Effect of mutations on acetohydroxyacid synthase (AHAS) function in Cyperus difformis L.
Xiaotong Guo ,Xiangju Li ,Zheng Li ,Licun Peng ,Jingchao Chen ,Haiyan Yu ,Hailan Cui#
1 State Key Laboratory for Biology of Plant Diseases and Insect Pests,Institute of Plant Protection,Chinese Academy of Agricultural Sciences,Beijing 100193,China
2 College of Resources and Environmental Sciences,China Agricultural University,Beijing 100193,China
Abstract Cyperus difformis L.is a troublesome weed in paddy fields and has attracted attention due to its resistance to acetohydroxyacid synthase (AHAS) inhibitors.It was found that the amino acid mutation in AHAS was the primary cause for the resistance of Cyperus difformis.However,the effect of different mutations on AHAS function is not clear in Cyperus difformis.To confirm the effect of mutations on AHAS function,six biotypes were collected,including Pro197Arg,Pro197Ser,Pro197Leu,Asp376Glu,Trp574Leu and wild type,from Hunan,Anhui,Jiangxi and Jiangsu provinces,China and the function of AHAS was characterized.The AHAS in vitro inhibition assay results indicated that the mutations decreased the sensitivity of AHAS to pyrazosulfuron-ethyl,in which the I50 (the half maximal inhibitory concentration) of wild type AHAS was 0.04 μmol L-1 and Asp376Glu,Pro197Leu,Pro197Arg,Pro197Ser and Trp574Leu mutations were 3.98,11.50,40.38,38.19 and 311.43 μmol L-1,respectively.In the determination of enzyme kinetics parameters,the Km and the maximum reaction velocity (Vmax) of the wild type were 5.18 mmol L-1 and 0.12 nmol mg-1 min-1,respectively,and the Km values of AHAS with Asp376Glu,Trp574Leu,Pro197Leu and Pro197Ser mutations were 0.38-0.93 times of the wild type.The Km value of the Pro197Arg mutation was 1.14 times of the wild type,and the Vmax values of the five mutations were 1.17-3.33-fold compared to the wild type.It was found that the mutations increased the affinity of AHAS to the substrate,except for the Pro197Arg mutation.At a concentration of 0.0032-100 mmol L-1 branched-chain amino acids (BCAAs),the sensitivity of the other four mutant AHAS biotypes to feedback inhibition decreased,except for the Pro197Arg mutation.This study elucidated the effect of different mutations on AHAS function in Cyperus difformis and provided ideas for further study of resistance development.
Keywords: acetohydroxyacid synthase (AHAS),mutation,enzyme function,Cyperus difformis
1.lntroduction
Cyperusdifformisis an annual highly self-pollinated weed and depends on seed reproduction with a large seed yield.It has been found in 47 countries in subtropical and warm temperate regions mainly distributed in most of China.It can cause a huge loss of rice yield with the serious occurrence ofC.difformis.To date,controllingC.difformisis still primarily to use herbicides such as pretilachlor,pyrazosulfuron-ethyl,bensulfuron methyl,MCPANa,bentazone,pyribenzoxim,penoxsulam,butachlor,bispyribac-sodium and halosulfuron methyl.From 1993 to 2021,the resistance ofC.difformisto bensulfuronmethyl,azimsulfuron,halosulfuron-methyl,ethoxysulfuron,bispyribac-sodium and imazamox,pyrazosulfuron-ethyl,imazosulfuron,cinosulfuron,cyclosulfamuron,penoxsulam,and imazapic was found in rice fields in many countries,including Australia,Italy,Brazil,Spain,South Korea,Greece,Turkey,the United States,India and China,where the resistance to acetohydroxyacid synthase (AHAS)inhibitors was more serious (Lietal.2020;Choudharyetal.2021;Heap 2022).
