Nature, 2009, 462(7271): 315–322
LIU F, LI J, LI J T,Effects of carbonate alkalinity stress on the survival, growth, reproduction, and immune enzyme activities of. Journal of Fishery Sciences of China, 2016, 23(5): 1137–1147 [柳飞, 李健, 李吉涛, 等. 碳酸盐碱度对脊尾白虾生存、生长、繁殖及免疫酶活性的影响. 中国水产科学, 2016, 23(5): 1137– 1147]
LOVETT D L, VERZI M P, CLIFFORD P D,Hemolymph levels of methyl farnesoate increase in response to osmotic stress in the green crab,. Comparative Biochemistry and Physiology A-Molecular and Integrative Physiology, 2001, 128(2): 299–306
LÜ J. Establishment of an efficient and cost-effective method for genome-wide DNA methylation profiling (MethylRAD-Seq) and its application in marine bivalves. Master´s Thesis of Ocean University of China, 2013 [吕佳. 高效低成本全基因组DNA甲基化检测技术(MethylRAD-Seq)的建立及其在海洋贝类中的应用. 中国海洋大学硕士研究生学位论文, 2013]
MAJEWSKI N, NOGUEIRA V, ROBEY R B,Akt inhibits apoptosis downstream of BID cleavage via a glucose- dependent mechanism involving mitochondrial hexokinases. Molecular and Cellular Biology, 2004, 24(2): 730–740
MARONDEDZE C, THOMAS L, LILLEY K S,Drought stress causes specific changes to the spliceosome and stress granule components. Frontiers in Molecular Biosciences, 2020, 6: 163
NIKINM A M. Chapter 15 – Interactions between natural environmental factors and toxicity. An Introduction to Aquatic Toxicology, 2014: 173–184
NOROUZITALLAB P, BARUAH K, VANDEGEHUCHTE M,Environmental heat stress induces epigenetic transgenerational inheritance of robustness in parthenogeneticmodel. FASEB Journal, 2014, 28(8): 3552–3563
QI Y F, WANG R J, JIN T C,Analyzing distribution patterns of epigenetic factor among different genome regions. Journal of Northeast Normal University (Natural Science), 2019, 51(1): 108–112 [齐云峰, 王仁俊, 金太成, 等. 表观遗传因子在不同基因组区域的分布模式研究. 东北师大学报(自然科学版), 2019, 51(1): 108–112]
RAJAN K C, YUAN M, YU Z,. Oyster biomineralisation under ocean acidification: From genes to shell. Global Change Biology, 2021, 27(16): 3779–3797
ROOT L, CAMPO A, MACNIVEN L,Nonlinear effects of environmental salinity on the gill transcriptome versus proteome ofmodulate epithelial cell turnover. Genomics, 2021, 113(5): 3235–3249
SAHA D, NORVIL A B, LANMAN N A,Simplified MethylRAD sequencing to detect changes in DNA methylation at enhancer elements in differentiating embryonic stem cells. Epigenomes, 2020, 4(4): 24
SARBASSOV D D, ALI S M, SABATINI D M. Growing roles for the mTOR pathway. Current Opinion in Cell Biology, 2005, 17(6): 596–603
SI L J, PAN L Q, ZHANG X,Evidence that dopamine is involved in neuroendocrine regulation, gill intracellular signaling pathways and ion regulation in. Journal of Experimental Biology, 2019, 222(15): jeb204073
SONG L, ZHAO Y, SONG YEffects of saline-alkaline water on growth performance, nutritional processing, and immunity in Nile tilapia (). Aquaculture, 2021, 544: 737036
SU H H, MA D M, ZHU H W,Transcriptomic response to three osmotic stresses in gills of hybrid tilapia (female ×hornorum male). BMC Genomics, 2020, 21(1): 110
TAPIA D, EISSLER Y, ESPINOZA J C,Inter-laboratory ring trial to evaluate real-time reverse transcription polymerase chain reaction methods used for detection of infectious pancreatic necrosis virus in Chile. Electronic Journal of Biotechnology, 2017, 28: 20–26
VARRIALE A, BERNARDI G. DNA methylation and body temperature in fishes. Gene, 2006, 385: 111–121
VENTURA T, BOSE U, FITZGIBBON Q P,CYP450s analysis across spiny lobster metamorphosis identifies a long sought missing link in crustacean development. Journal of Steroid Biochemistry and Molecular Biology, 2017, 171: 262–269
