Functional prediction of tomato PLATZ family members and functional verification of SlPLATZ17
Min Xu ,Zhao Gao ,Dalong Li ,Chen Zhang ,Yuqi Zhang,Qian He,Yingbin Qi,He Zhang,Jingbin Jiang,Xiangyang Xu#,Tingting Zhao#
1 College of Horticulture and Landscape Architecture,Northeast Agricultural University,Harbin 150030,China
2 Key Laboratory of Biology and Genetic Improvement of Horticultural Crops (Northeast Region),Ministry of Agriculture and Rural Affairs,Harbin 150030,China
Abstract PLATZ is a novel zinc finger DNA-binding protein that plays an important role in regulating plant growth and development and resisting abiotic stress.However,there has been very little research on the function of this family gene in tomatoes,which limits its application in germplasm resource improvement.Therefore,the PLATZ gene family was identified and analyzed in tomato,and its roles were predicted and verified to provide a basis for in-depth research on SlPLATZ gene function.In this study,the PLATZ family members of tomato were identified in the whole genome,and 19 SlPLATZ genes were obtained.Functional prediction was conducted based on gene and promoter structure analysis and RNA-seq-based expression pattern analysis.SlPLATZ genes that responded significantly under different abiotic stresses or were significantly differentially expressed among multiple tissues were screened as functional gene resources.SlPLATZ17 was selected for functional verification by experiment-based analysis.The results showed that the downregulation of SlPLATZ17 gene expression reduced the drought and salt tolerance of tomato plants.Tomato plants overexpressing SlPLATZ17 had larger flower sizes and long,thin petals,adjacent petals were not connected at the base,and the stamen circumference was smaller.This study contributes to understanding the functions of the SlPLATZ family in tomato and provides a reference for functional gene screening.
Keywords: tomato,PLATZ family,abiotic stresses,plant development
1.Introduction
Transcription factors (TFs) are a class of specific proteins that bind specific DNA sequences,which generally contain one or more specific DNA-binding domains.Target gene expression is regulated through specificcis-DNA sequences in target gene promoters (Honma and Goto 2001;Zhangetal.2007).Transcription factors interact with DNA promoters through their DNA-binding domain (Kouetal.2014).Typical DNA-binding domains in plant transcription factors include the AP2/EREBP domain (Riechmann and Meyerowitz 1998),MYB domain(Martin and Paz-Ares 1997),WRKY domain (Bakshi and Oelmuller 2014),zinc finger domain (Takatsuji 1998) and bZIP domain (Nantel and Quatrano 1996).
Zinc finger protein (ZFP) transcription factors account for approximately 15% of all transcription factors,which is a large family of plant transcription factors and is named for its finger-like structure and ability to bind zinc(Kielbowicz-Matuk 2012).ZFPs interact with nucleic acids in various forms,mainly binding with DNA but also with other proteins and RNA (Ciftci-Yilmaz and Mittler 2008).ZFPs generally function in the nucleus (Raikhel 1992;Nomanetal.2017).In addition to playing important roles in plant signal transduction (Zangetal.2015),growth and development (Yanetal.2014),and abiotic stress (Maetal.2016),some ZFPs have been shown to be related to antimicrobial immunity (Nietal.2010).
Plant AT-rich sequence and zinc binding (PLATZ)proteins are a novel class of zinc finger proteins initially,isolated from pea (Pisumsativum),a DNA-binding proteins unique to plants that contain conserved PLATZ domains and have been shown to nonspecifically bind A/T-rich sequences and inhibit transcription (Naganoetal.2001).Analysis of homologous proteins shows thatPLATZ1contains both cysteine and histamine residues and has two separated structural domains: C-x2-H-x11-Cx2-C-x(4-5)-C-x2-C-x(3-7)-H-x2-H and C-x2-C-x(10-11)-Cx3-C (Naganoetal.2001).However,these two isolated domains are different from the motifs previously identified for binding zinc,such as RING (C3HC4),LIM (C2HC5),and GATA finger (C-x2-C-x(17-18)-C-x2-C(C2C2)).Therefore,PLATZ encodes a new class of plant-specific zinc finger transcription factors.
ThePLATZexpression pattern differs among plant tissues.The mRNA expression level ofPLATZ1in pea is the highest in root tips and terminal buds but hardly detectable in other organs (Naganoetal.2001).ThePLATZgeneGL6(also known asSG6) is preferentially expressed in the panicle of rice in early development but not in the endosperm (Wangetal.2019;Zhou and Xue 2020).
Many studies have shown that PLATZ proteins play an important role in plant growth and development and in coping with abiotic stress.In sugarcane,the expression level of high-fiber genotype sugarcane is significantly different in mature and immature tissues,suggesting thatPLATZmay be a transcriptional regulator of secondary cell wall synthesis (Kasirajanetal.2018).In maize,thePLATZ1homologous gene is strongly induced by drought stress (González-Moralesetal.2016);thus,PLATZis a key factor regulating drought in maize (Zendaetal.2019).InArabidopsisthaliana,salt stress inducesAtPLATZ2transcription,and transgenic plants that compositionally overexpressAtPLATZ2show higher sensitivity to salt stress (Liuetal.2020).However,in-depth identification and analysis of thePLATZfamily in tomato has not been conducted.