AHAS is an herbicide target,and the key enzyme is involved in the biosynthetic pathway of branchedchain amino acids (BCAAs),which is feedback inhibited by BCAAs (Ray 1984;Tranel and Wright 2002;Yu and Powles 2014).Feedback inhibition by BCAAs has been reported to result in significant down-regulation of catalytic subunit and regulatory subunit coding genes (Shaoetal.2022).AHAS inhibitors have low toxicity and high selectivity,such as sulfonylamino-carbonyl-triazolinone,imidazolinone,pyrimidinylth-thiobenzoates,triazolinone and sulfonylurea and AHAS inhibitors are absorbent herbicides that can be absorbed by plant leaves and roots and accumulate in tissues to hinder the biosynthesis of BCAAs,thereby affecting protein synthesis,inhibiting cell mitosis (Chaleff and Mauvais 1984;Dale and Stidham 1984;Gerwicketal.1990;Whitcomb 1999;Beckie and Tardif 2012).
Mutations in AHAS that cause the change of AHAS structure to prevent or reduce binding to substrates or herbicides are the basis for weed resistance to AHAS inhibitors (Zhaoetal.2017;Gainesetal.2020).The mutation types of AHAS that have been reported include sites Ala122 (5 types),Pro197 (13 types),Ala205 (2 types),Phe206 (1 type),Asp376 (1 type),Arg377 (1 type),Trp574 (4 types),Ser653 (3 types),and Gly654(2 types) (Powles and Yu 2010;Fangetal.2022;Heap 2022).These substituted amino acids are located in the conserved functional domain of the catalytic subunit in AHAS.Mutations can cause reduced AHAS sensitivity leading to different resistance levels of weeds to various structure herbicides and affect the AHAS function including the activity,kinetics and feedback inhibition(Hwangetal.2001;Whaleyetal.2007;Yuetal.2008;Dengetal.2014).The kinetics and activity of extractable AHAS with specific mutations have been reported to increase,decrease or remain unchanged in various species.Therefore,AHAS function may also differ among species.
It has been reported that mutations have an effect on AHAS function,growth or competition inLoiliumrigidum,AlopecurusaequalisandDescurainiasophia,while there have been no correlational studies inC.difformis(Yuetal.2010;Yangetal.2018;Zhaoetal.2020).To confirm the effect of mutations on AHAS function inC.difformis,various mutations were collected to illuminate the differences in AHAS function by the whole plant bioassay,analysis ofinvitroAHAS activity and determination of kinetic parametersKm,Vmaxand feedback inhibition.
2.Materials and methods
2.1.Plant materials
The seeds collected from the fields in Hunan,Anhui,Jiangsu and Jiangxi provinces were confirmed to be mutated by preliminary screening of pyrazosulfuron-ethyl(Table 1).The seeds were obtained from single plant reproduced for two generations.During the reproduction process,5 plants were randomly selected for target gene detection in each generation to obtain homozygous seeds.The susceptible biotype also was reproduced through two generations.
The organic fertilizer and nutrient soil were mixed in a ratio of 1:4 and put into a 9 cm×9 cm×11 cm pot,in which the nutrient soil with pH 7-7.5 mainly contained 0.83% N,P,K and 15% organic matter.The seeds were soaked in 50 mg L-1ethephon for 12 h,washed with water,and sown into the pots above,covering a thickness of approximately 0.1 cm of soil.During the cultivation process,the seedlings were irrigated by water seepage from the bottom.
