DEP1、Gn1a和qSW5组合应用调控水稻穗部性状
温一博,陈淑婷,徐正进,孙健,徐铨
、和组合应用调控水稻穗部性状

1沈阳农业大学林学院,沈阳 110866;2沈阳农业大学水稻研究所,沈阳 110866
【】水稻是重要的粮食作物,为全球超过一半的人口提供主食。穗部性状是影响水稻产量的主要因素,挖掘调控穗部性状的优异基因组合,为提高水稻产量提供聚合育种策略。【】以弯穗型籼稻品种R99和直立穗型粳稻品种SN265构建的151个重组自交系为试材,应用Illumina测序平台对重组自交系和双亲进行全基因组重测序。结合表型数据与遗传图谱,对每穗粒数、一次枝梗着粒数、二次枝梗着粒数和粒型进行QTL分析,筛选QTL区间内的候选基因,应用基于三代测序组装的SN265和R99高质量基因组进行候选基因预测和序列比对,在重组自交系中筛选产量性状表现最好的基因组合,并在SN265遗传背景下应用CRISPR基因编辑技术对目标位点进行基因编辑。【】R99每穗粒数和二次枝梗着粒数显著多于SN265,SN265的一次枝梗着粒数显著高于R99,R99粒型细长,SN265粒型短圆。每个重组自交系平均测序深度为6.25×,R99和SN265的测序深度分别为30×和32×。获得1 456 445个高质量的SNP,利用划bin策略进行图谱构建,得到一个包含3 569个bins,平均长度为58.17 kb的遗传图。QTL分析在第9染色体检测到一个同时调控每穗粒数、一次枝梗着粒数和二次枝梗着粒数的QTL,在第1染色体鉴定到一个调控每穗粒数和二次枝梗着粒数的QTL,在第5染色体鉴定到一个调控粒型的QTL。候选基因预测和序列比对发现第9染色体的同时调控水稻一次和二次枝梗着粒数,第1染色体的主要调控水稻二次枝梗着粒数,第5染色体的主要调控粒型。在151个重组自交系中,对、和的不同组合进行分类并调查产量构成因素,发现Gn1a/DEP1/qSW5等位基因组合产量表现最好,Gn1a/DEP1/qSW5产量表现最差。对SN265的位点进行分子设计育种,获得2个独立的CRISPR基因编辑株系,通过调查其产量构成因素,发现基因编辑植株穗长显著变长,每穗粒数显著增加,进而显著增加单株产量。【】揭示了、和对每穗粒数和粒型的影响,明确了Gn1a/DEP1/qSW5为重组自交系中最佳基因组合,通过改良SN265的位点进一步提高了其单株产量。
水稻;高密度遗传图谱;每穗粒数;粒型;基因编辑
0 引言
【研究意义】水稻(L.)是重要的粮食作物,约为世界一半人口提供主粮,在全球粮食安全中也发挥重要作用[1]。水稻产量是一个复杂性状,主要由穗部性状控制。在过去的40年中,中国水稻的增产主要通过提高每穗粒数来实现[1],因此,剖析调控穗部性状的遗传机制对育种家提高水稻产量具有重要的意义。【前人研究进展】水稻的圆锥状花序由穗轴、枝梗、籽粒组成,且籽粒着生在枝梗上,枝梗着生于穗轴上。水稻穗的发育过程是复杂的,包括小穗分化、发育和退化等一系列生理过程。在生殖生长期,水稻茎尖分生组织转变为花序分生组织,分化为一次枝梗分生组织,二次枝梗分生组织在一次枝梗上相继产生,进一步分化为小穗分枝分生组织和侧穗分生组织。在同一时期,一次枝梗分生组织顶部分化为末端小穗分生组织[2-3]。这些分枝及其分化的穗状分生组织最终形成水稻穗的基本结构,决定每穗粒数。许多参与穗分枝形成的基因已被克隆,如、、/(/)、()、()、()、等参与顶端分生组织分化形成花序分生组织,进而形成一次枝梗和二次枝梗分生组织的分化过程[4-7];/(/)、()、()等参与枝梗分生组织到小穗分生组织的分化过程[8-11];/与()调控小穗分生组织到颖花分生组织的分化[12-16]。水稻粒形是穗部性状的重要组成部分,属于受多基因控制的数量性状,单个基因的效应值通常较小,受环境影响较大[17]。目前,已报道的粒形相关QTL位点约600个,随着水稻基因组测序和功能基因组研究的深入,已成功克隆近百个粒形相关基因[18-21]。这些基因通过多种途径调控水稻粒形,主要包括转录因子调控(如、、和[16, 22-31])、泛素途径(如、和[27-28, 32-33])、G蛋白途径(如、和[34-36]),以及激素水平控制粒形途径(如、、、和)等。