Long-noncoding RNAs as epigenetic regulators in neurodegenerative diseases
Paola Ruffo,Francesca De Amicis,Emiliano Giardina,Francesca Luisa Conforti,*
Abstract The growing and rapid development of high-throughput sequencing technologies have allowed a greater understanding of the mechanisms underlying gene expression regulation.Editing the epigenome and epitranscriptome directs the fate of the transcript influencing the functional outcome of each mRNA.In this context,non-coding RNAs play a decisive role in addressing the expression regulation at the gene and chromosomal levels.Long-noncoding RNAs,consisting of more than 200 nucleotides,have been shown to act as epigenetic regulators in several key molecular processes involving neurodegenerative disorders,such as Alzheimer’s disease,Parkinson’s disease,amyotrophic lateral sclerosis and Huntington’s disease.Long-noncoding RNAs are abundantly expressed in the central nervous system,suggesting that their deregulation could trigger neuronal degeneration through RNA modifications.The evaluation of their diagnostic significance and therapeutic potential could lead to new treatments for these diseases for which there is no cure.
Key Words: Alzheimer’s disease;amyotrophic lateral sclerosis;epigenetic mechanism;Huntington’s disease;long-noncoding RNAs;neurodegenerative disease;non-coding RNAs;Parkinson’s disease
From the Contents
Introduction 1243
Data Sources 1244
Long-Noncoding RNAs Epigenetic Regulation Mechanisms 1244
Long-Noncoding RNAs as Epigenetic Regulators in Alzheimer’s Disease 1244
Long-Noncoding RNAs as Epigenetic Regulation inParkinson’s Disease 1245
Long-Noncoding RNAs as Epigenetic Regulation in Amyotrophic Lateral Sclerosis 1246
Long-Noncoding RNAs as Epigenetic Regulation in Huntington’s Disease 1246
Long-Noncoding RNAs as Targets: Current Challenges in Alzheimer’s Disease/Parkinson’s Disease/Amyotrophic Lateral Sclerosis/Huntington’s Disease Therapy 1246
Conclusion 1246
Introduction
Genome size in all nucleated cells of an organism is identical but different genetic mechanisms allow the establishment of a differential gene expression.In 1492,C.H.Waddington introduced the term “epigenetic” to describe how the interactions between environment and genes determine phenotype development (Tronick and Hunter,2016).Genetic mechanisms explain how heritable traits arise from mutations in the DNA sequence,while epigenetic mechanisms describe how phenotypic changes occur independently of the underlying DNA sequence (Egger et al.,2004).Epigenetic changes ensure that differential expression patterns are inherited stably as cells divide by providing a form of cellular memory that is passed on to offspring.Furthermore,these mechanisms can ensure the stable inheritance of an active transcriptional state for some target genes or certain genomic regions.Alternately,they can rearrange the chromatin of some genomic regions adopting a completely condensed and therefore transcriptionally inactive form (Bird,2007).The most significant mechanisms for epigenetic labeling can be classified into three categories: DNA modifications (DNA methylation and hydroxy-methylation),histone modifications (histone methylation,acetylation,phosphorylation,ubiquitylation),and non-coding-mediated RNA modifications (ncRNA) (Wei et al.,2017).
Several studies have shown that ncRNAs play a decisive role in the epitranscriptomic alterations regulating expression at the gene and chromosomal level with consequent control of cell differentiation (Amaral et al.,2008;Costa,2008;Peschansky and Wahlestedt,2014;Wei et al.,2017;Figure 1).In particular,microRNA (miRNA) and short-interfering RNA(siRNA) are involved in the silent transcription gene,Piwi-interacting RNA(piRNA) performs the function of transposon repression DNA methylation,and long non-coding RNA (lncRNA) is involved in the genomic imprinting and inactivation of the X chromosome (Wei et al.,2017).