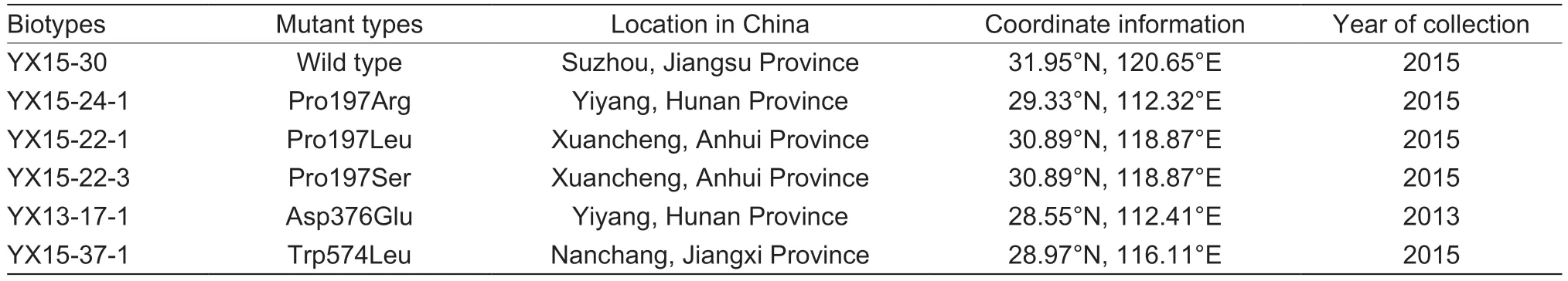
Table 1 The seed information of Cyperus difformis different biotypes
2.2.Whole-plant dose response bioassay
To evaluate the resistance levels of purified biotypes with specific mutations to pyrazosulfuron-ethyl,the six biotypes were taken as experimental materials to be measured by whole-plant response bioassay.The seedlings were thinned to 10 seedlings with uniform growth and distribution in each pot at the two-leaf stage.The biotypes with site 197,376 and 574 mutations were respectively treated with different doses (Table 2) of pyrazosulfuronethyl at the three-leaf stage.The moving-nozzle cabinet sprayer equipped with one TeeJet®XR8002 flat fan nozzle was used,and the spraying volume was 450 L ha-1at 0.275 MPa.The reduction biomass of dry weight was measured after 21 days of treatment.Three replicates were set for each treatment and repeated three times.
SigmaPlot (v.12.5) was used for nonlinear regression analysis according to the formulato calculate GR50of wild type and mutant biotypes,wherebdenotes the slope of the curve,Cdenotes the lower limit,Ddenotes the upper limit,GR50means the herbicide dose to reduce growth by 50% andYrepresents correspondingly the percentage of the control at herbicide doseX(Seefeldtetal.1995).The GR50of different biotypes was estimated according to the formula,and the estimated value of mutant biotypes was divided by the corresponding value of S biotypes to obtain the value of RI (R/S).
2.3.AHAS extraction and in vitro activity assays
Preparation of protein standard curve: 0,1,2,3,4,5 and 6 μL of bovine standard albumin (BSA) solution (1 mg mL-1) were added to centrifuge tubes,and the protein buffer was supplemented to 15 μL.A total of 285 μL of Coomash bright blue was added to each centrifuge tube,mixed and placed at room temperature for 5 min.A total of 200 μL were taken from each centrifuge tube and added to the elisa plate.The absorbance value at 595 nm was measured by a microplate reader.Threereplications were set for each treatment,and the standard curve was made with the concentration of BSA (mg mL-1)as the abscissa and the absorbance value at 595 nm as the ordinate.

Table 2 Doses of pyrazosulfron-ethyl in the whole-plant bioassay
A total of 2.85 mL of Coomassie bright blue solution was added to a 5-mL centrifuge tube,and the purified enzyme solution 100 μL was thoroughly mixed with elution buffer solution 900 μL.Then,150 μL of the mixed liquid was added to the prepared Coomash bright blue solution and allowed to react for approximately 6 min.Measuring the OD595value and calculating the concentration of different mutant AHAS were according to the established protein standard curve.
The AHAS activity was measured according to the method from Lietal.(2020).Plant leaves 4.5 g were taken from each biotype and stored at -80°C.The concentrations of pyrazosulfuron-ethyl (97%,Nanjing Luomaimei Biotechnology Co.,Ltd.,China) were 10-6,10-5,10-4,10-3,10-2,10-1,1,10,100,and 1,000 μmol L-1for the wild type and 10-3,10-2,10-1,1,10,100,1,000,and 10,000 μmol L-1for the mutant biotypes.Three replicates were set for each treatment and repeated three times.
The data obtained from theinvitroAHAS activity assay were converted into a control percentage.