WANG C L. Influence of high temperature stress on Hsp90 expression and CpG methylation in. Guangxi Sciences, 2019, 26(4): 424–429 [王翠丽. 高温胁迫对近江牡蛎Hsp90表达及CpG甲基化的影响. 广西科学, 2019, 26(4): 424–429]
WANG C N, DONG J, SHAO N S. Role of ADAM17 in transmembrane protein shedding. Journal of Medical Molecular Biology, 2016, 13(6): 345–349 [王超男, 董洁, 邵宁生. ADAM17在膜蛋白胞外结构域剪切机制中的作用. 医学分子生物学杂志, 2016, 13(6): 345–349]
WANG H, ZHONG J C, CHAI Z X,Advances of research on interactions between hypoxia and epigenetics. Journal of Southwest Minzu University, 2017, 43(6): 557–561 [王会, 钟金城, 柴志欣, 等.低氧与表观遗传学互作的研究进展. 西南民族大学学报, 2017, 43(6): 557–561]
WANG N, CHANG Y M, TANG R,Screening microsatellite markers associated with alkaline tolerance in. Journal of Fishery Sciences of China, 2015, 22(6): 1105–1114 [王楠, 常玉梅, 唐然, 等. 瓦氏雅罗鱼耐碱性状相关分子标记的筛选. 中国水产科学, 2015, 22(6): 1105–1114]
WANG S, LV J, ZHANG L L,MethylRAD: A simple and scalable method for genome-wide DNA methylation profiling using methylation-dependent restriction enzymes. Open Biology, 2015, 5: 150130
WANG X X, LI A, WANG W,Direct and heritable effects of natural tidal environments on DNA methylation in Pacific oysters (). Environmental Research, 2021, 197: 111058
WEN Z Z, ZUO S, CHEN M,. DNA methylation level of genomic DNA of. Progress in Fishery Sciences, 2021, 42(3): 46–54 [温争争, 左闪, 陈梦, 等. 刺参基因组DNA甲基化水平及模式对温度变化的响应. 渔业科学进展, 2021, 42(3): 46–54]
WU B, YANG A G, SUN X J,. Effects of acute temperature stress on genome-wide DNA methylation profiles in. Progress in Fishery Sciences, 2016, 37(5): 140–146 [吴彪, 杨爱国, 孙秀俊, 等. 急性温度胁迫对虾夷扇贝基因组DNA甲基化的影响. 渔业科学进展, 2016, 37(5): 140–146]
XU G L, BESTOR T H, BOURC'HIS D,Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature, 1999, 402(6758): 187–191
XU W L, WANG H Q, LI Y H. Distribution and aquaculture application of saline alkali water at home and abroad. China Fisheries, 2021(7): 50–53 [徐文龙, 汪惠庆, 李月红. 国内外盐碱水域分布及水产养殖应用. 中国水产, 2021(7): 50–53]
XU Y H, XU Y C, HOGSTRAND C,Waterborne copper exposure up-regulated lipid deposition through the methylation of GRP78 and PGC1alpha of grass carp. Ecotoxicology and Environmental Safety, 2020, 205: 111089
XUE B, ZHANG B, LI Z H,Methylation profile in mitochondrial genome of ridgetail white prawnin response to different developmental stages. Fisheries Science, 2017, 36(5): 628–633 [薛蓓, 张培, 李志辉, 等. 脊尾白虾不同发育期线粒体基因组甲基化特征分析. 水产科学, 2017, 36(5): 628–633]
YANG F Y, SUN L M, YANG X Q. Toxicity of carbonate alkalinity tojuveniles. Fisheries Science, 2004, 23(9): 3–6 [杨富亿, 孙丽敏, 杨欣乔. 碳酸盐碱度对南美白对虾幼虾的毒性作用. 水产科学, 2004, 23(9): 3–6]
YANG M S, ZHOU N, WANG Z G,Research progress on the role of transcription factor HIF-1α and its signal pathway in the pathogenesis. Biotechnology Bulletin, 2016, 32(8): 8–13 [杨梦思, 周娜, 王志钢, 等. 转录因子HIF-1α及其信号通路在疾病发生中的作用研究进展. 生物技术通报, 2016, 32(8): 8–13]
YANG Y, ZHENG Y, SUN L,. Genome-wide DNA methylation signatures of sea cucumberduring environmental induced aestivation. Genes, 2020, 11(9): 1020
YODER J A, WALSH C P, BESTOR T H. Cytosine methylation and the ecology of intragenomic parasites. Trends in Genetics, 1997, 13(8): 335–340
ZHANG J F. Discussion on ecological rehabilitation of salt-affected soils. Research of Soil and Water Conservation, 2008, 4: 74–78 [张建锋. 盐碱地的生态修复研究. 水土保持研究, 2008, 4: 74–78]
ZHANG X, LI Q, YU H,. Effects of air exposure on genomic DNA methylation in the Pacific oyster (). Journal of Fishery Sciences of China, 2017, 24(4): 690–697 [张鑫, 李琪, 于红, 等. 干露胁迫对太平洋牡蛎基因组DNA甲基化的影响. 中国水产科学, 2017, 24(4): 690–697]