【本研究切入点】数十年来,中国科学家在水稻穗部性状遗传基础、调控基因的定位与功能研究等方面取得了卓越成绩,但由于材料背景的限制,单一的杂交组合仅能鉴定到少数穗部性状相关QTL,单个QTL贡献率因试验材料和研究方法的不同往往表现出巨大差异。目前对穗部性状的研究还主要集中于单个功能基因的解析和应用,对每穗粒数和粒形调控基因的组合应用研究较少。【拟解决的关键问题】本研究以穗型和粒型差异显著的双亲所构建的重组自交系为试材,对每穗粒数、一次枝梗着粒数、二次枝梗着粒数和粒型进行QTL分析和候选基因功能鉴定,评价不同基因组合的产量构成因素,并对超级稻品种SN265进行分子设计基因编辑育种,为水稻优势等位基因聚合育种提供重要种质和基因资源。
1 材料与方法
1.1 供试水稻材料
R99为典型的弯穗型籼稻品种,SN265为中国第一个直立大穗型粳型超级稻品种。以SN265和R99为双亲配制杂交组合,采用单粒传法套袋自交12代,获得包含151个株系的稳定遗传重组自交系。亲本和重组自交系于2021年春季种植于沈阳农业大学水稻所试验田(123°E,41°N),每个株系按照3行×6株规模种植一小区,株行距均为20 cm,单苗插植,常规栽培管理。
1.2 表型考察
抽穗后45 d,每小区取中部5株收获,充分晒干后,考察单株穗数。取长势均匀的5穗,统计一次枝梗着粒数、二次枝梗着粒数、每穗粒数和结实率。随机取500粒饱满籽粒称重统计千粒重。最后以Microsoft Excel 2016计算各材料每株的各性状平均值、标准差,并对各性状进行两尾等方差检验,使用GraphPad Prism 8进行作图。
1.3 全基因组测序和遗传图谱构建
选取插秧后3周龄植株的幼嫩叶片,采用CTAB法提取DNA,送北京百迈克生物科技有限公司进行高通量测序分析。参照Li等[37]报道的重测序手段,采用“滑动窗口”法构建R99和SN265的重组自交系群体的遗传图谱。利用划bin策略得到3 569个bins,平均长度为58.17 kb的遗传图谱。采用R/qtl的CIM方法进行QTL定位,采用mqmpermutation命令进行排列组合1 000次的LOD阈值(=0.05)确定,当实际求得的LOD值大于LOD阈值时,就认为该区段存在1个QTL,其置信区间为LOD峰值向下1个LOD值单位的区间[38]。
1.4 CRISPR/Cas9基因编辑
以超级稻品种SN265为遗传背景材料进行CRISPR基因编辑。通过华南农业大学亚热带农业生物资源保护与利用国家重点实验室刘耀光院士团队开发的基因编辑工具包CRISPR-GE(http://skl.scau.edu. cn/)进行靶位点的设计,应用BLAST比对日本晴参考基因组确认靶点的特异性(https://rapdb.dna. affrc.go.jp/tools/blast)。基因编辑靶点序列和引物合成,以及测序服务均由华大基因完成。参照Li等[37]方法构建基因编辑载体以及基因编辑植株的遗传转化和筛选。
2 结果
2.1 双亲穗部性状差异显著
R99穗型松散,SN265穗型紧凑。R99每穗粒数显著多于SN265。进一步调查一次枝梗和二次枝梗,发现SN265的一次枝梗着粒数显著多于R99,而R99的二次枝梗着粒数显著多于SN265。此外,R99和SN265的粒型也存在显著差异,R99籽粒细长,籽粒长宽比超过2.5,而SN265的籽粒较为短圆,籽粒长宽比约为2(图1)。
2.2 遗传图谱构建与QTL分析
应用Illumina测序平台对R99和SN265为亲本构建的重组自交系和双亲进行全基因组重测序,每个重组自交系平均测序深度为6.25×,R99和SN265的测序深度为30×和32×。得到1 456 445个高质量的SNP,利用划bin策略进行图谱构建,得到3 569个bins,平均长度为58.17 kb的遗传图谱[37]。利用R/qtl软件对重组自交系群体的每穗粒数、一次枝梗着粒数、二次枝梗着粒数和粒型进行QTL分析。获得2个控制每穗粒数的QTL,分布在第1和第9染色体,其LOD值分别为7.8和12.6,表型贡献率分别为16.8%和28.1%。随后,把每穗粒数拆分成一次枝梗着粒数和二次枝梗着粒数分别进行QTL分析。结果显示,第9染色体的QTL同时控制一次枝梗着粒数和二次枝梗着粒数,而第1染色体的QTL只调控二次枝梗着粒数。在第5染色体检测到一个LOD值为18.5,贡献率为42.1%的主效粒型QTL(图2)。