The protein standard curve of the protein standard was drawn by Sigmaplot 12.5,the AHAS enzyme activity is calculated according to the acetoin production catalyzed per milligram of enzyme per unit time.
2.4.Enzyme kinetics parameter determination of AHAS
According to the method described by Yuetal.(2010),a slight change was used to determineKmandVmax(Kmindicates the affinity between AHAS and the substrate sodium pyruvate;Vmaxdenotes the reaction rate of AHAS when it is completely saturated with the substrate sodium pyruvate).Sodium pyruvate was not contained in the extraction and reaction buffers,and sodium pyruvate was added separately at concentrations of 0.39-25 mmol L-1.The independent experiment was conducted with two technical replications.Finally,according to the Michaelis-Menten kinetic equation:v=Vmax[S]/(Km+[S]),theKmandVmaxvalues of different biotypes ofC.difformiswere calculated,where [S] represents the concentration of sodium pyruvate,andvcorresponds to the reaction velocity at different concentrations of sodium pyruvate.Three replicates were set for each treatment and repeated three times.
SPSS (v.25.0) was used to conduct one-way ANOVA on the data obtained from feedback inhibition,KmandVmax,and the significance of the difference between treatments was determined with the minimum significant difference (P<0.05).One-way ANOVA means that only one factor changed in the assay.
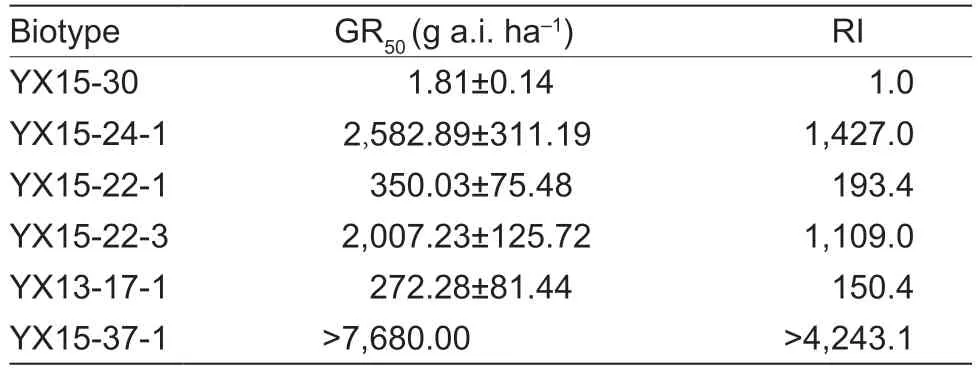
Table 3 The GR50 values and RI of Cyperus difformis biotypes to pyrazosulfuron-ethyl1)
2.5.Feedback inhibition of BCAAs on AHAS
In the measurement of BCAAs feedback inhibition on AHAS,the effect of different concentrations of BCAAs on AHAS activity was determined.In the experiment,the extraction buffer,resuspension buffer and elution buffer did not change,but the herbicide was replaced by leucine,isoleucine and valine.The BCAAs concentrations in this experiment were 0.0032-100 mmol L-1.Three replicates were set for each treatment and repeated three times.
The effect of the branched-chain amino acids on the activity of different mutant AHAS was calculated and compared by Sigmaplot 12.5.
3.Results
3.1.Whole-plant dose response assays
The results of the whole-plant bioassay showed that all the mutant biotypes exhibited higher resistance to pyrazosulfuron-ethyl than the wild type.In particular,YX15-37-1 (Trp574Leu) was assumed to have the highest level of resistance to pyrazosulfuron-ethyl,and all plants survived at 7,680 g a.i.ha-1.After 21 days of pyrazosulfuron-ethyl treatment,the wild type was killed absolutely at 15 g a.i.ha-1,and the RI of YX13-17-1 (Asp376Glu),YX15-22-1 (Pro197Leu),YX15-22-3(Pro197Ser) and YX15-24-1 (Pro197Arg) were 150.4,193.4,1,109.0 and 1,427.0,respectively (Table 3).
