Virtual screening, molecular docking and identif ication of umami peptides derived from Oncorhynchus mykiss
Wenzhu Zho, Lijun Su, Shitong Huo, Zhipeng Yu,*, Jinrong Li, Jingo Liu
a School of Food Science and Engineering, Hainan University, Haikou 570228, China
b College of Food Science and Engineering, Bohai University, Jinzhou 121013, China
c Lab of Nutrition and Functional Food, Jilin University, Changchun 130062, China
Keywords:Oncorhynchus mykiss Umami peptides Electronic tongue Molecular docking
A B S T R A C T Oncorhynchus mykiss is delicious and contains abundant f lavor substances. However, few studies focused on umami peptides of O. mykiss. In the current work, umami peptides derived from O. mykiss were identif ied using virtual screening, molecular docking, and electronic tongue analysis. First, the O. mykiss protein was hydrolyzed using the PeptideCutter online enzymolysis program. Subsequently, water-soluble and toxicity screening were performed by Innovagen and ToxinPred software, respectively. The potential peptides were docked with umami receptor T1R1/T1R3. Furthermore, taste properties of potential peptides were validated by electronic tongue. Docking results suggested that the three tetrapeptide EANK, EEAK, and EMQK could enter the binding pocket in the T1R1 cavity, wherein Arg151, Asp147, Gln52, and Arg277 may play key roles in the production of umami taste. Electronic tongue results showed that the umami value of EANK, EEAK,and EMQK were stronger than monosodium glutamate. This work provides a new insight for the screening of umami peptides in O. mykiss.
1. Introduction
Umami, as one of the f ive basic taste, can bring people a sense of pleasure and is an essential taste requirement of the human. Umami taste could be attributed to the organic acids, free amino acids,nucleotides, organic bases, Maillard reaction products and some peptides [1]. Umami peptides, as a new umami substance proposed in recent years, can interact with umami receptors on human taste buds to exhibit certain umami characteristics, and enhance the original f lavor of food through synergistic effect [2]. Umami taste is initiated by the interaction between umami substances and umami receptors located in taste buds. At present, two main umami receptors have been identif ied: heterodimer T1R1/T1R3 and taste-metabotropic glutamate receptor 4 (mGluR4). Both of which are C-type G-protein coupled receptors (GPCR), consists of an N-terminal venus f lytrap domain (VFTD),an extracellular cysteine-rich domain (CRD), and a seven transmembrane domain (TMD), wherein VFTD is mainly responsible for the recognition of umami ligand [3,4]. The T1R1/T1R3 signal-mediated played crucial role in the production of umami taste [5], and could response to most amino acids [6]. Hence, T1R1/T1R3 is considered to be the optimal umami receptor. T1R1/T1R3 was used to docking various umami peptides reported, indicating that Glu or Asn played an important role in the production of umami taste [7]. Previous work has focused on used molecular docking and dynamic simulation to study the binding relationship between umami hexavalent peptides and T1R1/T1R3, and showed that charged amino acids and hydrophobic amino acids had important contributions to the production of umami peptide [7]. Therefore, strengthening the research on the recognition mechanism of umami receptors T1R1/T1R3 can deepen the understanding of the structure characteristics of umami peptides, and provide a theoretical basis for screening, identification and design of new umami peptides. Previous research has focused on the use of ultrafiltration, gel filtration chromatography, and reversedphase high performance liquid chromatography to isolation and identification of umami peptides [8-10]. The isolation and preparation of umami peptides are mainly based on experiments, which requires a lot of labor and cost. Therefore, the virtual screening method has been applied, which can replace the traditional umami peptide screening method to some extent.
Based on advances in computer science research, the screening and identification of umami peptides is becoming more convenient.The PeptideCutter online enzymolysis program can be used to predict the peptide sequence released from some representative protein [11].Molecular docking refers to the mutual recognition of two molecules through structural complementary and energy matching. This is a useful tool for complex interactions between receptors and ligands and virtual screening [12]. Currently, molecular docking has been widely used to simulate the interaction between ligand and receptor [7,8,13],and has been proven to be an important tool for computer-assisted receptor-ligand binding to predicting umami peptides.Oncorhynchus mykissis delicious and containsabundant flavor substances, which was first introduced into China from North Korea. The nebulin fromO. mykisscontains a large number of amino acid, and many peptides with varieties of bioactivities have been identified by hydrolysis. It may be used as a potential source of umami peptides.