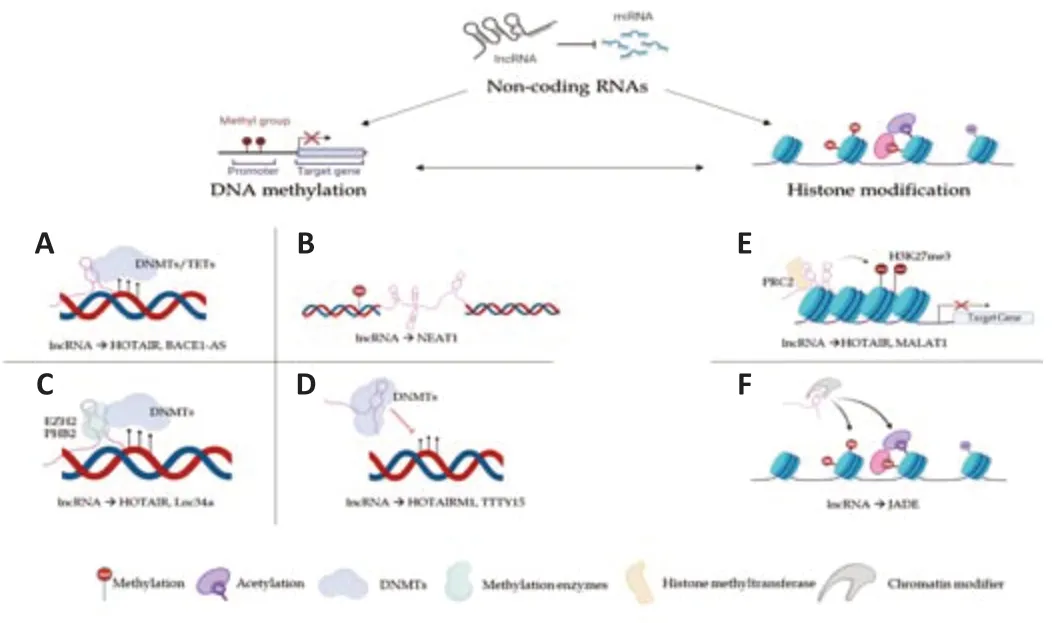
Figure 1|Mechanisms for epigenetic labeling: function of ncRNAs.
Epitranscriptome and epigenome alterations also play a key role in neuronal aging in a not yet defined way,although the importance of these processes in the genesis of neurons and the consolidation of memory is known(Creighton et al.,2020).Different scientific evidence shows the presence of global and gene-specific epigenetic changes at the peripheral and brain levels in patients with overt neurodegenerative diseases (NDs),such as Alzheimer’s disease (AD) (Coppede,2021),Parkinson’s disease (PD) (Rathore et al.,2021) and amyotrophic lateral sclerosis (ALS) (Coppede,2020).In the tissues of ND patients,both short RNAs and lncRNAs were found to be deregulated (Giulia Gentile 2022),candidates for these molecules as probable therapeutic biomarkers (Huaying et al.,2020).The results ofin vitrostudies on the targeting of epigenetic signs in ND show improvements in synaptic plasticity,reduction of disease progression,and motor and cognitive functions(Coppede,2022).
LncRNAs are non-coding RNAs longer than 200 nucleotides.They have a specific tissue expression and are copiously expressed in the central neuron system where they participate in various biological processes,such as epigenetic regulation,programmed cell death,synaptic activity,and inflammatory response processes (Ruffo et al.,2021).Misregulation of lncRNAs is directly related to the dysregulation of the pathogenesis of some diseases,including NDs (Zhang and Wang,2021).Specifically,several lncRNAs have been found to be involved in organ development (Rayner and Liu,2016),differentiation (Chen and Zhang,2016),synaptic formation (Maag et al.,2015),learning and memory (Gudenas and Wang,2015) as well as cell senescence at different stages of the cell cycle (Puvvula,2019).Several lncRNAs play a role in cellular senescence and organism aging through cell cycle regulation.Among these,many studies have shown thatMALAT1(Metastasis-Associated Lung Adenocarcinoma Transcript 1) is a cell cycle regulator causing cycle arrest in the G1 or G1/S phase by improving the senescence phenotype (Tripathi et al.,2013).Another aging-related lncRNA isTUG1(Taurine upregulated gene 1) that blocks p53-mediated growth and apoptosis as well as modifies gene expression of theHOX(Homeobox) gene family,resulting in aging (Khalil et al.,2009).LncRNAs are also involved in chromatin modulation leading to senescence and aging process.Among these,ANRASSF1is an lncRNA-AS that potentially reduces the transcription of the tumor suppressor gene,Ras-associated domain-containing protein 1A (RASSF1A),involved in cell cycle arrest and apoptosis (Beckedorff et al.,2013).Aging is also associated with the production of senescence-associated secretory phenotype factors that facilitate inflammation and the onset of agerelated diseases.Many lncRNAs participate in innate immunity such asFIRRE(Functional intergenic repeating RNA element),a recently discovered lncRNA controlled byNF-κB(nuclear factor kappa-light-chain-enhancer of activated B cells) signaling in macrophages.This lncRNA impacts nuclear architecture through interaction withhnRNP-U(Heterogeneous Nuclear Ribonucleoprotein U) and positively regulates several inflammatory genes expression (Lu et al.,2017).
In this review,following a general introduction to the lncRNA epigenetic regulation mechanisms,we will provide a brief overview of the role of lncRNAs whose epigenetic regulatory mechanisms have previously been known in AD,PD,ALS,and Huntington’s disease (HD),discussing those eligible as therapeutic markers.