3.2.AHAS in vitro inhibition assays and enzyme activity
The I50values of the wild type and mutant biotypes with various mutations were significantly different under the pyrazosulfuron-ethyl treatment (Table 4).The wild type YX15-30 was strongly inhibited,with an I50value of 0.04 μmol L-1.In contrast,biotypes with mutations were significantly less sensitive to pyrazosulfuron-ethyl.Amongthe mutations,YX15-37-1 (Trp574Leu) had the lowest sensitivity to pyrazosulfuron-ethyl.The RI values of YX15-22-1 (Pro197Leu),YX15-22-3 (Pro197Ser),YX15-24-1(Pro197Arg) and YX13-17-1 (Asp376Glu) were 287.5,954.8,1,009.5 and 99.5,respectively.As shown in Fig.1,the reaction curve of YX15-24-1 (Pro197Arg) decreased significantly after 10 μmol L-1and was lower than those of YX15-22-1 (Pro197Leu) and YX15-22-3 (Pro197Ser).

Fig.1 The response curve of acetohydroxyacid synthase(AHAS) in vitro activity in wild type and mutant biotypes(Pro197Arg,Pro197Ser,Pro197Leu,Asp376Glu,Trp574Leu)to pyrazosulfuron-ethyl.Each point is the mean±SE (n=3) of the three assays.

Table 4 The I50 and enzyme activity values of different biotypes in Cyperus difformis1)
The AHAS enzyme activity from the mutated biotypes was significantly higher than that from the wild type,except for YX15-22-1 (Pro197Leu).The AHAS activity of mutant biotypes was 3-68% higher than that of the wild type in the study.The AHAS activities of YX15-24-1(Pro197Arg) and YX15-37-1 (Trp574Leu) were 68 and 3%,respectively,higher than that of the wild type.For YX15-22-1 (Pro197Leu),the activity was significantly lower than that of the wild type,which was consistent withD.sophiareported in 2018 (Yangetal.2018).Therefore,the mutation of site 197 from Pro to Leu may lead to decreased activity compared to the wild type.Therefore,comparing the wild type with four other mutated biotypes,it was obvious that the AHAS activity ofC.difformismay be affected by mutations.
3.3.AHAS kinetics connected to the specific mutations
TheKmvalue of the wild type was 5.18,and different mutant biotypes significantly changed the affinity between AHAS and sodium pyruvate.TheKmvalues of YX13-17-1 (Asp376Glu),YX15-22-1 (Pro197Leu),YX15-37-1(Trp574Leu) and YX15-22-3 (Pro197Ser) decreased significantly and were 0.38-0.93 times of the wild type.However,theKmvalue of YX15-24-1 (Pro197Arg) was significantly higher than that of the wild type (Table 5).TheVmaxvalue of the wild type was 0.12,and the five mutant biotypes were 2.42-3.33 times higher than that of the wild type,indicating that the above mutations resulted in higher AHAS-specific activity.There was no significant difference inVmaxbetween YX15-22-1 (Pro197Leu) and wild type YX15-30.
3.4.Sensitivity of mutant AHAS to BCAAs feedback inhibition
At concentrations of 0.0032-100 mmol L-1,isoleucine and valine showed significant differences in the feedback inhibition of five mutant AHAS,while leucine displayed significant differences in the feedback inhibition of AHAS at concentrations of 0.08 and 2 mmol L-1.Compared with the wild type YX15-30,the sensitivity of AHAS with only Pro197Arg (YX15-24-1) to feedback inhibition by BCAAs was enhanced,but the sensitivity of the other four mutantbiotypes was decreased.Under BCAAs treatment,the AHAS activity of different biotypes was compared with that of the wild type (Fig.2).The inhibition rates of leucine,valine and isoleucine on the activity of the five mutant biotypes were 40-80%,44-57% and 19-47% at 0.4 mmol L-1BCAAs,respectively.At the concentration of 2 mmol L-1,the inhibition rates were 48-81%,50-63% and 38-62%,respectively.Therefore,different AHAS biotypes were more sensitive to feedback inhibition by leucine.