2.3 候选基因序列比对
通过数据库比对发现第1染色体上调控每穗粒数和二次枝梗着粒数的QTL与已报道的编码细胞分裂素降解酶位置重合[6],第9染色体控制每穗粒数、一次枝梗着粒数和二次枝梗着粒数的QTL与已经报道的G蛋白伽马亚基位置重合[7],第5染色体控制粒型的QTL与已报道的油菜素内酯信号传导的新型正调因子位置重合[33, 39-40]。应用基于三代测序组装的SN265和R99高质量基因组[37, 41],比对双亲、和的基因序列发现,与R99相比,SN265在位点上游5 kb存在一个1 212 bp的缺失,该1 212 bp缺失通过调控的表达量进而调控籽粒大小[33]。SN265在的3′端有一段637 bp的序列被12 bp序列所替换,使蛋白缺失了C端的Cys富集区域,该突变能促进细胞分裂,降低穗颈节长度并使稻穗变密、枝梗数增加、每穗籽粒数增多,从而促进水稻增产。SN265在和位点均为优势等位基因,而R99在位点的第1个外显子处6 bp的插入和2个SNP(C/G和G/A),以及第4个外显子处的1个SNP(G/T),导致其蛋白产物与粳稻品种产生差异,引起花序分裂组织中细胞分裂素的积累,因而增加每穗粒数,最终导致产量提高(图3)。

A:株型;B:穗型;C:粒型;D:每穗粒数;E:一次枝梗着粒数;F:二次枝梗着粒数;G:籽粒长宽比。*:P<0.05
2.4 不同DEP1、Gn1a和qSW5组合的产量表现
为了阐明、和的不同基因组合对水稻产量表现的影响,根据3个基因的等位基因型,将151个重组自交系分为8个类型(图4)。通过比对其每穗粒数、千粒重和单株产量,发现Gn1a/DEP1等位基因的组合每穗粒数表现最佳,Gn1a/DEP1和Gn1a/DEP1次之,Gn1a/DEP1组合的每穗粒数最少。千粒重主要受基因型调控,含有qSW5的4种类型千粒重普遍显著高于含有qSW5等位基因4种类型。总之,Gn1a/DEP1/qSW5因每穗粒数和千粒重的优势体现出最好的产量表现,Gn1a/DEP1/qSW5则因为每穗粒数和千粒重的劣势产量表现最差(图4)。
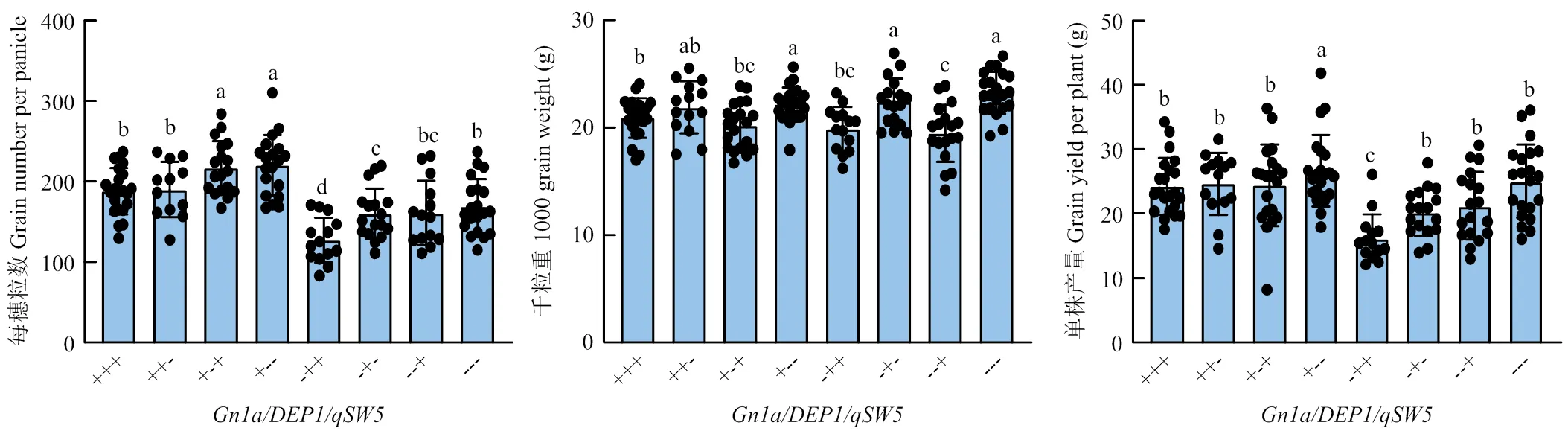
+:R99基因型;-:SN265基因型。不同字母表示差异显著(P<0.05)。下同
2.5 CRISPR/Cas9基因编辑聚合优势基因型
基于2.4结果,Gn1a/DEP1/qSW5有最好的产量表现,而超级稻品种SN265的基因型仅与Gn1a/DEP1/qSW5在位点上存在差别,在粳稻中花11遗传背景下对进行Knock-out突变,可以显著增加每穗粒数[42-43]。因此,对SN265进行基因设计育种,在的第一个外显子设计PAM序列,进行基因编辑,在T0筛选阳性植株,在T1进行测序,鉴定到CR-1和CR-2 2个纯合突变株系,分别缺失了1和2 bp,造成移码突变(图5)。2个基因编辑突变体穗长较SN265穗长显著增长,每穗粒数显著增加(图5)。基因编辑植株与SN265的结实率、穗数和千粒重差异不显著,因为每穗粒数的增加,基因编辑植株的单株产量显著高于SN265(图5)。综上,Gn1a/DEP1/qSW5基因编辑植株较SN265体现出更好的单株产量表现。
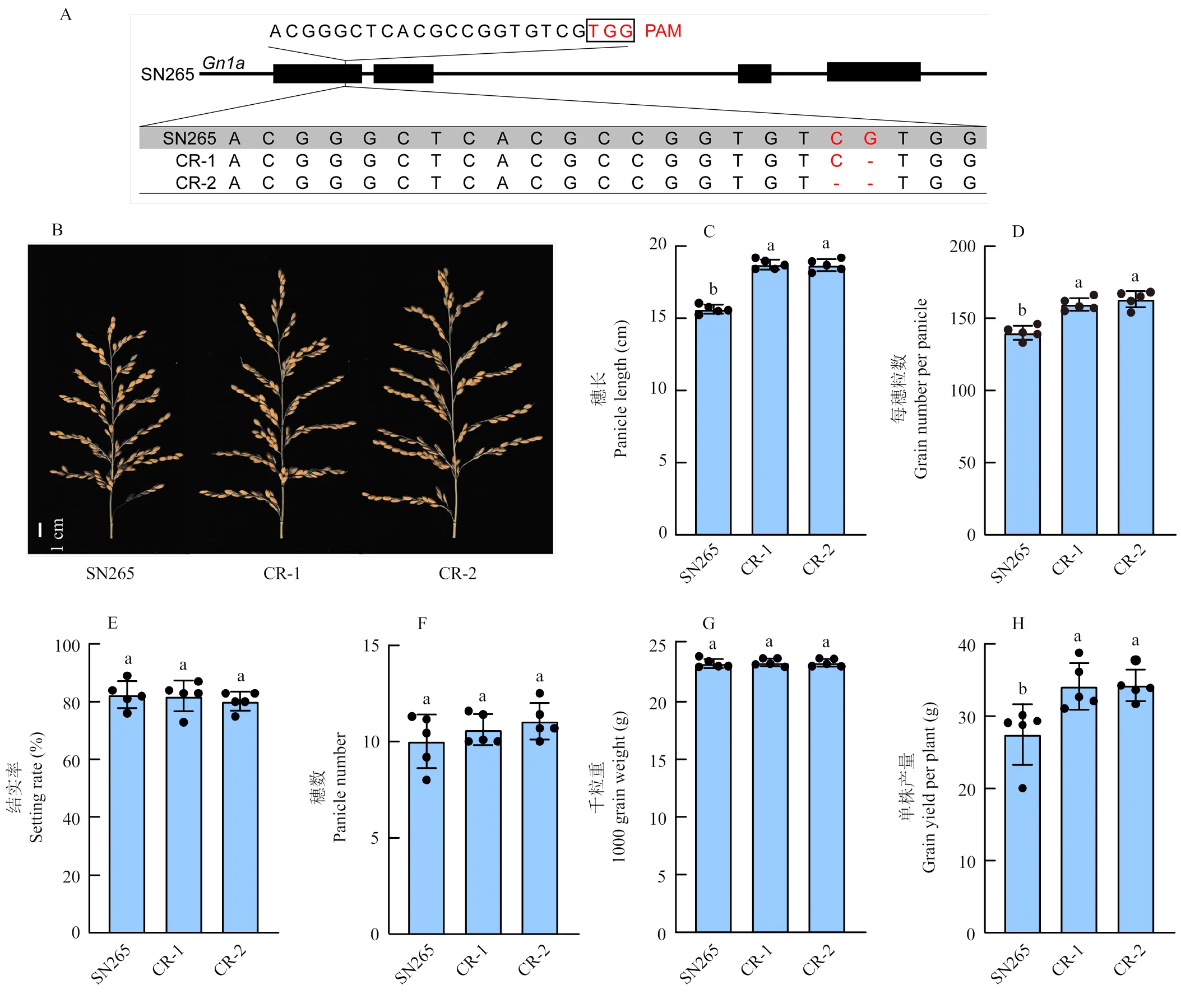
A:CRISPR基因编辑的PAM序列和突变体序列;B:SN265和基因编辑植株的穗型;C:SN265和基因编辑植株的穗长;D:SN265和基因编辑植株的每穗粒数;E:SN265和基因编辑植株的结实率;F:SN265和基因编辑植株的穗数;G:SN265和基因编辑植株的千粒重;H:SN265和基因编辑植株的单株产量
3 讨论
3.1 优异等位基因聚合