Data Sources
Database: https://pubmed.ncbi.nlm.nih.gov/ (May 20,2022) was searched.The first research was conducted using keywords: lncRNAs AND neurodegenerative diseases AND Alzheimer’s disease AND Parkinson’s disease AND amyotrophic lateral sclerosis.Subsequently,we have selected only the papers in which the mechanisms of lncRNAs epigenetic regulation were discovered.Furthermore,the selected lncRNAs underwent a further filter:only those eligible as therapeutical markers were discussed.
Long-Noncoding RNAs Epigenetic Regulation Mechanisms
The ability to be variable,flexible,and structurally changeable makes lncRNAs the main characters of genomic dynamism,thus resulting in significant components in epigenetic regulation.This can occur according to different mechanisms that allow the classification of lncRNAs into several classes:guides,dynamic scaffolds,molecular recall,signal,and enhancers (Figure2;Balas and Johnson,2018).In particular,lncRNAs classified as guides can bind chromatin-modifying proteins and target complexes formed at specific genomic locations by reprogramming the epigenetic state.By implementing this,lncRNAs change gene expression to cis or trans (Figure 2A;Asadi et al.,2021).LncRNAs acting as dynamic scaffolds play a structural role in assembling different complexes and recruiting transcriptional enzymes.Following the assembly phase,one can witness their activation or repression.This underlines the importance of lncRNAs in the transcription process(Figure2B;Fang and Fullwood,2016).LncRNAs that act as molecular recall are likely negative regulators in both transcriptional processing and chromatin modification.The lncRNAs belonging to this category can sponge miRNAs by creating a competing endogenous RNA network to inhibit their binding to the target mRNA (Figure 2C;Asadi et al.,2021).LncRNAs perform a signal function and are expressed at a precise moment and in a specific position in response to external stimuli.Some of these are regulatory,while others are simply products of transcription,with the act of regulatory initiation,elongation,or termination.Signal lncRNAs are known to interact with chromatin-modifying enzymes such as histone methyltransferase to silence their target genes by blocking their transcription or through the formation of heterochromatin (Figure 2D;Wang and Chang,2011).Other lncRNAs exert function as enhancers,i.e.,activating gene transcription by serving as the cisregulatory molecules.Furthermore,recent studies have demonstrated that many enhancer elements can be transcribed and produce RNA molecules,which are termed enhancer RNAs (Figure 2E;Kim et al.,2015).
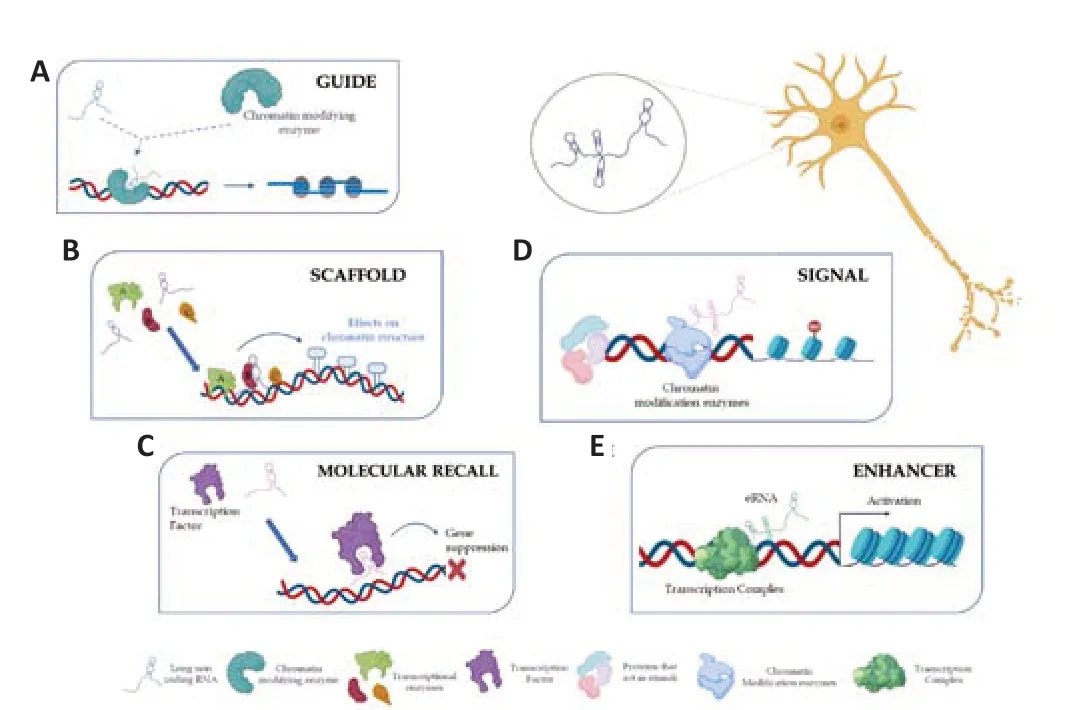
Figure 2|Function and mechanism of lncRNAs in transcriptional regulation.