Fig.2 The sensitivity determination to feedback inhibition of branched amino acids in six Cyperus difformis mutations (A,D,G,J and M were treated with isoleucine;B,E,H,K and N were treated with leucine.C,F,I,L and O were treated with valine).AHAS,acetohydroxyacid synthase.

Table 5 Km and Vmax values of the different biotypes of Cyperus difformis1)
4.Discussion
4.1.Effect of mutations on the resistance level and sensitivity of in vitro AHAS activity to pyrazosulfuronethyl
To date,C.difformishas evolved resistance to pyrazosulfuron-ethyl,and the resistance mechanism is due to mutations in AHAS causing changes in the 3D structure.The decreased ability of the combination of AHAS and related herbicides is primarily accountable for resistance to AHAS inhibitors,and differences exist in resistance levels among mutations (Ntoanidouetal.2016;Fangetal.2022).Cyperusdifformisfrom America,South Korea and China has developed different resistance levels to AHAS inhibitors caused by different mutations(Yongetal.2010;Tehranchianetal.2015).
In this study,the resistance level of the Trp574Leu biotype was the highest among the five mutations,which was consistent withRaphanusraphanistrum,Amaranthus palmeriandRapistrumrugosumreported (Yuetal.2012;Larranetal.2017;Ntoanidouetal.2018).However,it differed from the Asp376Glu biotype inD.sophiaand the Pro197Ala biotype inConyzacanadensis,which had the highest resistance (Zhengetal.2011;Zhangetal.2017).The Asp376Glu biotype resistance level was the lowest in the mutant biotypes,and the Pro197Arg biotype had the highest resistance level among the Pro197 mutations in this study.Therefore,the resistance caused by mutations was not only dependent on the mutation sites but also related to the specific amino acid mutation.Currently,the most common mutation is the site 197Pro,which has been substituted by 13 kinds of amino acids.
In this study,pyrazosulfuron-ethyl inhibited wild type AHAS activity,and mutant biotypes were at least 99-fold more insensitive than wild type.Such AHAS activity decreased in response to herbicides based on target mutations has been reported inC.difformisfrom California(Busietal.2006).The AHAS I50values of different mutant biotypes were significantly different.Mutations may limit the binding of herbicides to AHAS,resulting in reduced AHAS sensitivity,and the AHAS sensitivity of all mutant biotypes was lower than that of wild type.
4.2.Effect of mutations on AHAS enzyme activity
It has been reported that higher AHAS enzyme activity is unlikely to be caused by overexpressed AHAS genes but may be due to increased enzyme stability (Yuetal.2003).According to some reports,AHAS is composed of catalytic subunits and regulatory subunits,while the regulatory subunit has no activity and can greatly stimulate the activity of the catalytic subunit,especially sensitive to feedback inhibition by BCAAs (Lee and Duggleby 2001,2002;Yangetal.2018).The extracted AHAS activity of mutations may increase with the improved stability of catalytic subunits,regulatory subunits or cofactor binding (Yuetal.2010).It has been reported that the increased activity of AHAS may be connected with the cost of resistance due to the toxic effects of excessive amino acid synthesis (Vila-Aiubetal.2009).Different resistance mutations have effects on the binding between cofactors thiamine diphosphate (ThDP)on catalytic subunits and FAD inArabidopsisthaliana(Chang and Duggleby 1998;Yuetal.2010).Then,Kim proposed that existing “C terminals” and “moving circuits”in the crystal structure of yeast AHAS are accountable for the binding or stabilization of the active dimer and the cofactor ThDP (Kimetal.2004).Both the binding site with herbicide and the active site are located in the interface of the catalytic subunit.This result suggested that the binding site with herbicides may change the geometry of the active site to alter the catalytic activity (Duggleby and Pang 2000).The activity of mutant AHAS in some weeds decreased or remained unchanged (Ashigh and Tardif 2007;Yangetal.2018;Zhaoetal.2020).For example,the Ala205Val mutation in yeast AHAS resulted in reduced extractable AHAS activity compared to the wild type,and Pro197 is the part of the binding channel between yeast AHAS and substrate,and it will produce a resistant enzyme to herbicide after 197 site mutation (Dugglebyetal.2003).Different mutations will produce enzymes with diverse characteristics (Prestonetal.2006).For example,Pro197His/Thr substitutions resulted in reduced extractable activity in eastern black nightshade AHAS,and Pro197Ser substitution significantly reduced the extractable activity in yeast AHAS (Dugglebyetal.2003;Ashigh and Tardif 2007).However,the Pro197Ser mutation did not change extractable AHAS activity inA.thaliana(Mouradetal.1995).In this study,only the extracted AHAS with Pro197Leu was lower than the wild type,which was not consistent with the description of Yuetal.(2010) inR.raphanistrum,but consistent withD.sophia(Yangetal.2018).Some researchers proposed a relationship model between catalytic subunits and regulatory subunits by studying the structure of AHAS that there may exist leucine and isoleucine/valine binding sites on the regulatory subunit and signals can be transmitted to the catalytic subunit to regulate AHAS activity when one site is bound (Lee and Duggleby 2002).We found that leucine had the strongest feedback inhibition on AHAS activity in the following study,resulting in decreased AHAS activity.