In this study,whole-genome identification and functional prediction of thePLATZgene family in tomato were carried out using bioinformatics techniques.Firstly,the functions of the gene family were predicted based on a preliminary analysis of exon intron organization,motif composition and promoterciselements,and further functional prediction was conducted by expression pattern analysis according to RNA seq data.Secondly,SlPLATZgenes with significantly differential expression patterns under abiotic stresses (cold,drought and salt) and in special tissues were screened out,and these genes may function in response to the conditions used for screening.Thirdly,importantSlPLATZgenes were obtained after the prediction of genomic and transcriptional levels,and their functions were verified by experiment-based analysis.Our results will help to elucidate the general functions of the tomatoSlPLATZfamily and provide useful gene resources for breeding.
2.Materials and methods
2.1.Whole-genome identification of the SlPLATZ family
Tomato genome and gene annotation files were downloaded from Solanaceae Genomics Network(https://solgenomics.net/) (Fernandez-Pozoetal.2015).The HMM file for the conserved domain of thePLATZtranscription factor family (PF04640) was downloaded from Pfam (http://pfam.xfam.org/).The HMM search program (http://hmmer.janelia.org/) was used to identify all possible tomato genomePLATZmembers of the family of transcription factors.Then,we further screened members of the tomatoPLATZfamily by NCBI-Blastp.Finally,NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd/) was used to confirm tomatoPLATZmembers among transcription factors.Next,Expasy (https://web.expasy.org/) (Ramsby and Makowski 2005) was used to predict isoelectric point (pI) and molecular weights (MWs) of all SlPLATZ proteins,and WoLF PSORT (https://wolfpsort.hgc.jp/) was employed to predict subcellular localization (Hortonetal.2007).
2.2.Gene structure and conserved domain analysis
The structure and conserved domains of the tomatoSlPLATZgenes were analyzed by NCBI-CDD and visualized in TBtools (Chenetal.2020).To further understandSlPLATZfunctions,protein sequences were submitted to MEME (http://meme-suite.org/tools/meme)(Baileyetal.2009) to identify the order ofSlPLATZtranscription factors in tomato.The parameters were as follows: The number of repetitions was set to zero or one,the maximum number of motifs was set to 10 (Baileyetal.2009),and other parameters were the default.The positions of conserved domains were predicted using Jalview software,and exon-intron structures were displayed in TBtools (Chenetal.2020).
2.3.Cis-regulatory element (CRE) analysis
CREs were selected from the sequence 2,000 bp upstream of the start codon of allSlPLATZgenesviathe PlantCare database (http://bioinformatics.psb.ugent.be/webtools/ plantcare/html/) (Lescotetal.2002).Analysis ofcis-acting components was performed to predict their possible functions.
2.4.Expression pattern analysis based on RNA-seq analysis
Based on RNA-seq data,the expression patterns ofSlPLATZgenes under cold,drought,and salt stresses and tissue specificity were analyzed.The original RNA-seq data were obtained from the NCBI database;the accession numbers related to the responses to cold,drought and salt stresses are GSE148887,GSE148539,GSE148353,respectively.The tissuespecific transcriptome data entry number is GSE33507,and only RNA-seq data of AC(CK) was used.The fragments per kilobase per million (FPKM)-normalized data of significantly expressed genes (FPKM>1) were log2transformed.Multiple experiments were performed to visualize the expression pattern and tissue-specific expression pattern ofSlPLATZgenes in heatmap 4.0.
2.5.Plant material and treatments
Ailsa Craig (AC) tomato material was planted as the experimental material at the experimental station of Northeast Agricultural University,China.All seedlings were grown under a diurnal temperature of 20-25°C:13-15°C,the light period is 16 h and darkness period is 8 h,light intensity 35,000 lx and relative humidity of 45%.When the tomato seedlings had grown to the five-leaf monophase,they were cultured in 20% PEG6000 (drought stress) and 0.20 mol L-1NaCl (salt stress) solutions for 12 h;whole seedlings were subjected to cold stress at 4°C for 12 h.Perform qRT-PCR test after AC plant stress treatment.Ten seedlings were selected for each treatment,and all treatments were repeated three times.Leaves ofSlPLATZ17-silenced plants were collected at 0,1.5,3,6 and 12 h after drought,salt and cold stresses for subsequent determination of physiological indices,immediately frozen in liquid nitrogen and stored at -80°C.