The purpose of this study was to screen and identify novel umami peptides from nebulin inO. mykissvia virtual screening and molecular docking. First,O. mykissprotein was hydrolysed via the PeptideCutter online enzymolysis program. Then water-soluble and toxicity screening were performed by Innovagen and ToxinPred software, respectively. Next the selected peptides were docked with umami receptor T1R1/T1R3. Finally the potential peptides were verified by electronic tongue. In addition, the molecular mechanism of the umami peptides and umami receptor T1R1/T1R3 was revealed.This work may be helpful for discovering new umami peptides from fish resource.
2. Materials and methods
2.1 Chemical reagents
Ethyl alcohol, hydrochloric acid, potassium chloride, potassium hydroxide, tartaric acid, monosodium glutamate (MSG) were purchased from commercial sources. All chemicals and solvents used in the experiment were of analytical grade.
2.2 In silico hydrolysis of nebulin in O. mykiss
The protein sequence of nebulin (GenBank accession number:XP_021453115) was obtained from the NCBI (https://www.ncbi.nlm.nih.gov/).In silicohydrolysis of nebulin was performed by two specific enzymes of pepsin (pH 1.3) (EC: 3.4.23.1) and trypsin(EC: 3.4.21.4) using the PeptideCutter online enzymolysis program,available at http://web.expasy.org/peptide_cutter [14]. Previous research showed that the length of the peptide chain is closely related to the umami properties of the peptides. Small molecular peptides play critical roles in the production of umami properties [15]. In addition, short peptides with a molecular mass of 500–1 000 u have the strongest umami taste, which plays a key role in enhancing the umami [16]. Therefore, tripeptides, tetrapeptides, pentapeptides, and hexapeptides were collected for the following research.
2.3 Prediction of toxicity and solubility of peptides
Toxicity remains the major concern in the development of peptides. In addition, good water solubility is the key to the normal metabolism of peptidesin vivo.Therefore, toxicity and solubility of peptides should be predicted byin silicoapproach. The program ToxinPred was performed for predicting toxicity of these peptidesin silico(http://www.imtech.res.in/raghava/toxinpred/) (accessed November 26th, 2019) [17]. In addition, solubility of these peptides were estimated [11] (http://www.innovagen.com/ and http://www.imtech.res.in/raghava/toxinpred/) (accessed November 26th, 2019).
2.4 Molecular docking of potential peptides and umami receptor T1R1/T1R3
Molecular docking was performed according to the previous method reported by Dang et al. [2], with a slight modification. The crystal structures of T1R1/T1R3 were selected for molecular docking with peptides. Then the T1R1/T1R3 were prepared by removing water and adding hydrogen atoms. The peptides structures were drawn in Discovery Studio (DS) 2017 client software. The docking of peptides and T1R1/T1R3 umami receptor were performed using CDOCKER protocol by software DS 2017. The energy of the peptides were minimized by the CHARMm force field. Docking pocket coordinatesx: 26.348 230,y: –4.815 884, andz: 18.701 097 with a radius of 22 Å. The molecular docking was evaluated according to the docking energy, and the optimal docking poses was retained to display the two-dimensional figures.
2.5 Peptide preparation
The selected peptides were synthesized by Nanjing Yuanpeptide Biotech Co., Ltd. (Nanjing, Jiangsu, China). The purity and molecular mass of the synthetic peptides were determined by high performance liquid chromatography and mass spectrometry [18], respectively.
2.6 Electronic tongue determination of taste characteristics
Taste properties of the potential peptides were validated by the SA402B electronic tongue (INSENT, Kanagawa, Japan). The instrument includes five taste probes based on artificial bimolecular membranes and two reference electrode. The five test sensors were applied to gather taste information. The five test sensors were named as AAE, CT0, CA0, C00 and AE1, which represent umami, salty,sour, bitter and astringent tastes, respectively. The reference probes include the positive and negative reference probes. When testing, the odorless sample containing 30 mmol/L KCl and 0.3 mmol/L tartaric acid was used as a reference. MSG was selected as the positive control in this experiment. Then three cleaning operations were performed,the sensor was cleaned in the positive and negative cleaning solution for 90 s, after which they were cleaned in the other two reference solutions for 120 s. After cleaning, balanced in reference solution for 30 s and then 30 s for measurement. Then the sensor was cleaned twice quickly, and the aftertaste value was measured in the reference solution, the measurement completed for one time [19]. Each sample was measured 4 times, and the first data were deleted, due to the first measurement value instability.