水稻育种实践表明增加每穗粒数是提高水稻产量的最有效途径之一。近几十年来,水稻每穗粒数的研究取得了很大进展,已成功克隆了多个影响每穗粒数的基因,这些影响每穗粒数的基因可以作为育种家的潜在种质资源。分子标记辅助选择是基因聚合的有效工具[44],如的3型等位基因和OsSPL14等位基因通过重复回交聚合,显著增加每穗粒数[45]。和的聚合可以通过增加每穗粒数来提高水稻产量[46]。因为一些控制每穗粒数的基因在水稻中产生其他不良影响,例如延迟水稻抽穗[47],使得水稻籽粒变小[9],减少分蘖数[31, 48],直立穗型等位基因有降低千粒重的趋势[49-50]。如何平衡这些性状与每穗粒数之间的关系仍然是育种中亟待解决的问题。本研究发现同时调控水稻一次枝梗着粒数和二次枝梗着粒数,主要调控二次枝梗着粒数,qSW5等位基因通过增加粒宽提高千粒重,可减轻所引起的千粒重降低,、和优势等位基因的组合应用可以获得最优的产量表现。
3.2 水稻分子设计育种
分子设计育种技术体系不断实践与完善对促进作物育种技术发展有重要作用,近年来越来越多的水稻产量构成因素调控基因被克隆,其分子调控网络也被深入解析,加上CRISPR基因编辑技术的广泛应用,分子设计育种开始应用到水稻生产实践当中[51-52],以为核心的理想株型设计育种[53],以为核心的品质改良设计育种都已初见成效[54]。本研究针对中国第一个直立大穗型超级稻品种,根据试验结果尝试对其位点进行基因编辑以期进一步增加SN265的每穗粒数。本研究证实了SN265的编辑植株每穗粒数显著增加,显著提高了其单株产量。但是水稻作为群体作物,单株产量的提高并不能保证其单位面积产量增产。超级稻SN265的直立穗型有助于改善群体受光结构,适应高密度密植[55],其编辑植株穗长增加,是否会削弱其对高密度栽培的适应能力还需进一步研究,与其配套的栽培技术也将是今后研究的方向之一。在水稻种质资源中发现有利基因或等位基因并在不同遗传背景下对其功能进行评估,以及通过分子标记辅助选择和基因编辑技术阐明这些有利等位基因之间的遗传互作及其对产量的聚合效应是今后水稻分子设计育种的两项主要任务。
4 结论
阐明了、和对一次枝梗着粒数、二次枝梗着粒数和粒型的影响,发现了Gn1a/DEP1/qSW5为重组自交系中最佳基因组合,通过改良SN265的位点增加每穗粒数,从而进一步提高了其单株产量。
[1] LI G L, ZHANG H L, LI J J, ZHANG Z Y, LI Z C. Genetic control of panicle architecture in rice. The Crop Journal, 2021, 9(3): 590-597.
[2] WANG Y H, LI J Y. Molecular basis of plant architecture. Annual Review ofBiology, 2008, 59: 253-279.
[3] WANG B, SMITH S M, LI J Y. Genetic regulation of shoot architecture. Annual Review ofBiology, 2018, 69: 437-468.
[4] OIKAWA T, KYOZUKA J. Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. The Plant Cell, 2009, 21(4): 1095-1108.
[5] TABUCHI H, ZHANG Y, HATTORI S, OMAE M, SHIMIZU-SATO S, OIKAWA T, QIAN Q, NISHIMURA M, KITANO H, XIE H, FANG X H, YOSHIDA H, KYOZUKA J, CHEN F, SATO Y. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. The Plant Cell, 2011, 23(9): 3276-3287.