LncRNAs,impacting gene function through transcriptional and epigenetic regulation,include also RNA transcribed from the opposite strength of protein-coding genes or sense strand derived mRNA,called lncRNA-AS(antisense lncRNA) (Gagliardi et al.,2018).Precisely this characteristic allows the formation of particular structural configurations capable of addressing the action of lncRNA in a specific and selective manner (MacDonald and Mann,2020).There are several mechanisms of lncRNA-AS action,but these will not be presented in this review.
Long-Noncoding RNAs as Epigenetic Regulators in Alzheimer’s Disease
AD is the most common cause of progressively disabling degenerative dementia with onset in presenile age characterized by the formation of β-amyloid plaques and neurofibrillary tangles resulting in brain atrophy and neuronal death.The etiology of the disease is largely unknown and,in most cases,occurs sporadically.The best-known genetic risk factor is the inheritance of the ε4 allele of Apolipoprotein E (APO-E).Between 40% and 60% of people with the disease have at least one APOE-ε4 allele.Although the hereditary genes that cause familial AD are rare,they have been identified as being associated with the processing or production of amyloid-beta (Soria Lopez et al.,2019).
BACE1(β-site amyloid precursor protein cleavage enzyme 1,chr.11q23.3,OMIM * 604252) encodes for an enzyme responsible for β-amyloid plaque formation through the cleavage of the amyloid beta precursor protein.In mouse models,BACE1has been shown to regulate voltage-gated sodium channels that control neuronal activity involved in the pathophysiology of AD.In particular,BACE1expression is localized to the presynaptic terminals surrounding the amyloid plaques showing thatBACE1-deficient mice have a healthy phenotype and suppressed β-amyloid production (Sayad et al.,2022).Deregulation ofBACE1plays an important role in the initial phase of the disease by initiating the process of toxicity (Hampel et al.,2021).The action of several miRNAs,such as miR-34a-5p,miR-125b-5p,miR-15b,and miR-149,inhibit the expression ofBACE1,reduce amyloid accumulation and improve neuronal damage (Sayad et al.,2022).BACE1-AS(BACE1antisense RNA,11q23.3,OMIM * 614263) lncRNA acts in the post-transcriptional regulation ofBACE1.BACE1-ASis an antisense transcript that appears to be elevated in patients with AD,determines a molecular change promoting β-secretase synthesis,and can function as competing endogenous RNA capable of avoiding the degradation ofBACE1mRNA (Faghihi et al.,2008).The aggregation and the formation of a duplex betweenBACE1-ASandBACE1increase the stability of the transcript,underlining how the lncRNA determines an upregulation of mRNA and BACE1 protein.TheBACE1-AS/miR-485-5p/BACE1axis demonstrates the involvement of this lncRNA in the pathogenesis of AD through the post-transcriptional regulation ofBACE1(Faghihi et al.,2010).Furthermore,He and collaborators demonstrated thatBACE1-ASavoidsBACE1transcript degradation by acting against miR-29b-3p/miR-107/miR-124-3p/miR-485-5p/miR-761 (He et al.,2020).TheBACE1-AS/miR-132-3p axis is involved in the neuroprotective process induced by berberine,a protoberberine isoquinoline alkaloid that limits the formation of β-amyloid plaques and intracellular neurofibrillary tangles (Cai et al.,2016).In particular,berberine treatment downregulatesBACE1-ASin AD,modulating the function of miR-132-3p whose accumulation alleviates the neuronal damage induced by β-amyloid (Ge et al.,2020).
SNHG1(Small Nucleolar RNA Host Gene 1,chr.11q12.3) is an lncRNA involved in neuronal damage induced by the presence of β-amyloid plaques through the axisSNHG1/miR-137/KREMEN1(Kringle Containing Transmembrane Protein 1) andSNHG1/miR-361-3p/ZNF217(Zinc Finger Protein 217) (Sabaie et al.,2021).SNHG1may play a positive role in the progression of NDs as theSNHG1knockdown can promote cell survival mitigating the progression of AD.The study results conducted by Gao et al.(2020) showed that resveratrol exerts a neuroprotective effect by acting on theSNHG1/miR-361-3p/ZNF217axis.In particular,the expression ofSNHG1is negatively regulated by resveratrol that inhibits the sponging of miR-361-3p whose overexpression determinesZNF217silencing with consequent inhibition of cell injury (Gao et al.,2020).