2.7 Statistical analysis
Data were expressed as means ± SD and subjected to one-way analysis of variance (ANOVA). The software SPSS 19 was used for the main data processing of this study. Values ofP< 0.05 were considered as statistically.
3. Results and discussions
3.1 In silico screening of peptides with umami taste
Nebulin inO. mykisscontained 6 883 amino acids in total, and 1 918 peptides were obtained after virtual enzymolysis by pepsin and trypsin. Peptides have numerous advantages including high biological activity, low production cost, strong specificity, and strong penetration. However, toxicity remains the major concern in the development of peptides [17]. In addition, good water solubility is the key to the normal metabolism of peptidesin vivo.Therefore, toxicity and solubility of peptides should be predicted byin silico. The results showed that 332 peptides have good water solubility and non-toxic,especially 93 tripeptides, 102 tetrapeptides, 71 pentapeptides, and 53 hexapeptides. Most tripeptides and tetrapeptides have good water solubility and non-toxic. This may be the length of the peptide chain is closely related to the water solubility of the peptide. As the length of the peptide chain increases, its water solubility decreases.
3.2 Molecular docking analysis of potential peptides and T1R1/T1R3
In recent research, molecular docking technology has been used to study umami peptides and taste receptors T1R1/T1R3 to reveal the taste mechanism of umami ingredient. Researches showed that the T1R1 subunit was mainly responsible for the identification of umami substances, while the T1R3 subunit was responsible for other auxiliary functions [20]. Peptides were docked with the T1R1 subunit. The docking energy of the peptides with T1R1 was shown in Table 1. Molecular docking results showed that lower ‘CDocker-Energy’ represents a more affinity between peptides and T1R1/T1R3.Among the 332 peptides with good water-soluble and non-toxic properties, the 20 peptides had the lowest docking energy. Therefore,20 peptides ENQK, DYEK, EEAK, DVEK, DMGK, EEGK, DFNK,DHVK, EANK, DFHK, SEVK, EMQK, DYQK, NEVK, DSSAK,DWDK, QSDK, ENTK, AYEK, ASGEK were selected according to CDocker enegry value. The CDocker enegry value of these 20 peptides increased successively, among which the lowest CDocker Enegry value is ENQK and the highest CDocker enegry value is ASGEK, with the value of –119.089 and –104.807 kcal/mol. The CDocker enegry value of the known umami peptides QEEL, LEQL,ESLA, INEL, EESLA, and VVGET to T1R1 were -95.389, –91.955,–88.292, –82.795, –89.564, and –75.024 kcal/mol, respectively.The CDocker enegry value of known umami peptides are all higher than ASGEK, indicating that the selected 20 peptides have a strong affinity with T1R1/T1R3. Umami properties were mainly through the interaction between acidic and basic groups, and usually contain glutamate and aspartic acid residues [21]. In addition, small peptides with glutamic acid (E) at the N-terminus of the polypeptide have a stronger umami taste. Thus, three tetrapeptides EANK, EEAK, and EMQK were selected and synthesized for further study.