4.3.Effect of mutations on AHAS kinetics
It is well known that the target mutation can result in AHAS 3D structure changes by affecting the binding between AHAS and substrates (Yuetal.2010;Zhaoetal.2020).According to the reports,kinetic changes may have a negative effect on plant growth,and this may be caused by the influence of enzyme affinity with substrate (Purrington and Bergelson 1999;Menegatetal.2016;Duetal.2019).In this study,the affinity of the substrate to AHAS increased in four mutations,and only Pro197Arg reduced the affinity between substrate and AHAS compared with the wild type.There also existed unchanged affinity between substrate and AHAS with mutations;for example,the affinity of Pro197Leu and Pro197Ser mutations inD.sophiawere unchanged (Yangetal.2018).The Pro197Ser mutation in AHAS caused lowerKmand higherVmaxcompared to the wild type inMyosotonaquaticum(Liuetal.2017).The Pro197Ala/Gln mutations inL.rigidumAHAS and the Trp574Leu substitution inAlopecurusaequalisAHAS produced lowerKmand higherVmaxthan the wild type (Yuetal.2010;Zhaoetal.2020).Pro197Arg mutation inL.rigidumresulted in higherKmandVmax,while Pro197Tyr resulted in higherKmand lowerVmax(Yuetal.2010).InD.sophia,Pro197Leu/His/Ser/Thr,Asp376Glu and Trp574Leu mutations all had lowerKmandVmax(Yangetal.2018).Therefore,each specific resistance mutation needs to be analyzed on the basis of species.Mutated AHAS can still work normally due to indistinctly unchanged affinity between AHAS and substrates,and many resistance mutations have no significant adverse effects on the functionality of AHAS (Mccourtetal.2006).
In this study,theKmvalue did not change drastically,but the Pro197Arg mutation slightly increasedKm.The difference from other mutations is required to confirm whether the adverse impacts on plant physiology and biochemistry.It has been reported that it will result in insufficient or excessive synthesis of BCAAs or impaired AHAS activity to affect the growth or reproduction of plants if resistance mutations cause changes in the binding between the substrate and AHAS (Vila-Aiubetal.2009).Therefore,AHAS function may also differ among species.
4.4.Effect of mutations on the sensitivity of AHAS to feedback inhibition
AHAS in plants is sensitive to all branched chain amino acids,and reducing the sensitivity of AHAS to feedback inhibition will increase the synthesis of branched chain amino acids so that weeds can still survive under the treatment of AHAS inhibitors (Eberleinetal.1997,1999;Prestonetal.2006).However,AHAS feedback inhibition by BCAAs studies have been relatively limited.It has been demonstrated that the sensitivity of AHAS with Pro197His,Pro197Thr and some mutations to feedback inhibition by BCAAs was significantly reduced (Eberleinetal.1997,1999;Prestonetal.2006;Ashigh and Tardif 2007,2009).Pro197Ser,Trp574Leu mutations inA.thalianaand in transgenic tobacco have unaltered sensitivity to feedback inhibition (Hattorietal.1995;Mouradetal.1995).In this study,the sensitivity of the Pro197Arg mutation to feedback inhibition was significantly increased,while the other four mutations were reduced.It may be that the regulatory subunit containing binding sites for leucine and isoleucine/valine endows AHAS with sensitivity to feedback inhibition,and various mutations may diversely influence the regulatory subunit (Lee and Duggleby 2001).