NEAT1(Nuclear Paraspeckle Assembly Transcript 1,chr.11q13.1,OMIM #612769) is an lncRNA involved in epigenetic regulation in AD through chemical modifications of long-term memory formation processes.In particular,NEAT1knockdown with siRNA regulatesc-Fos(human Fos proto-oncogene) promoter methylation by H3K9me2,an epigenetic modification to the DNA packaging protein histone H3,increasing gene expression and improving long-term memory (Butler et al.,2019).Furthermore,upregulatedNEAT1regulates the interaction betweenPINK1(PTEN-induced kinase 1) andNEDD4L(ubiquitinprotein ligase nedd4-like) with consequent ubiquitination and degradation ofPINK1which causes an increase in the accumulation of β-amyloid and cognitive decline (Huang et al.,2020).This lncRNA acts through the miR-124/BACE1axis with consequent promotion of the development of AD (Zhao et al.,2019).In particular,Zhao and collaborators observed that the downregulation ofNEAT1expression contributed to the deposition of β-amyloid,inhibiting the expression of multiple endocytosis-related genes,through the interaction with the P300/CBP complex (Zhao et al.,2019).
SOX21-AS1(SOX21 Antisense Divergent Transcript 1,chr.13q32.1) is an lncRNA that plays a similar role toNEAT1: they both act against miR-107 which is found to be deregulated in brain tissue of AD patients.In particular,SOX21-AS1knockdown attenuates neuronal apoptosis and mitigates oxidative stress in AD by sponging miR-107,reducing Aβ-induced neuronal damage (Xu et al.,2020).Furthermore,silencing of this lncRNA could act by upregulating the expression of FZD3/5 (frizzled class receptor) and following activation of the Wnt signaling pathway (Zhang et al.,2019a).
LoNA(long nucleolus-specific lncRNA) is a recently discovered lncRNA that works by reducingNCL(nucleolin) transcription and 2’-O-methylation rRNA(ribosomal RNA) methylation resulting in a reduction of activeFBL(fibrillarin),a component of a nucleolar small nuclear ribonucleoprotein (snRNP)particle thought to participate in the first step in processing preribosomal RNA.A downregulation ofLoNAcauses an increase in rRNA concentrations,improving synaptic activity and long-term memory (Asadi et al.,2021).In vivostudies suggest thatLoNAplays a key role in NDs by representing a probable therapeutic target for the treatment of AD (Li et al.,2018).
MALAT1(metastasis-associated lung adenocarcinoma transcript 1,chr.11q13.1,OMIM * 607924),known asNEAT2,regulates neuronal and synaptic activity by modulating the expression of genes involved in the formation and maintenance of synapses (Asadi et al.,2021).Emerging evidence suggests a neuroprotective function ofMALAT1through the inhibition of neuroinflammation.Indeed,MALAT1knockdown promotes neuronal apoptosis and represses neurite growth while the overexpression of this lncRNA determines a modification of the inflammatory picture,favoring neuritic growth and preventing apoptosis in AD,through the interaction with miR-125b (Ma et al.,2019;Lan et al.,2021).In addition,the study conducted by Zhuang and collaborators demonstrated thatMALTA1/miR-125b can be considered prognostic and predictive markers of AD and that the correlation between this axis andFOXQ1(forkhead box Q1),PTGS2(prostaglandinendoperoxide synthase 2) andCDK5(cyclin-dependent kinase 5) could provide important information for the therapies of AD (Zhuang et al.,2020).
A newly discovered lncRNA involved in AD is MAPT-AS1,located within the antisense strand of theMAPTpromoter region,which plays a critical role in the formation and maintenance of microtubules.Several evidences show thatMAPTupregulation causes an increase in events related to neurodegeneration(Adams et al.,2009).Recently,de Silva et al.(2018) showed that delivery ofMAPT-AS1vectors into the hippocampus of mouse models via adenoassociated virus,resulted in a decrease in tau levels,suggestingMAPT-AS1as a potential therapeutic approach to treating AD (Rohan de Silva,2006).
Long-Noncoding RNAs as Epigenetic Regulation in Parkinson’s Disease
PD is a neurodegenerative disease age-related that predominantly affects dopaminergic neurons in a specific area of the brain: the substantia nigra(Strafella et al.,2021).The etiology of the disease is not entirely clear,although scientists believe that the cause is a combination of genetic and environmental factors.Familial cases of PD can be caused by mutations in theLRRK2(Leucine-rich repeat kinase 2),PARK7(Parkinsonism associated deglycase),PINK1(PTEN-induced kinase 1),PRKN(Parkin RBR E3 Ubiquitin Protein Ligase),orSNCA(Synuclein Alpha) genes,or by alterations in genes not yet identified (Cherian and Divya,2020).