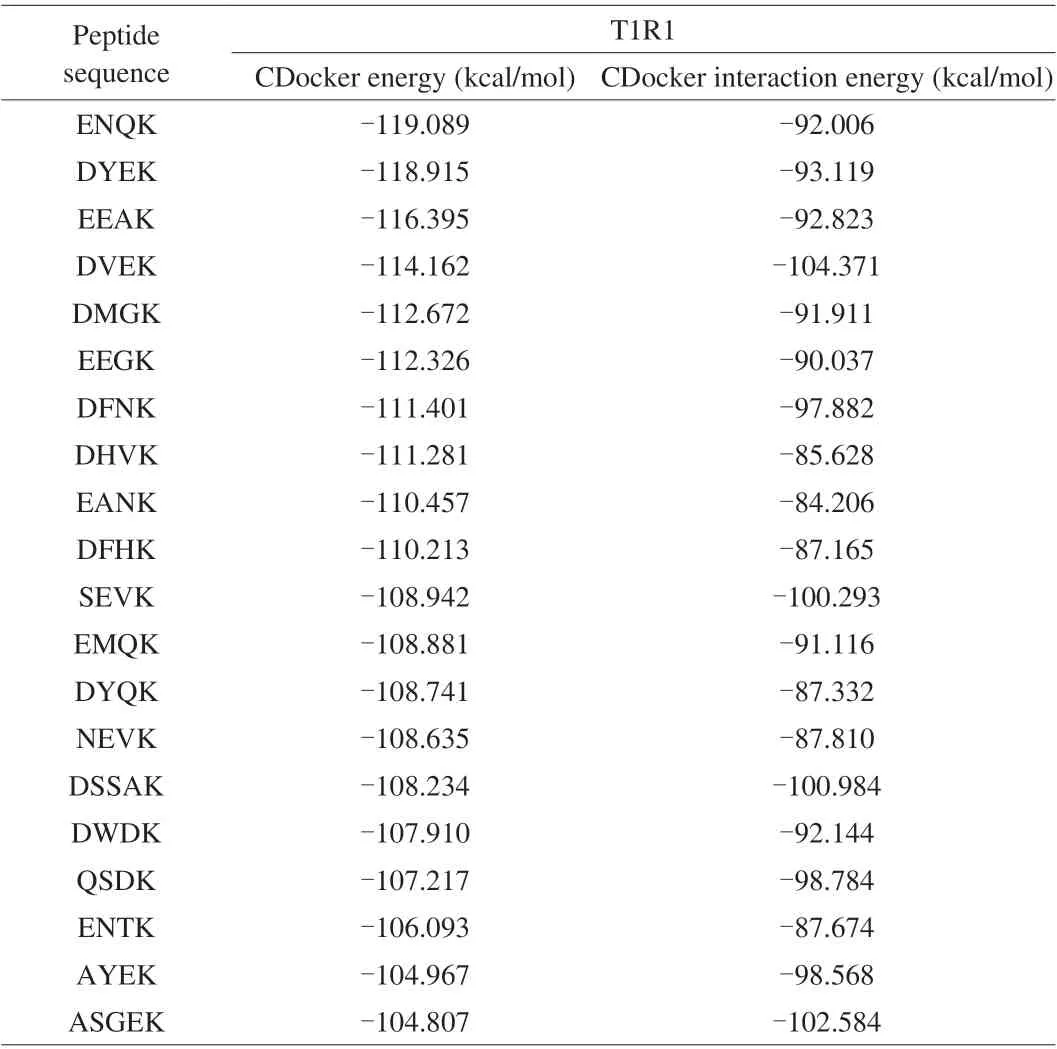
Table 1 Molecular docking results of peptides.
3.3 Electronic tongue evaluation of umami characteristics
The potential peptides were synthesized and taste properties were verified by electronic tongue. The umami values of EANK, EEAK,EMQK, and MSG were -5.38, –5.38, –4.48, and –6.33, respectively.The umami value of all the three tetrapeptides were stronger than the same concentration MSG. MSG is used as a positive control, and its main component is the sodium salt of glutamic acid, which is used as a flavor or taste enhancer in food. Richness is the umami aftertaste [22].Aftertaste is the taste intensity of the taste immediately felt after the food is taken out of the mouth. Unifying characteristic of aftertaste is that it is felt after a food is either swallowed or spat out [23]. Richness values were 0.67, 0.70, 0.77, and 0.70, respectively. The richness values of these three tetrapeptides are similar to MSG, indicating that the peptides have good umami aftertaste.
3.4 Interaction mechanism of umami peptides and T1R1/T1R3
Peptide EANK, EEAK, and EMQK was analyzed by molecular docking using Discovery Studio 2017 software. The active site sphere was (26.348 230, –4.815 884, and 18.701 097). The molecular docking results of three tetrapeptides and umami receptor T1R1/T1R3 were shown in Figs. 1–3. The interactions between ligands and surrounding residues were observed, including salt bridges,attractive charge, conventional hydrogen bond, carbon-hydrogen bond and pi-cation. Fig.1B shows that the residues, ARG277,GLU52, ARG307, and SER248 formed hydrogen bonds with ligand EANK; in addition, ASP218, ARG151, and GLU70 formed electrostatic interactions with EANK. The ligand EEAK interacted with the surrounding residues, ARG277, GLU278, ARG307,SER148, and THR149 via hydrogen bond; LYS328, ASP147, and ARG151 through electrostatic interactions (Fig. 2B). The ligand EMQK via the hydrogen bond interactions with ARG151, GLU52,HIS398, ARG307, THR149, LYS328, and TYR220; electrostatic interactions with ASP147, GLU301, and GLU70 (Fig. 3B).Among hydrogen bonding, hydrophobic interactions, electrostatic interactions and van der Waals interactions, hydrogen bonding and electrostatic interactions are often basic factors for the interaction between ligands and receptors [7,24]. Thus, in this work hydrogen bonding and electrostatic interactions were important interaction forces. In addition, Amino acid residues Arg277, Arg151, Asp147,and Gln52 were always the docking locations between three umami peptides and T1R1. Moreover, Arg151 appeared with the highest frequency, followed by Asp147 and Gln52. In addition, the docking results of umami peptides DDD, EGS, LYSE, EAGIQ, EEDGK with T1R1 were consistent with our results. This suggested that the amino acid residues Arg277, Arg151, Asp147, and Gln52 may play critical roles in the production of umami taste.