2.6.Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from samples using TRIzol.cDNA synthesis was performed using the M-MLVRTase Kit (TaKaRa,Dalian,China).qRT-PCR was performed using the iQ5 system.TheActin-7 gene(Solyc11g005330.1.1) was used as an internal control.For each qRT-PCR,the mixture consisted of 10 μL SYBR®Green Master Mix,0.5 μL each of forward primer and reverse primer,1 μL of diluted cDNA and 8 μL of ddH2O in a 20 μL reaction.The reaction was carried out as follows: 95°C for 5 min,followed by 40 cycles of 94°C for 5 s,60°C for 15 s and 72°C for 10 s;three biological replicates were included.The 2-ΔΔCTmethod (Livak and Schmittgen 2001) was used for data analysis,andActin-7 was used as a reference gene.Information about the primers is shown in Table 1.Three replicates were performed for each time point.
2.7.Generation of SlPLATZ17-silenced plants and abiotic stress treatments
TheSlPLATZ17fragment was converted from RNA extracted from AC leaves to cDNA using VIGS-SlPLATZ17(Table 1).The target sequence ofSlPLATZ17was thencloned into tobacco rattle virus RNA2 (TRV2)viaa cloning kit (Vazyme,Nanjing,China).The fusion plasmid TRV2-SlPLATZ17was introduced intoAgrobacteriumtumefaciensGV3101,and GV3101 was infiltrated into the expanded leaves of 4 AC lines (Velásquezetal.2009) to obtainSlPLAZT17-silenced plants.There were three experimental replicates.Then,SlPLATZ17gene expression was measured by qRT-PCR,and plants showing more than a 50% decrease in gene expression were selected for subsequent abiotic stress treatment experiments.

Table 1 All primers used in this experiment
2.8.Physiological measurements
Leaf samples were collected randomly from CK,CKTRV2 andSlPLATZ17-silenced seedlings before and after drought and salt stresses.Each sample included three replicates.The superoxide dismutase (SOD) activity was determined following the methods of Beyer and Fridovich (1987),and the activity of peroxidase (POD)was determined according to the methods of Chance and Maehly (1955).The proline (Pro) content in leaves was estimated by the methods of Claussen (2005) and the malondialdehyde (MDA) content by the methods of Cervillaetal.(2007).
Reactive oxygen species (ROS) were detected using the nitroblue tetrazolium (NBT) (Bournonville and Díaz-Ricci 2011) and 3,3´-diaminobenzidine (DAB) staining methods (Rameletal.2009).Blade conductivity was measured with a DS-II conductivity meter.
2.9.Generation of transgenic plants and developmental observations
Primers forSlPLATZ17were designed using Primer Premier 5.0 software,and then the CDS of theSlPLATZ17gene was cloned.The CDS was inserted into the pB121-EYFP vector to construct an overexpression plasmid.Transgenic plants were produced byAgrobacteriummediated transformation (Ecketal.2006).After that,kanamycin was used to screen the transgenic lines and verified by genomic detection PCR and qRT-PCR.TheSlPLATZ17primers used for PCR and qRT-PCR are shown in Table 1.
TheSlPLATZ17-overexpressing lines showed natural growth,and T2-generationSlPLATZ17-overexpressing seeds and AC (WT) were then simultaneously seeded in seed germination bags to observe their root growth and flower development in the later stage.
2.10.Statistical analysis
Statistically significant differences between CK,CK-TRV2 andSlPLATZ17-silenced plants in terms of the measured parameters were tested by Duncan’s multiple range tests.Asterisks (*and**) were used to denote significant differences (P≤0.05 and ≤0.01),respectively.All experimental data were statistically processed by SPSS v10.0 software.
3.Results
3.1.ldentification and physical property analysis of the SlPLATZ family in tomato
A total of 19 PLATZ family members (designatedSlPLATZ1toSlPLATZ19) were identified using a bioinformatics approach.Their basic information is presented in Table 2.The predicted molecular weights ranged from 17,055.98 Da (SlPLATZ3) to 29,157.37 Da (SlPLATZ13);SlPLATZ3 (146 aa) had the shortest sequence and SlPLATZ13 (255 aa) the longest.The isoelectric points varied from 6.64 (SlPLATZ11) to 9.48 (SlPLATZ10),and the aliphatic indices varied from 64.96 (SlPLATZ17) to 81.83 (SlPLATZ7).The hydrophilicity indices ranged from -0.709 (SlPLATZ10)to -0.272 (SlPLATZ1).The instability indices vary from 31.79 (SlPLATZ3) to 69.76 (SlPLATZ19),and those of SlPLATZ3/4 were lower than 40,indicating that these genes are relatively stable.The subcellular prediction results showed all proteins resided in the nucleus.
3.2.Function prediction by gene structure analysis
The results of motif analysis are shown in Fig.1.Each protein sequence contains a different number of motifs(1-10),and each motif is present only once.The number and location of motif members contained in the first set of genes are highly similar,indicating that they may have similar biological functions.Most PLATZ proteins contain motif 1,motif 2,motif 4 and motif 7.The most closely related members of the evolutionary tree share similar motifs,showing that they are highly conserved.For example,motif 9 is present only in group three members (Fig.1-B),indicating that motif 9 is specific to some PLATZ proteins.Batch-Web CD-Search Tool was used to analyze conserved domain of SlPLATZ proteins,and we found that the 19SlPLATZgenes all contain the conservedPLATZdomain;the conservedPLATZdomain of the SlPLATZ18 protein is preceded by a Bbox domain(Fig.1-C),which is reported to play vital roles in stress tolerance (Liuetal.2019).