[6] ASHIKARI M, SAKAKIBARA H, LIN S Y, YAMAMOTO T, TAKASHI T, NISHIMURA A, ANGELES E R, QIAN Q, KITANO H, MATSUOKA M. Cytokinin oxidase regulates rice grain production. Science, 2005, 309(5735): 741-745.
[7] HUANG X Z, QIAN Q, LIU Z B, SUN H Y, HE S Y, LUO D, XIA G M, CHU C C, LI J Y, FU X D. Natural variation at the DEP1 locus enhances grain yield in rice. Nature Genetics, 2009, 41(4): 494-497.
[8] OOKAWA T, HOBO T, YANO M, MURATA K, ANDO T, MIURA H, ASANO K, OCHIAI Y, IKEDA M, NISHITANI R, EBITANI T, OZAKI H, ANGELES E R, HIRASAWA T, MATSUOKA M. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nature communications, 2010, 1: 132.
[9] KOMATSU M, CHUJO A, NAGATO Y, SHIMAMOTO K, KYOZUKA J. FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development, 2003, 130(16): 3841-3850.
[10] ZHU Q H, HOQUE M S, DENNIS E S, UPADHYAYA N M. Ds tagging of BRANCHED FLORETLESS 1 (BFL1) that mediates the transition from spikelet to floret meristem in rice (L). BMC Plant Biology, 2003, 3: 6.
[11] YOSHIDA A, SASAO M, YASUNO N, TAKAGI K, DAIMON Y, CHEN R, YAMAZAKI R H, TOKUNAGA H, KITAGUCHI Y, SATO Y, NAGAMURA Y, USHIJIMA T, KUMAMARU T, IIDA S, MAEKAWA M, KYOZUKA J. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(2): 767-772.
[12] LEE D Y, LEE J, MOON S, PARK S Y, AN G. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. The Plant Journal, 2007, 49(1): 64-78.
[13] LEE D Y, AN G. Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. The Plant Journal, 2012, 69(3): 445-461.
[14] JEON J S, JANG S, LEE S, NAM J, KIM C, LEE S H, CHUNG Y Y, KIM S R, LEE Y H, CHO Y G, AN G. leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. The Plant Cell, 2000, 12(6): 871-884.
[15] AGRAWAL G K, ABE K, YAMAZAKI M, MIYAO A, HIROCHIKA H. Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Molecular Biology, 2005, 59(1): 125-135.