Fig. 1 Docking for the interaction of EANK peptide with T1R1/T1R3.(A) The 3D hydrogen bonds surface plot at the binding site. (B) A twodimensional plan of the interaction of EANK peptide and T1R1/T1R3.
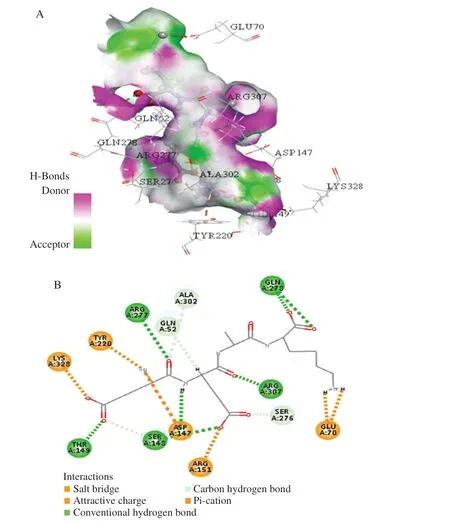
Fig. 2 Docking for the interaction of EEAK peptide with T1R1/T1R3.(A) The 3D hydrogen bonds surface plot at the binding site. (B) A twodimensional plan of the interaction of EEAK peptide and T1R1/T1R3.
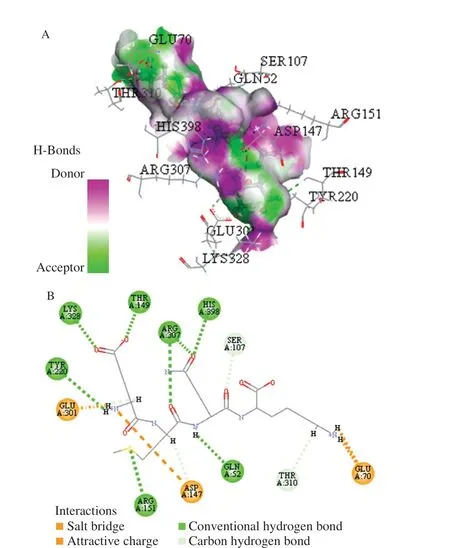
Fig. 3 Docking for the interaction of EMQK peptide with T1R1/T1R3.(A) The 3D hydrogen bonds surface plot at the binding site. (B) A twodimensional plan of the interaction of EMQK peptide and T1R1/T1R3.
4. Conclusions
Three novel umami peptides, EANK, EEAK, and EMQK were identified from the nebulin fromO. mykiss. Through the prediction of taste characteristics and the electronic tongue evaluation of the synthetic peptides verified that those peptides were umami peptides.The molecular docking results indicated that the peptides EANK,EEAK, and EMQK could enter the binding pocket in the T1R1 cavity, wherein Arg151, Asp147, Gln52, and Arg277 may play critical roles in the production of umami taste. The hydrogen bonding and electrostatic interactions were crucial interaction forces for the interaction between ligands and receptors. This work is helpful to understand the delicious taste mechanism of theO. mykissand the discovery of new umami peptides from fish resources.
Conflicts of interest
The authors have declared no conflict of interest.
Acknowledgements
This paper was supported by The National Key R&D Program of China (2019YFD0901702).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://doi.org/10.1016/j.fshw.2022.07.026.
- 食品科学与人类健康(英文)的其它文章
- Wine, beer and Chinese Baijiu in relation to cardiovascular health:the impact of moderate drinking
- Comparative analysis of physicochemical properties, ginsenosides content and α-amylase inhibitory effects in white ginseng and red ginsen
- Monitoring and identif ication of spoilage-related microorganisms in braised chicken with modif ied atmosphere packaging during refrigerated storage
- Effect of cooking processes on tilapia aroma and potential umami perception
- Formation mechanisms of ethyl acetate and organic acids in Kluyveromyces marxianus L1-1 in Chinese acid rice soup
- Volatile prof ile and multivariant analysis of Sanhuang chicken breast in combination with Chinese 5-spice blend and garam masala