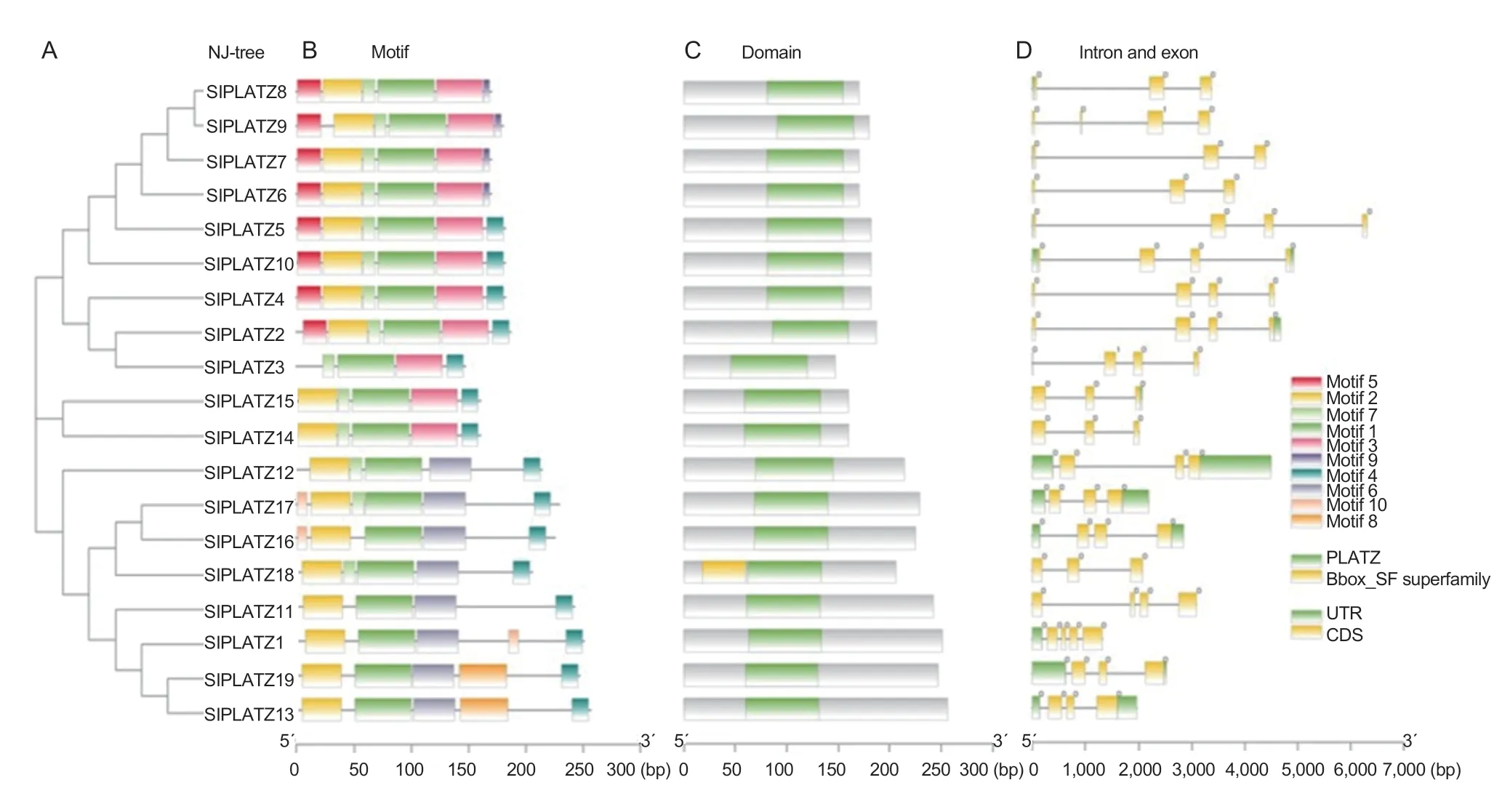
Fig.1 The NJ tree,motif,domain and intron and exon of PLATZ in tomato.A,the NJ tree was constructed by MEGA 7.0 software using PLATZ family members.B,motif positions of tomato PLATZ family members.Ten motifs are displayed in different colors.C,domain positions of tomato PLATZ family members;green represents the PLATZ conserved domain,and yellow represents the Bbox domain.D,intron and exon analysis of 19 predicted tomato PLATZ family members.Green represents the untranslated region(UTR),and yellow represents the coding sequence (CDS).The scale at the bottom shows the length of the motif,exon and intron.
Studying introns and exons can provide more insight into the evolution and function of thePLATZfamily members in tomato.The results are shown in Fig.1-D.AllSlPLATZgenes contain exons and introns,the number of which ranges from 1 to 5.The 3´ UTR length of Group five subfamily members is longer than that of the other subfamilies (Fig.1-D).Overall,structural analysis of PLATZ members provides reliable materials for functional prediction.