SNHG1,also known aslinc00057,is a recently discovered lncRNA in PD.Several studies have shown that this lncRNA is involved in different molecular and cellular mechanisms directly related to the phenotype of the disease.The main biological function ofSNHG1is to promote the ubiquitination of α-synuclein and regulate its toxicity through theSNHG1-miR15b-5p-SIAH1-linc-p21-miR-1277-5p axis (Xu et al.,2018).Specifically,SNHG1promotes the aggregation and toxicity of α-synuclein by targeting miR15b-5p.This interaction activates the gene SIAH1 (Siah E3 Ubiquitin Protein Ligase 1) (Chen et al.,2018) regulated by the lncRNA linc-p21 which,in turn,sponges miR-1277-5p and indirectly increases the expression of α-synuclein to suppress viability and activate apoptosis (Xu et al.,2018).Moreover,SNHG1promotes neuroinflammation through the miR-7/NLRP3(NLR Family Pyrin Domain Containing 3) axis (Rasheed et al.,2021) and,when upregulated,promotes 1-methyl-4-phenylpyridinium (MPP+) induced cytotoxicity through miR-153-3p sponging.This positive ion has been related to chemical reactions causingin vitroPD-like cytotoxic cellular events (Shishido et al.,2019).SNHG1is also involved in the autophagic process by regulating the expression of p27/mTOR(mammalian target of rapamycin kinase) through competitive interaction with miR-221/222 members (Qian et al.,2019).
The above-discussedNEAT1is also involved in PD and it has been strengthened by a recent study discussing contradictory results on its upregulation as part of a protective or a damaging mechanism (Boros et al.,2021).Yang et al.(2018) observed thatNEAT1promotes the autophagic process induced by MPTP (1-methyl 4-phenyl 1,2,3,6-tetrahydropyridine)by stabilizingPINK1(Yan et al.,2018).In a more recent work by Sun et al.(2021),it was observed that downregulated NEAT1 binds miR-1301-3p and inhibits the expression of GJB1 (gap junction protein beta 1) resulting in decreased α-induced NLRP3 inflammation.Zhou et al.(2020) demonstrated that an upregulation of theNEAT1lncRNA can contribute to the neuronal damage created by MPP+via theNEAT1-miR-1277-5p-ARHGAP26(Rho GTPase Activating Protein 26) (Zhou et al.,2021a).Despite several studies suggest a protective role ofNEAT1downregulation in PD progression (Boros et al.,2021),no data fromin vitroandin vivomodels studies are available to confirm a direct role of this lncRNA in the pathogenesis of the disease (Boros et al.,2021).
HOTAIR(Hox Transcript Antisense RNA,chr.12q13.13,OMIM * 611400) is an lncRNA whose functional role is well known in oncogenetic processes while its role in PD is unclear.This lncRNA is associated with humanHOXloci and,by targeting the repressivePRC2,can modify the status of methylation of H3K27me,and manipulate gene expression patterns throughout the genome.The molecular pathogenesis of PD is hypothesized to be associated with mitochondrial dysfunction and activation of the apoptotic cascade.MPP+-induced neuronal death,mediated by loss of mitochondrial membrane potential,is attenuated byHOTAIRknockdown,through the reduction of the cell death protease activity of Caspase 3 (Wang et al.,2017).Zhao and collaborators demonstrated thatHOTAIRsponging miR-874-5p is involved in the neuronal damage generated by MPP+(Zhao et al.,2020).In the same year,a study by Zhang and collaborators showed thatHOTAIRinhibits the activation ofNLRP3-mediated pyroptosis,a form of lytic and inflammatory cell death,in mouse models of PD through the miR-326/ELAVL1(ELAV Like RNA Binding Protein 1) axis (Zhao et al.,2020;Zhang et al.,2021).Furthermore,this lncRNA tagged miR-126-5p andRAB3IP(RAB3A Interacting Protein) resulting in increased disease progression (Rasheed et al.,2021).
SNHG14(Small Nucleolar RNA Host Gene 14,chr.15q11.2,OMIM * 616259) is a lncRNA potentially involved in the loss of dopaminergic neurons,capable of regulating miR-133b which,in turn,can control the expression of α-synuclein.TheSNHG14/miR-133b complex determines a downregulation of α-synuclein and,therefore,could mitigate the symptoms (Zhang et al.,2019b).Since α-synuclein is the major component of Lewy bodies,then changes in α-synuclein influence the development of PD.In addition,SNHG14sponge miR-214-3p tagsKLF4(Kruppel Like Factor 4) (Zhou et al.,2020).The latter favors the effects of MPP+by delaying cell proliferation and programmed cell death (Chen et al.,2013).Based on these findings,theSNHG14/miR-214-3p/KLF4axis could therefore be considered a therapeutic target (Zhou et al.,2020).
The expression of α-synuclein is also regulated by the lncRNATP53COR1(Tumor Protein P53 Pathway Corepressor 1,Chr.6p21.2,OMIM * 616343),also known aslincRNA-p21.Specifically,this lncRNA sponges miR-1277-5p and determines the activation of α-synuclein with consequent promotion of programmed cell death and repression of cell viability (Xu et al.,2018).Furthermore,thelincRNA-p21/miR-625 axis determines the inhibition of the neuronal damage generated by MPP+(Ding et al.,2019).