[16] LIU Q, HAN R X, WU K, ZHANG J Q, YE Y F, WANG S S, CHEN J F, PAN Y J, LI Q, XU X P, ZHOU J W, TAO D Y, WU Y J, FU X D. G-protein βγ subunits determine grain size through interaction with MADS- domain transcription factors in rice. Nature Communications, 2018, 9(1): 852.
[17] TAN Y F, XING Y Z, LI J X, YU S B, XU C G, ZHANG Q F. Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theoretical & Applied Genetics, 2000, 101(5/6): 823-829.
[18] HUANG R Y, JIANG L R, ZHENG J S, WANG T S, WANG H C, HUANG Y M, HONG Z L. Genetic bases of rice grain shape: so many genes, so little known. Trends in Plant Science, 2013, 18(4): 218-226.
[19] ZUO J R, LI J Y. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annual Review of Genetics, 2014, 48: 99-118.
[20] 丁膺宾, 张莉珍, 许睿, 王艳艳, 郑晓明, 张丽芳, 程云连, 吴凡, 杨庆文, 乔卫华, 兰进好. 基于染色体片段置换系的野生稻粒长QTL-的精细定位. 中国农业科学, 2018, 51(18): 3435-3444.
DING Y B, ZHANG L Z, XU R, WANG Y Y, ZHENG X, ZHANG L F, CHENG Y L, WU F, YANG Q W, QIAO W H, LAN J H. Fine mapping of grain length associated QTL, qGL12 in wild rice (L.) using a chromosome segment substitution line. Scientia Agricultura Sinica, 2018, 51(18): 3435-3444. (in Chinese)
[21] 张亚东, 梁文化, 赫磊, 赵春芳, 朱镇, 陈涛, 赵庆勇, 赵凌, 姚姝, 周丽慧, 路凯, 王才林. 水稻RIL群体高密度遗传图谱构建及粒型QTL定位. 中国农业科学, 2021, 54(24): 5163-5176.
ZHANG Y D, LIANG W H, HE L, ZHAO C F, ZHU Z, CHEN T, ZHAO Q Y, ZHAO L, YAO S, ZHOU L H, LU K, WANG C L. Construction of high-density genetic map and QTL analysis of grain shape in rice RIL population. Scientia Agricultura Sinica, 2021, 54(24): 5163-5176. (in Chinese)
[22] WANG S K, WU K, YUAN Q B, LIU X Y, LIU Z B, LIN X Y, ZENG R Z, ZHU H T, DONG G J, QIAN Q, ZHANG G Q, FU X D. Control of grain size, shape and quality by OsSPL16 in rice. Nature Genetics, 2012, 44(8): 950-954.
[23] WANG S K, LI S, LIU Q, WU K, ZHANG J Q, WANG S S, WANG Y, CHEN X B, ZHANG Y, GAO C X, WANG F, HUANG H X, FU X D. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nature Genetics, 2015, 47(8): 949-954.
[24] WANG C S, TANG S C, ZHAN Q L, HOU Q Q, ZHAO Y, ZHAO Q, FENG Q, ZHOU C C, LYU D F, CUI L L, LI Y, MIAO J S, ZHU C R, LU Y Q, WANG Y C, WANG Z Q, ZHU J J, SHANGGUAN Y Y, GONG J Y, YANG S H, WANG W Q, ZHANG J F, XIE H A, HUANG X H, HAN B. Dissecting a heterotic gene through GradedPool-Seq mapping informs a rice-improvement strategy. Nature Communications, 2019, 10(1): 2982.
[25] XIONG H Y, YU J P, MIAO J L, LI J J, ZHANG H L, WANG X, LIU P L, ZHAO Y, JIANG C H, YIN Z G, LI Y, GUO Y, FU B Y, WANG W S, LI Z K, ALI J, LI Z C. Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiology, 2018, 178(1): 451-467.
[26] YU J P, XIONG H Y, ZHU X Y, ZHANG H L, LI H H, MIAO J L, WANG W S, TANG Z S, ZHANG Z Y, YAO G X, ZHANG Q, PAN Y H, WANG X, RASHID M A R, LI J J, GAO Y M, LI Z K, YANG W C, FU X D, LI Z C. OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biology, 2017, 15(1): 28.
[27] SONG X J, HUANG W, SHI M, ZHU M Z, LIN H X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nature Genetics, 2007, 39(5): 623-630.
[28] SHI C L, REN Y L, LIU L L, WANG F, ZHANG H, TIAN P, PAN T, WANG Y F, JING R N, LIU T Z, WU F Q, LIN Q B, LEI C L, ZHANG X, ZHU S S, GUO X P, WANG J L, ZHAO Z C, WANG J, ZHAI H Q, CHENG Z J, WAN J M. Ubiquitin specific protease 15 has an important role in regulating grain width and size in rice. Plant Physiology, 2019, 180(1): 381-391.
[29] WANG S K, WU K, YUAN Q B, LIU X Y, LIU Z B, LIN X Y, ZENG R Z, ZHU H T, DONG G J, QIAN Q, ZHANG G Q, FU X D. Control of grain size, shape and quality by OsSPL16 in rice. Nature Genetics, 2012, 44(8): 950-954.