Table 2 Physicochemical properties of tomato PLATZ transcription factor family
3.3.Function prediction by promoter cis-element analysis
To study the potential roles of tomato PLATZ transcription factors,we used PlantCare to analyze the promoter sequence of the tomatoPLATZgenes (2,000 bp upstream of the CDS) (Fig.2).Allcis-elements detected could be mainly classified into two types: phytohormone-responsive elements (ABRE,P-box,TGA-box,MeJA,SA,MBS) and abiotic stress response elements (ARE,LTR,STRE) (Suetal.2021).
EachSlPLATZgene contains a large number of different types and quantities ofcis-elements.ABRE and ARE are the most obvious and participate in the response to abscisic acid and anaerobic induction,respectively.This indicates thatSlPLATZgenes may be induced or inhibited by abiotic stress,participate in plant stress resistance,and play multiple roles in biological processes.
3.4.Expression pattern analysis and important response gene screening based on RNA-seq data
As shown in Fig.3,7 genes were expressed in response to cold stress (Fig.3-A),8 genes were expressed in response to drought stress (Fig.3-B),and 8 genes were expressed in response to salt stress (Fig.3-C).

Fig.3 Expression profiles and Venn diagram of SlPLATZ genes under three different abiotic stresses and tissue specificity of family members.A,cold stress.B,drought stress.C,salt stress.D,upregulated response mode.E,downregulated response mode.F,tissue specificity.
We counted the family members with upregulated(Fig.3-D) and downregulated (Fig.3-E) response patterns under three types of stress and found that two genes were coupregulated and two codownregulated.Pairwise comparisons showed that salt stress and low temperature stress had the most common expression patterns of genes.However,the number of genes shared by the salt stress,drought stress and low temperature stress groups was the same among the upregulated expression genes.These results indicate thatSlPLATZgenes are involved in abiotic stress responses but that not all members participate in the response.
In the tissue-specific expression analysis,a total of 9SlPLATZfamily members were found in flowers,roots,leaves,and fruits.In general,these genes were mainly highly expressed in roots and flowers (Fig.3-F).Among them,theSlPLATZ11,SlPLATZ17andSlPLATZ18genes were highly expressed in roots and flowers,though the other genes were only highly expressed in root or flower tissues.The results suggested that this gene family may play an important role in the development of tomato roots and flowers.
Based on the above expression pattern analysis,theSlPLATZ17gene was upregulated under all three stresses,with the highest expression level.Tissue specificity analysis results showed that the expression level of this gene was higher in tomato flowers and roots and was significantly different from that in other tissues.It is speculated that this gene may be closely related to stress resistance and plant growth and development.Therefore,we selected this gene for further experimentalbased functional verification.
3.5.Validation of the SlPLATZ17 expression pattern by qRT-PCR
To verify the expression pattern of theSlPLATZ17gene based on RNA-seq data,qRT-PCR was used to study the transcription levels of theSlPLATZ17gene under different treatments and in different tissues.The detailed expression pattern of theSlPLATZ17gene is shown in Fig.4.SlPLATZ17gene expression was upregulated under the three stress treatments.Expression of theSlPLATZ17gene was significantly upregulated in the early stage of the three stress treatments,reaching a peak at 1.5 h,and then decreased gradually but was always higher than before treatment.Induced expression of theSlPLATZ17gene was more significant under drought and salt stresses,up to approximately 8 times that before treatment.The expression pattern of theSlPLATZ17gene was similar under drought and salt stress.It is speculated that this gene may have similar regulatory functions under drought and salt stresses.In addition,the tissue-specific test results ofSlPLATZ17showed that the expression level in roots and flowers was 25-30 times that in stems,leaves and fruits,indicating that the RNA-seq results were reliable and that the function ofSlPLATZ17may be related to the abiotic stress response and flower and root development.

Fig.4 Expression pattern of the SlPLATZ17 gene under cold (A),drought (B),and salt (C) treatments and in different tissues (D).Data are presented as mean±SE (n=3).* (P≤0.05) and ** (P≤0.01) indicate a significant differences compared to the control group at 0 h (A,B,C) and the control group stalk (D),analyzed by one-way ANOVA.
3.6.SlPLATZ17 function verification under abiotic stress
The VIGS method was chosen to obtain quick and efficient observations.Plants withSlPLATZ17expression levels 50% lower than CK were selected for further experiments.As shown in Fig.5-A,the plants showed no significant changes before and after cold stress,with no effect on the cold response.After 3 h of drought treatment,the leaves of plants in whichSlPLATZ17was silenced began to wither,and their stems began to curl;the other plants showed no obvious symptoms (Fig.5-B).After 1.5 h of salt treatment,theSlPLATZ17-silenced plants were completely inverted with bent stems,whereas other plants showed only slightly curled stems (Fig.5-C).In conclusion,with the extension of drought and salt stress,the leaf wilt,drying phenomenon and stem curvature of theSlPLATZ17-silenced group were more serious than those of the other groups.Therefore,we further measured the physiological indices of tomato plants under drought and salt stress.SOD and POD activities as well as MDA and Pro contents were determined to explain the decrease in drought resistance and salt tolerance ofSlPLATZ17-silenced plants.