AL049437is an lncRNA with evident neuroprotective functions.As demonstrated by Zhang et al.,this lncRNA reduces the presence ofTNF-α(Tumor necrosis factor α),IL-6(Interleukin 6),and reactive oxygen species in MPP+models (Zhang et al.,2020).Transcriptomic analysis performed by Ni et al.(2017),highlighted a high number of deregulated lncRNAs in PD patients,underlining that the lncRNAsAL049437andAK021630are involved in the pathogenetic mechanisms of the disease (Ni et al.,2017).To our knowledge,no other studies have investigated the role ofAL049437in PD and further research is needed to understand theAL049437mechanism of action in the pathogenesis of the disease.
Long-Noncoding RNAs as Epigenetic Regulation in Amyotrophic Lateral Sclerosis
ALS is a neurodegenerative disease caused by the progressive loss of motor neurons resulting in weakness and paralysis of the voluntary muscles.The main clinical feature of ALS is the involvement of upper and lower motor neurons in multiple regions of the brain stem and spinal cord.Although important progress has been made in recent times in understanding etiopathology,ALS still remains an unknown disease in many respects (Masrori and Van Damme,2020).10% of individuals affected by this pathology show a genetic driver that allows the distinction between familial ALS and sporadic ALS cases (Wijesekera and Leigh,2009).There is extensive heterogeneity in the genetic causes of familial ALS,but the forms of familial ALS and sporadic ALS have similarities in their pathological and clinical characteristics,suggesting a convergence of cellular and molecular events leading to motor neuron degeneration (Grad et al.,2017).To date,more than 100 genes have been associated with ALS (https://alsod.ac.uk/),but only four of them are linked to a significant percentage of ALS cases includingSOD1(superoxide dismutase 1),C9orf72,TARDBP(TAR DNA Binding Protein),andFUS(fused in sarcoma) (Ungaro et al.,2021).
NEAT1_2is a lncRNA with an important structural role in nuclear paraspeckles(Fox et al.,2002).TDP-43 and FUS proteins bindNETA1_2to properly create these structures.Mutations in these proteins result in functional defects in nuclear paraspeckles.Furthermore,the role ofNEAT1_2as a disease biomarker is highlighted as it is poorly expressed in motor neurons of healthy subjects while it is highly expressed in motor neurons of ALS patients(Nishimoto et al.,2013).However,stresses can cause NEAT1_2-induced paraspeckle formation in the nucleus,but it is not yet known the mechanism involved inNEAT1_2lncRNA upregulation during the early phase of ALS(Nishimoto et al.,2013).NEAT1_2can bind RBPs (RNA binding protein) in the nuclei of motor neurons by regulating their expression (Nishimoto et al.,2013) and allowing a greater understanding of lncRNAs in ALS as both prognostic and therapeutic markers.
C9Orf72-ASis an lncRNA-AS that contains the reverse repeat sequence of the causative hexanucleotide of ALS disease.The function of theC9Orf72sense transcript is better known than that of the antisense transcript which could lead to the formation of polypeptides and RNA foci (Freibaum and Taylor,2017;Mizielinska et al.,2017).The production of these foci causes the RBPs sequester and an indirect regulation of gene expression.Greater knowledge of these transcripts may prove to be an alternative strategy for targeting repeatedC9Orf72expansions and reversing the transcriptional changes typical of the disease (Chen and Chen,2020).
Long-Noncoding RNAs as Epigenetic Regulation in Huntington’s Disease
HD is an autosomal dominant neurodegenerative disease clinically characterized by movement disorders,progressive dementia,and psychiatric and behavioral disorders.An increase in the copy number of CAG trinucleotide repeats in the first exon of the HTT (Huntingtin) gene,that encodes polyglutamine,causes HD (Jimenez-Sanchez et al.,2017).Specifically,an increase in the size of the CAG segment leads to the production of an abnormally long version of the huntingtin protein.The stretched protein is cut into smaller,more toxic fragments that bind and build up in neurons,disrupting the normal functions of these cells.Dysfunction and eventual death of neurons in certain areas of the brain underlie the signs and symptoms of pathology (Walker,2007).
TUG1(Taurine Upregulated Gene 1,22q12.2,OMIM * 614971),highly expressed in the mammalian brain,appears to be involved in neurodegenerative diseases in various physiological processes such as those regulating genes at epigenetic,post-transcriptional,and post-translational regulation (Guo et al.,2020).A study conducted by Khalil et al.(2009),showed thatTUG1binds to the PRC2 epigenetic regulatory complex and its knockdown causes widespread changes in gene expression and in particular in the upregulation of cell cycle genes.Interestingly in the HD context,TUG1serves as a direct downstream transcriptional repressor of p53 (known to be up-regulated in HD),acting as a survival factor in neurons (Khalil et al.,2009).However,further studies are needed to identify the mechanism of action of this lncRNA at the neuronal level.Different other lncRNAs,such asMEG3(Maternally Expressed Gene 3,14q32.2,OMIM *605636),NEAT1,andXIST(X Inactivation-Specific Transcript,Xq13.2,OMIM *314670),have shown to contribute to HD pathogenesis.Some studies suggest that these lncRNAs can interact with many miRNAs as miR-9,miR-125b,miR-132,miR-146a,miR-150,miR-221,and miR-222,potentially reducing the efficiency of binding to their target mRNAs and thus promoting the development of the disease (Zhou et al.,2021b).