ZIMMER A M, MANDIC M, HONG M Y,Use of a carbonic anhydrase Ca17a knockout to investigate mechanisms of ion uptake in zebrafish (). American Journal of Physiology-Renal Physiology, 2021, 320(1): R55–R68
Effects of Saline-Alkaline Water Environment on DNA Methylation of
QIN Zhen1,2, LI Jitao2①, LI Mingdong2, WANG Jiajia2, GE Qianqian2, LIU Ping2, LI Jian2
(1. National Experimental Teaching Demonstration Center of Aquatic Science, Shanghai Ocean University, Shanghai 201306, China; 2. Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences; Key Laboratory of Sustainable Development of Marine Fisheries, Ministry of Agriculture and Rural Affairs, Laboratory for Marine Fisheries Science and Food Production Processes, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, Shandong 266071, China)
The global levels of soil and water salinization and alkalinization are increasing with the influence of climate and topography changes, as well as other natural and human factors. Saline-alkaline water (SAW) all over the world has specific characteristics such as high alkalinity, high pH, and complex water quality types, which inhibits the survival and culture of common aquatic animals. The ridgetail white prawnis an economically important marine shrimp with many advantages, such as widely environmental adaptability, rapid growth, and good reproductive capability. It is potentially suitable for large-scale culture in SAW; however, its adaptability to this environment remains unclear. Exploring theadaptability mechanism to SAW will help to guide culture management for marine crustaceans. In this study, the DNA methylomes of thegill tissue cultured in SAW and normal seawater (SW) were analyzed and the impact on gene regulation was investigated by MethylRAD sequencing. The results showed 2 347 003 and 416 176 methylations at the CG and CWG sites (W = A or T), respectively. Comparing the SAW and SW groups, the CG and CWG loci in the SAW group increased slightly, indicating that SAW induced more DNA methylation in the gill cells that activated or inhibited pathways and played a crucial role in the environmental changes adaption. Methylation was prevalent in the exon, intron, splice site, and upstream and downstream regions of thegill genes, as well as in the intergenic regions. DNA methylation sites were mostly distributed in the Genebody. The DNA methylation distribution curve peaked in the downstream sequence of the transcription start site and upstream sequence of the transcription termination site. The methylation label frequency was significantly higher in these regions in relation to other sequences. A total of 8805 differential methylation sites (DMSs) were screened, including 8189 CG DMSs and 616 CWG DMSs. Obviously, the CG DMS was significantly higher than the CWG DMS. The intergenic and intron regions accounted for a large proportion of the DMS observed. Overall, the DMS showed a higher trend in the genic downstream regions of the gene relative to upstream regions. The Gene Ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) based on the DMS showed enrichment of genes involved in the "development, heterochrony, and protein disables isomerase activity," which played a role in the CG level down-regulation. In addition, "incubation involved in sorocarp development" and "nucleus and double-stranded RNA binding" were molecular functions up-regulated by the CG methylations. The down-regulated genes with CWG methylation