Fig.5 Silencing SlPLATZ17 reduced drought and salt stress tolerance in tomato plants.Phenotypes of CK,CK-TRV2 and SlPLATZ17-silenced plants under cold stress (A),drought stress (B) and salt stress (C).
After drought and salt treatment,the activities of SOD,POD and the contents of MDA,Pro increased in all plants.The activities of SOD,POD and the content of Pro inSlPLATZ17-silenced plants were lower than those in the other two groups,but the MDA content ofSlPLATZ17-silenced plants was higher than that in the other two groups (Fig.6-A).

Fig.6 Physiological and biochemical changes in control and gene-silenced group plants under drought and salt stresses.A,superoxide dismutase (SOD) and peroxidase (POD) activities;and proline (Pro) and malondialdehyde (MDA) contents.B,reactive oxygen species staining.C,relative conductivity.Data are presented as mean±SE (n=3).* (P≤0.05) and ** (P≤0.01) indicate a significant differences compared with CK at each time point,analyzed by one-way ANOVA.
Moreover,DAB and NBT staining under drought and salt stresses were assessed (Fig.6-B).The overall degree of DAB staining in the control plants in the two groups was lower than that in the VIGS-SlPLATZ17genesilenced plants,with almost no plants showing staining at 0 h.At 0 h after NBT staining,the leaf base of droughtstressed leaves was stained,as were the veins of saltstressed leaves.With the extension of stress treatment,the DAB and NBT staining area in all plants increased,and the degree of staining deepened.At 12 h,the degree of staining of leaves under salt and drought stresses was more serious than that of the other two groups,and the whole leaves were stained.
Under drought and salt stresses,relative electrical conductivity in the three groups increased,but relative electrical conductivity in VIGS-SlPLATZ17gene-silenced plants was higher than that in the other two groups during the whole treatment period (Fig.6-C).
3.7.Functional verification of SlPLATZ17 in tomato plant development
Considering that it takes a long time to observe the entire plant development process,we chose the method of establishing stable overexpression lines for verification.We cloned theSlPLATZ17gene and constructedSlPLATZ17-overexpressing transgenic lines using AC material as background.The clonedSlPLATZ17gene sequence was consistent with the sequence information in the genome,without mutation.The T2-overexpressing line OE2 was obtained by genome amplification and fluorescence quantitative expression analysis,and itsSlPLATZ17gene expression was 87 times higher than that of wild type,which could be used for phenotypic observation (Fig.7-A).
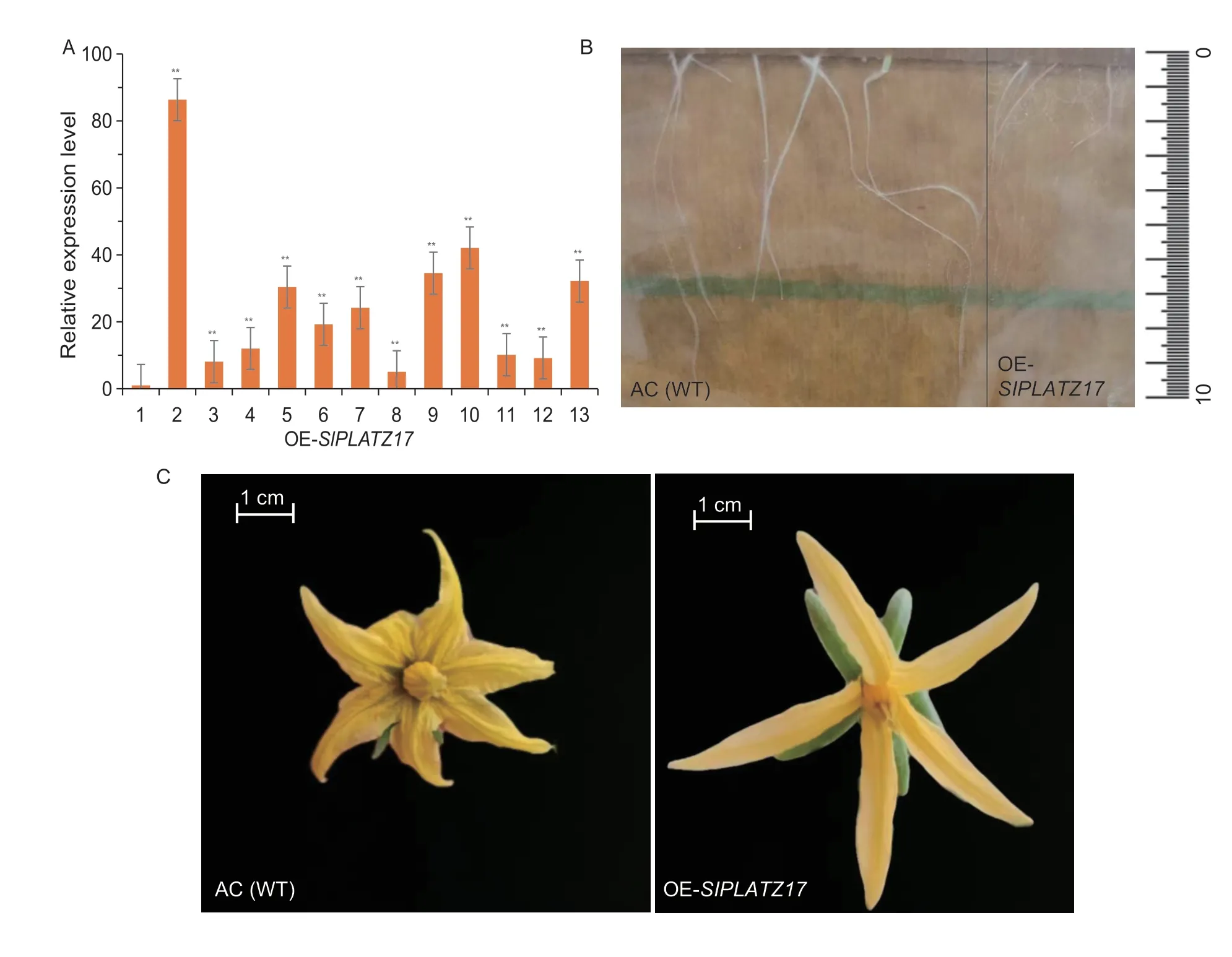
Fig.7 The overexpression of SlPLATZ17 gene making the root system of T2-generation plants shorter and the inflorescence larger.A,expression of the SlPLATZ17 gene in overexpressed plants.1,control plant;2-13,SlPLATZ17-overexpressing plants.Phenotypic observation of the growth and development of roots (B) and flowers (C) of overexpressing T2-generation plants and AC wild-type plants.Data are presented as mean±SE (n=3).* (P≤0.05) and ** (P≤0.01) indicate significant differences compared with 1 (AC),analyzed by one-way ANOVA.