Long-Noncoding RNAs as Targets: Current Challenges in Alzheimer’s Disease/Parkinson’s Disease/Amyotrophic Lateral Sclerosis/Huntington’s Disease Therapy
To date,the studies performed on lncRNAs leave open the possibility of using these as new biomarkers for the diagnosis and clinical therapy of NDs.In the clinical context,the detection of circulating lncRNA has been widely described in many body fluids (plasma,urine,saliva,blood,and CSF samples)from NDs patients,indicating their potential role as prognostic and diagnostic biomarkers (Feng et al.,2018;Gagliardi et al.,2018;Fan et al.,2019;Fotuhi et al.,2019;Cheng et al.,2021;Yu et al.,2022).However,limitations due to the lncRNA detection and/or disease-specificity limit their use for early disease diagnosis.
lncRNAs can be targeted by multiple approaches in the nucleus and cytoplasm including: knockdown of pathogenic RNAs by using siRNAs or antisense oligonucleotides,modulation of lncRNA genes by steric blockade of the promoter or by using genome-editing techniques as CRISPR/Cas9.Another interesting approach involves the use of natural antisense transcripts (NATs):lncRNAs that act as repressors of specific genomic loci.In particular,socalled “antagoNATs”,antisense oligonucleotides that target NATs have shown exciting results for gene reactivation in the CNS (Modarresi et al.,2012).
Currently,various siRNA or antisense oligonucleotide-based therapies involved in pathogenesis have been approved by the Food and Drug Administration (FDA) and the European Medicines Agency (EMA) (Winkle et al.,2021).Despite nobody lncRNA-based therapeutics having entered the clinic,there are many ongoing studies for the evaluation of lncRNAs as targets and/or therapeutic systems,especially in oncological pathologies (Zhou et al.,2021b).In our context,several lncRNAs with key roles in NDs could be considered therapeutic targets.InAdditional Table 1,we provide a detailed list of the lncRNAs,previously described in the text,associated with AD/PD/ALS/HD.
Despite lncRNAs targeting are receiving considerable attention,understanding of their function and effects is incomplete,and there are still some limitations to their clinical application.First,the sensitive detection and tissue diseasespecificity of lncRNA are challenging and require more comprehensive studies in the future.Moreover,clinical trial results about lncRNAs are lacking so far and many questions still need to be answered beyond the identification and the choice of the disease-specific lncRNA reference,from the system used to deliver lncRNA in targeting NDs to the safety of its application in the human body.
Conclusion
In recent decades,thanks to advances in high-throughput sequencing technologies and whole-genome and transcriptome studies,it has been possible to discover new mechanisms of variability involved in the regulation of gene expression.
In recent years,various research groups have pointed out that lncRNAs play a major role in central nervous system development through epigenetic and translational modifications.The emerging role of lncRNAs in NDs (AdditionalTable 1),suggests that their dysregulation could trigger neuronal death through as yet unexplored RNA-based regulatory mechanisms that deserve further investigation.The evaluation of their diagnostic significance and their therapeutic potential could also address the development of new treatments for diseases for which a cure is not yet available.
Author contributions:Conceptualization: PR,EG,FLC;methodology,resources,writing—original draft preparation: PR;writing—review and editing: PR,FDA,EG,and FLC.All authors have read and agreed to the published version of the manuscript.
Conflicts of interest:The authors declare no conflicts of interest.
Availability of data and materials:All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open access statement:This is an open access journal,and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License,which allows others to remix,tweak,and build upon the work non-commercially,as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional file:
Additional Table 1: List of lncRNAs associated with the pathogenesis of NDs and eligible as therapeutic targets.
- 中国神经再生研究(英文版)的其它文章
- Neuro faces of beneficial T cells: essential in brain,impaired in aging and neurological diseases,and activated functionally by neurotransmitters and neuropeptides
- Profiling neuroprotective potential of trehalose in animal models of neurodegenerative diseases:a systematic review
- Cdk5 and aberrant cell cycle activation at the core of neurodegeneration
- Recent advancements in noninvasive brain modulation for individuals with autism spectrum disorder
- Vicious cycle of lipid peroxidation and iron accumulation in neurodegeneration
- Cell-based therapeutic strategies for treatment of spinocerebellar ataxias: an update