[30] MIURA K, IKEDA M, MATSUBARA A, SONG X J, ITO M, ASANO K, MATSUOKA M, KITANO H, ASHIKARI M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nature Genetics, 2010, 42(6): 545-549.
[31] JIAO Y Q, WANG Y H, XUE D W, WANG J, YAN M X, LIU G F, DONG G J, ZENG D L, LU Z F, ZHU X D, QIAN Q, LI J Y. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics, 2010, 42(6): 541-544.
[32] WENG J F, GU S H, WAN X Y, GAO H, GUO T, SU N, LEI C L, ZHANG X, CHENG Z J, GUO X P, WANG J L, JIANG L, ZHAI H Q, WAN J M. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell research, 2008, 18(12): 1199-1209.
[33] LIU J F, CHEN J, ZHENG X M, WU F Q, LIN Q B, HENG Y Q, TIAN P, CHENG Z J, YU X W, ZHOU K N, ZHANG X, GUO X P, WANG J L, WANG H Y, WAN J M. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nature Plants, 2017, 3: 17043.
[34] TAKANO-KAI N, JIANG H, KUBO T, SWEENEY M, MATSUMOTO T, KANAMORI H, PADHUKASAHASRAM B, BUSTAMANTE C, YOSHIMURA A, DOI K, MCCOUCH S. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics, 2009, 182(4): 1323-1334.
[35] MIAO J, YANG Z F, ZHANG D P, WANG Y Z, XU M B, ZHOU L H, WANG J, WU S J, YAO Y L, DU X, GU F F, GONG Z Y, GU M H, LIANG G H, ZHOU Y. Mutation of RGG2, which encodes a type B heterotrimeric G protein gamma subunit, increases grain size and yield production in rice. Plant Biotechnology Journal, 2019, 17(3): 650-664.
[36] SUN S Y, WANG L, MAO H L, SHAO L, LI X H, XIAO J H, OUYANG Y D, ZHANG Q F. A G-protein pathway determines grain size in rice. Nature Communications, 2018, 9(1): 851.
[37] LI X K, WU L, WANG J H, SUN J, XIA X H, GENG X, WANG X H, XU Z J, XU Q. Genome sequencing of rice subspecies and genetic analysis of recombinant lines reveals regional yield- and quality-associated loci. BMC Biology, 2018, 16(1): 102.
[38] JIANG S K, YANG C, XU Q, WANG L Z, YANG X L, SONG X W, WANG J Y, ZHANG X J, LI B, LI H Y, LI Z G, LI W H. Genetic dissection of germinability under low temperature by building a resequencing linkage map inrice. International Journal of Molecular Sciences, 2020, 21(4): 1284.
[39] SHOMURA A, IZAWA T, EBANA K, EBITANI T, KANEGAE H, KONISHI S, YANO M. Deletion in a gene associated with grain size increased yields during rice domestication. Nature Genetics, 2008, 40(8): 1023-1028.
[40] DUAN P G, XU J S, ZENG D L, ZHANG B L, GENG M F, ZHANG G Z, HUANG K, HUANG L J, XU R, GE S, QIAN Q, LI Y H. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Molecular Plant, 2017, 10(5): 685-694.
[41] WANG Y, LI F C, ZHANG F, WU L, XU N, SUN Q, CHEN H, YU Z W, LU J H, JIANG K, WANG X C, WEN S Y, ZHOU Y, ZHAO H, JIANG Q, WANG J H, JIA R Z, SUN J, TANG L, XU H, HU W, XU Z J, CHEN W F, GUO A P, XU Q. Time-ordering japonica/geng genomes analysis indicates the importance of large structural variants in rice breeding. Plant Biotechnology Journal, 2023, 21(1): 202-218.
[42] LI M R, LI X X, ZHOU Z J, WU P Z, FANG M C, PAN X P, LIN Q P, LUO W B, WU G J, LI H Q. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Frontiers in plant science, 2016, 7: 377.
[43] LI M R, PAN X P, LI H Q. Pyramiding of,, andexhibits complementary and additive effects on rice yield. International Journal of Molecular Sciences, 2022, 23(20): 12478.
[44] ASHIKARI M, MATSUOKA M. Identification, isolation and pyramiding of quantitative trait loci for rice breeding. Trends in Plant Science, 2006, 11(7): 344-350.
[45] KIM S R, RAMOS J M, HIZON R J M, ASHIKARI M, VIRK P S, TORRES E A, NISSILA E, JENA K K. Introgression of a functional epigeneticWFPallele into elite indica rice genomes greatly improved panicle traits and grain yield. Scientific Reports, 2018, 8(1): 3833.
[46] WANG Y, ZHAI L Y, CHEN K, SHEN C C, LIANG Y T, WANG C C, ZHAO X Q, WANG S, XU J L. Natural sequence variations and combinations of GNP1 and NAL1 determine the grain number per panicle in rice. Rice, 2020, 13(1): 14.
[47] XUE W Y, XING Y Z, WENG X Y, ZHAO Y, TANG W J, WANG L, ZHOU H J, YU S B, XU C G, LI X H, ZHANG Q F. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics, 2008, 40(6): 761-767.