Seeds of OE2 and WT were placed in seed germination bags for root development observation.The root system of OE2 was obviously shorter and less developed than that of WT plants (Fig.7-B).There were obvious differences in flower phenotypes.Compared with WT,OE2 has a larger flower size and long,thin petals,the adjacent petals were not connected at the base,and the stamen circumference was smaller (Fig.7-C).It is suggested thatSlPLATZ17regulates root and flower development in tomato.
4.Discussion
Since thePLATZgene was defined in tomato,genomewide identification ofPLATZgenes has been conducted in many species;15PLATZgenes have been identified in rice (Wangetal.2019),with 17 in maize (Wangetal.2018),24 in turnip,9 in cotton and 12 inArabidopsis(Zhangetal.2018).This study identified 19PLATZgenes in tomato.The tomato genome (900 Mb) (Satoetal.2012) is 2.1 times larger than that of rice (430 Mb)(Saegusa 1999),and 3.2 times of turnip (283.8 Mb) (Wangetal.2011),indicating that the number ofPLATZgenes is relatively conserved in different plant genomes.In this study,it was found that the 19SlPLATZgenes all contain conserved PLATZ domains,and similar results were found in a study of maize.Among the 17 members of maize,the expression of 15 ZmPLATZs has been confirmed in variant tissues (Wangetal.2018).The conserved PLATZ domain of theSlPLATZ18gene includes a preceding Bbox domain,which also exists in maize (Wangetal.2018) and alfalfa (Lietal.2023).These results indicate that the Bbox domain may undergo coevolution with special PLATZ members in some species and participate in the regulation of plants in stress environments,thereby enhancing or reducing the stress resistance of plant systems.
Adverse conditions such as low temperature,salinity and drought are limiting factors in plant growth and development.Many studies have shown that thePLATZgene family can affect the ability of plants to resist abiotic stress (Huangetal.2008;Kumarietal.2015;Zhangetal.2018).Plant hormones are the main regulatory factors involved in plant growth,development,and stress responses (Sirkoetal.2021).Under stress,abscisic acid rapidly accumulates in plants in response to abiotic stress (Yoshidaetal.2015).In this study,we found by analyzing the sequence structure of the gene interior and promoter region that mostSlPLATZfamily members contain multiple promoter elements related to stress resistance.An ABRE promoter element was predicted to exist in 10SlPLATZgenes and was associated with the response to abscisic acid,which may indirectly regulate plant stress resistance.It was found in alfalfa research that this promoter may be related to plant drought and salt resistance (Lietal.2023).STRE was predicted to exist in 9SlPLATZgenes,and was closely related to the plant stress response;its presence in the tomato BES1 gene family is reported to have an impact on plant stress resistance (Suetal.2021).The presence of promoter elements such as ABRE and STRE provides a structural basis forSlPLATZfamily members to exert stress resistance regulatory functions and demonstrates the possible regulatory modes of this gene family in stress response regulation.