were enriched for the "regulation of transcription and DNA template" process, while the up-regulated genes were enriched for the "epithelial cell migration and open trail” system. These two processes were induced incultured in saline-alkali water. The key genes in the HIF-1 signal and spliceosome pathways, such as,, and, play an important role in response to saline-alkaline stress; therefore, the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis based on DMS of the DEG was performed. In addition, 158 CG methylated genes (MGs) were detected in DEGs from shrimps exposed to SAW, among which 77 and 81 were up- and down-regulated, respectively. Moreover, 94 CWG MGs were differentially expressed, from which 33 and 61 were up- and down-regulated, respectively. GO enrichment analysis of the CG MGs showed significant enrichment of the "muscle organ development" process; CWG MGs were significantly enriched for the "vesicle-m transport" and "membrane mediated zinc binding plasma transport" processes. This indicated thatwas damaged by the saline-alkaline environment, but the shrimp may adapt to this stress by adjusting the ion balance. The KEGG enrichment analysis indicated that lipid metabolism and signal transduction pathways may play crucial roles in thegill tissue response to saline-alkaline stress. Moreover, changes in lipid peroxidation and physiological metabolism may be caused by long-term saline-alkaline stress. The energy metabolism pathway was significantly enriched and many different genes in the lipid metabolism pathway were expressed in the stressed. Therefore, it was speculated that the changes in DNA methylation level might play an important role in response to saline-alkaline stress. Overall, the results showed that a series ofphysiological activities related to environmental adaptation was activated by SAW. In addition, a small number of methylated loci were negatively correlated with gene expressions, which indicated a complex relationship between DNA methylation and gene regulation. Although,, andshowed hypomethylation, the corresponding differentially MGs showed a significantly increased expression level in a saline-alkaline environment. Therefore, genomic DNA methylation may promote gene expression under saline-alkaline stress, including the,, andgenes. This study analyzed the DNA methylation levels in gill tissue ofcultured with SAW and provided information that will further elucidate the molecular mechanisms involved in crustacean adaptation to saline-alkaline environment.
; Saline-alkaline water environment; DNA methylation; Differentially expressed gene
LI Jitao, E-mail: lijt@ysfri.ac.cn
10.19663/j.issn2095-9869.20220310001
S917.4
A
2095-9869(2022)04-0033-18
*国家重点研发计划课题(2019YFD0900404-03)、国家自然科学基金项目(32072974)、财政部和农业农村部: 国家现代农业产业技术体系和中国水产科学研究院基本科研业务费项目(2020TD46)共同资助 [This work was supported by National Key Research and Development Program of China (2019YFD0900404-03), National Natural Science Foundation of China (32072974), China Agriculture Research System of MOF and MARA, and Central Public-Interest Scientific Institution Basal Research Fund, CAFS (2020TD46)]. 秦 桢,E-mail: 1285946211@qq.com
李吉涛,研究员,E-mail: lijt@ysfri.ac.cn
2022-03-10,
2022-04-07
http://www.yykxjz.cn/
秦桢, 李吉涛, 李明栋, 王佳佳, 葛倩倩, 刘萍, 李健. 盐碱水环境对脊尾白虾基因组DNA甲基化的影响. 渔业科学进展, 2022, 43(4): 33–50
QIN Z, LI J T, LI M D, WANG J J, GE Q Q, LIU P, LI J. Effects of saline-alkaline water environment on DNA methylation of. Progress in Fishery Sciences, 2022, 43(4): 33–50
(编辑 冯小花)