[48] MIURA K, IKEDA M, MATSUBARA A, SONG X J, ITO M, ASANO K, MATSUOKA M, KITANO H, ASHIKARI M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nature Genetics, 2010, 42(6): 545-549.
[49] WU L, WANG X D, YU Z W, CUI X, XU Q. Simultaneous improvement of grain yield and quality through manipulating two type C G protein gamma subunits in rice. International Journal of Molecular Sciences, 2022, 23(3): 1463.
[50] LI X B, TAO Q D, MIAO J, YANG Z F, GU M H, LIANG G H, ZHOU Y. Evaluation of differential qPE9-1/DEP1 protein domains in rice grain length and weight variation. Rice, 2019, 12(1): 5.
[51] ZENG D L, TIAN Z X, RAO Y C, DONG G J, YANG Y L, HUANG L C, LENG Y J, XU J, SUN C, ZHANG G H, HU J, ZHU L, GAO Z Y, HU X M, GUO L B, XIONG G S, WANG Y H, LI J Y, QIAN Q. Rational design of high-yield and superior-quality rice. Nature Plants, 2017, 3: 17031.
[52] QIAN Q, GUO L B, SMITH S M, LI J Y. Breeding high-yield superior- quality hybrid super-rice by rational design. National Science Review, 2016, 3(3): 283-294.
[53] SONG X G, MENG X B, GUO H Y, CHENG Q, JING Y H, CHEN M J, LIU G F, WANG B, WANG Y H, LI J Y, YU H. Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nature Biotechnology, 2022, 40: 1403-1411.
[54] HUANG L C, LI Q F, ZHANG C Q, CHU R, GU Z W, TAN H Y, ZHAO D S, FAN X L, LIU Q Q. Creating novel Wx alleles with fine-tuned amylose levels and improved grain quality in rice by promoter editing using CRISPR/Cas9 system. Plant Biotechnology Journal, 2020, 18(11): 2164-2166.
[55] FEI C, YU J H, XU Z J, XU Q. Erect panicle architecture contributes to increased rice production through the improvement of canopy structure. Molecular Breeding, 2019, 39: 128.
Combination of,, andregulates the panicle architecture in rice

1College of forestry, Shenyang Agricultural University, Shenyang 110866;2Rice research institute of Shenyang Agricultural University, Shenyang 110866
【】Rice is an important food crop, providing staple food for more than half of the world’s population. Panicle traits are the main factors affecting rice yield. Discover the elite haplotype of the panicle regulation gene, and provide important germplasm and gene resources for pyramiding breeding. 【】In this study, recombinant inbred lines (RILs) derived from a cross between SN265 and R99 were re-sequenced through high-throughput sequencing. QTL analysis and candidate gene identification were conducted on the grain number on the primary branch, the grain number on the secondary branch, and the grain shape. The sequences of candidate genes were compared using the long-read sequence assemblies of SN265 and R99. The combination of candidate genes that can maximize grain yield was selected among RILs. Finally, the super rice variety SN265 was improved using CRISPR/Cas9 gene editing technology. 【】The R99 had significantly more grain number per panicle and grain number on the secondary branch, whereas SN265 had significantly more grain number on the primary branch. The grain of R99 is slender, and the grain of SN265 is short and round. The RILs were sequenced with approximately 6.25-fold depth. For parent lines, 30.0-fold depth and 32.0-fold depth data were generated for R99 and SN265, respectively. Subsequently, a bin map was constructed by 1456445 high-quality SNPs. The genetic map containing 3 569 recombinant blocks, with an average length of 58.17 kb. The QTL analysis detected a QTL on Chr.9 for grain number per panicle and grain number on both primary and secondary branch, a QTL on Chr.1 for grain number per panicle and grain number on the secondary branch, a QTL on Chr.5 for grain shape. The candidate gene prediction and sequence comparison showed thatregulated the grain number on both primary and secondary branches of rice,mainly regulated the grain number on secondary branches of rice, andmainly regulated the grain shape. The yield of the combination ofGn1a/DEP1/qSW5alleles showed an advantage in yield performance among the RILs. We further conducted a molecular design breeding to SN265 by knocking out thelocus using CRISPR/Ca9 gene editing technology, and the grain number per panicle of the transgenic plants increased significantly compared to that of SN265. 【】This study used RILs derived from a XI/GJ cross and high-throughput sequencing technology to conduct QTL analysis of rice panicle traits, revealed the effects of,andon grain number per panicle and grain shape, and clarified thatGn1a/DEP1/qSW5was the best gene combination in RILs. The yield per plant was further improved by knocking out thelocus of SN265. This study provided important germplasm and gene resources for pyramiding breeding with elite alleles.
rice; high density genetic map; grain number; grain shape; gene editing

2022-10-21;
2022-11-14
国家自然科学基金(32071982)
温一博,E-mail:wenyibo@syau.edu.cn。通信作者徐铨,E-mail:kobexu34@syau.edu.cn。通信作者孙健,E-mail:sunjian811119@syau.edu.cn
(责任编辑 李莉)