PLATZplays an important role in plant growth and development (Kasirajanetal.2018;Wangetal.2018;Chaoetal.2019).In this study,theSlPLATZgeneis significantly expressed in both tomato roots and flowers,indicating that its gene function may be involved in the growth and development of tomato roots and flower organs.We found significant differences in the development of roots and flowers compared to wild-type AC in one overexpression line.Soybean GmPLATZ17 shows a higher gene expression level in root and nodule tissues and shows the highest transcription level in the root system (Zhaoetal.2022).High levels of GhPLATZ1 transcripts are detected in roots,stems,and cotyledons of cotton,while lower levels are detected in leaves and seeds (Zhangetal.2018),which is consistent with the results of tissue-specific experiments conducted in this study.These results indicate thatPLATZgenes are significantly expressed in the roots of different species,indicating that they have potential function in root development.
In addition,compared with wild-type plants,the flowers of plants overexpressingSlPLATZ17were larger.The dominant ORESARA15 mutant ofArabidopsisthalianaexhibits larger leaf size and a longer leaf lifespan (Kimetal.2018).ThePLATZgene family may also play a role in cotton fiber initiation and elongation (Handeetal.2017).These results indicate that somePLATZgenes may play certain roles in regulating the expansion or elongation of plant organs.
In one overexpression line examined in this study,it was found that the development of flowers and roots overexpressingSlPLATZ17significantly differed from that in the wild type.This indicates that theSlPLATZ17gene does indeed have an impact on the development of tomato roots and flowers.However,not all flower phenotypes on an individual plant of an overexpression line showed changes.The observed large flower phenotypes occurred at certain proportions (approximately 30 to 50% mutant large flower phenotypes on the 2nd to 5th spikes).Research shows that flower development and fertility are coordinately regulated by endogenous developmental signals,including the phytohormones jasmonates (JAs),auxin,and gibberellin,and environmental cues (Huangetal.2023).Therefore,there have other complex factors involved in the regulation of flower development bySlPLATZ17.In addition,during the construction of overexpression lines,we found that most of the lines had weak growth potential,a low seed setting rate,and a low overexpression efficiency of theSlPLATZ17gene.Therefore,only one OE2 line was used in the flower development observation experiment,which resulted in a lack of sufficient evidence for the phenotypic results.Therefore,in future research,a batch of overexpression lines will be reconstructed,and genome editing lines will be constructed to verify the developmental morphology of both flowers and roots in a two-way manner to conduct comprehensive research on the function ofSlPLATZ17.
Based on results obtained in this study,we propose a simple model diagram (Fig.8).SlPLATZgene family has an impact on plant stress response and growth and development (Fig.8-A).As shown in Fig.8-B,theSlPLATZ17silencing reduces plant resistance to salt and drought stresses.The Fig.8-C shows that the overexpression ofSlPLATZ17in plants affects their growth and development,resulting in shorter roots and larger flowers.

Fig.8 The SlPLATZ gene family and SlPLATZ17 gene having an impact on plant growth,development,and stress resistance.A,functions involved by the SlPLATZ gene family.B,SlPLATZ17 gene silencing reduces plant drought and salt resistance.C,overexpression of SlPLATZ17 gene affects the development of plant roots and flowers.
5.Conclusion
In this study,we obtained 19PLATZfamily genes through the whole-genome mining of tomatoes.Functional predictions were obtained for theseSlPLATZgenes at the transcription and gene structure levels,and the comprehensive screening of important response genes was conducted using database resources.Finally,experiments and analyses were conducted to verify the functionality ofSlPLATZ17.The results indicate that the downregulation ofSlPLATZ17expression reduced the drought resistance and salt tolerance of tomato plants and that overexpression affected the development of tomato roots and flowers.However,the phenotype is influenced by both the environment and genes,and the specific causes require further research to confirm.
Acknowledgements
We thank the support from the National Natural Science Foundation of China (32102390 and 32072589),the China Agriculture Research System(CARS-23-A11),the Heilongjiang Provincial Natural Science Foundation of China (YQ2021C013),and the Northeast Agricultural University Scholars Program(20XG28),China.
Declaration of competing interest
The authors declare that they have no conflict of interest.
 Journal of Integrative Agriculture2024年1期
Journal of Integrative Agriculture2024年1期
- Journal of Integrative Agriculture的其它文章
- Advances in DNA methylation and its role in cytoplasmic male sterility in higher plants
- Assessment of molecular markers and marker-assisted selection for drought tolerance in barley (Hordeum vulgare L.)
- 3D genome organization and its study in livestock breeding
- SUPER WOMAN 2 (SPW2) maintains organ identity in spikelets by inhibiting the expression of floral homeotic genes OsMADS3,OsMADS58,OsMADS13,and DROOPING LEAF
- Genetic dissection and validation of a major QTL for grain weight on chromosome 3B in bread wheat (Triticum aestivum L.)
- Coupling of reduced inorganic fertilizer with plant-based organic fertilizer as a promising fertilizer management strategy for colored rice in tropical regions