In this study,Trp574Leu and Asp376Glu were more sensitive to feedback inhibition.However,the relationship between the regulatory subunit and the catalytic subunit is not clear,and the increased or decreased sensitivity to feedback inhibition induced by mutations cannot be elucidated.It was reported that variations in AHAS sensitivity to feedback inhibition by BCAAs may change the concentration of amino acids in plants,while it will not be obvious until the concentration of BCAAs reaches 0.4 mmol L-1,and the impacts on plant physiology are still unclear(Yangetal.2018).In general,mutations in AHAS may have effects on growth,with the mutated gene persisting or becoming fixed in untreated populations or populations that have ceased to be selected (Jasieniuketal.1996;Vila-Aiubetal.2015).Current studies have found that it had no adverse effects on plant growth resulting from the increased activities conferred by Pro197Ser and Trp574Leu,which contradicted the effects of Pro197Ser onA.thalianagrowth (Purrington and Bergelson 1999;Yuetal.2010).In resistantAmaranthus,it has been revealed that Trp574Leu is related to plant growth,but its effects on AHAS function are unknown (Tardifetal.2006).
5.Conclusion
In summary,each homozygote for Trp574Leu,Asp376Glu,Pro197Ser,Pro197Arg and Pro197Leu mutations ofC.difformisbiotypes were generated to investigate the impacts of the above mutations on AHAS function by measuring AHAS activity,kinetics,and sensitivity to feedback inhibition compared with the wild type AHAS.The resistance conferred by mutations to pyrazosulfuronethyl was exhibited,and it also had a few impacts on AHAS function (activity,kinetics,feedback inhibition).AHAS with Trp574Leu had almost unchanged extractable AHAS activity,but AHAS with Trp574Leu obviously altered the kinetics.Only the Pro197Leu mutation decreased extractable AHAS activity and inapparently increasedVmax,while other mutations obviously altered the kinetics.Notably,Pro197Arg significantly increased theKmfor sodium pyruvate and the sensitivity to feedback inhibition by BCAAs.In this study,AHAS carrying Pro197Ser demonstrated the least effect on kinetics compared with other mutations so that it may increase the frequency of evolution in the future on account of the subtle effect on enzyme kinetics.The regulatory subunit and catalytic subunit of the plant AHAS with a clear 3D structure can greatly illustrate how resistance mutations affect the catalytic subunit to change the enzyme activity and feedback inhibition;therefore,it is of great significance to study the AHAS structure.
Acknowledgements
This research was funded by the National Natural Science Foundation of China (31972281).
Declaration of competing interest
The authors declare that they have no conflict of interest.
 Journal of Integrative Agriculture2024年1期
Journal of Integrative Agriculture2024年1期
- Journal of Integrative Agriculture的其它文章
- Advances in DNA methylation and its role in cytoplasmic male sterility in higher plants
- Assessment of molecular markers and marker-assisted selection for drought tolerance in barley (Hordeum vulgare L.)
- 3D genome organization and its study in livestock breeding
- SUPER WOMAN 2 (SPW2) maintains organ identity in spikelets by inhibiting the expression of floral homeotic genes OsMADS3,OsMADS58,OsMADS13,and DROOPING LEAF
- Genetic dissection and validation of a major QTL for grain weight on chromosome 3B in bread wheat (Triticum aestivum L.)
- Coupling of reduced inorganic fertilizer with plant-based organic fertilizer as a promising fertilizer management strategy for colored rice in tropical regions
