Establishment of an indirect immunofluorescence assay for the detection of African swine fever virus antibodies
Wan Wang ,Zhenjiang Zhang ,Weldu Tesfagaber ,Jiwen Zhang ,Fang Li ,Encheng SunLijie Tang,Zhigao BuYuanmao Zhu#,Dongming Zhao#
1 State Key Laboratory for Animal Disease Control and Prevention,National African Swine Fever Para-reference Laboratory,National High Containment Facilities for Animal Diseases Control and Prevention,Harbin Veterinary Research Institute,Chinese Academy of Agricultural Sciences,Harbin 150069,China
2 College of Veterinary Medicine,Northeast Agricultural University,Harbin 150069,China
Abstract African swine fever (ASF) continues to cause enormous economic loss to the global pig industry.Since there is no safe and effective vaccine,accurate and timely diagnosis of ASF is essential to implement control measures.Indirect immunofluorescence assay (IFA) is a gold standard serological method recommended by the World Organization for Animal Health (WOAH).In this study,we used primary fetal kidney cells to establish a wild boar cell line (BK2258) that supported the efficient replication of ASF virus (ASFV) SD/DY-I/21 and showed visible cytopathic effect (CPE).Moreover,using BK2258,we established a sensitive and specific IFA for ASFV antibody detection.To standardize and evaluate the performance of this assay,we used serum samples from pigs infected with the low virulent genotype I SD/DY-I/21 and genotype II HLJ/HRB1/20,and immunized with the vaccine candidate HLJ/18-7GD,field samples,and negative serum samples.The IFA reacted with the ASFV-positive sera and displayed bright fluorescence foci.There was no non-specific green fluorescence due to cellular senescence or other cell damage-causing factors.Compared to a commercial indirect enzyme-linked immunosorbent assay(iELISA),ASFV antibodies were detected 1-4 days earlier using our IFA.The detection limits of the IFA and iELISA for the same ASFV-antibody positive serum samples were 1:25,600 and 1:6,400,respectively,indicating that the IFA is more sensitive than iELISA.The newly established IFA was highly specific and did not cross-react with sera positive for six other important porcine pathogens (i.e.,Classical swine fever virus (CSFV),Porcine reproductive and respiratory syndrome virus (PRRSV),Porcme circovirus type 2 (PCV2),Pseudorabies virus (PRV),Foot-and-Mouth disease virus type O (FMDV/O),and Porcine epidemic diarrhea virus (PEDV)).This study thus provides a sensitive,specific,and reliable detection method that is suitable for the serological diagnosis of ASF.
Keywords: African swine fever,antibody,IFA,serological method
1.lntroduction
African swine fever (ASF) is a highly contagious and lethal disease of domestic pigs and wild boars (Susscrofa).In recent years,ASF has caused considerable alarm among pig farmers due to its significant threat to agricultural development worldwide.The clinical outcomes and gross pathological lesions of ASF are highly variable,ranging from hyperacute or acute infection to a subclinical or chronic infection,depending on the virus virulence,route of exposure,infectious dose,and host characteristics (Beltrán-Alcrudoetal.2017;WOAH 2019).The disease spreads rapidly among pig populations either through direct animalto-animal contact or indirectly,viaswill feed or contaminated fomites like clothes,footwear,equipment,food waste,and bedding (Viñuela 1985;Penrith and Vosloo 2009).
ASF is caused by African swine fever virus (ASFV),which is a large,double-stranded DNA virus with a genomic size ranging from 170-194 kb that encodes 150-200 proteins (Costardetal.2009;Chapmanetal.2011).ASFV is the sole member of the genusAsfiviruswithin the familyAsfarviridaeand is the only known DNA arbovirus that replicates and transmits through a tick vector (Kleiboekeretal.1998).Currently,there are 24 genotypes of ASFV based on the sequence of its B646L gene,which encodes the capsid protein p72 (Bastosetal.2003),and eight ASFV serotypes based on the hemagglutinin CD2-like protein (CD2v) and C-type lectin(Malogolovkinetal.2015).
ASFV was first introduced into China in August 2018,and within a few months,the epidemic spread to most of the 34 provinces and regions (Geetal.2018;Zhaoetal.2019).Since then,ASFV has remained prevalent in China.Recent epidemiological surveys have shown that new strains of ASFV with different virulence are emerging and circulating in China (Sunetal.2021a,b).Low virulent ASFVs often lead to subacute or chronic disease,and the infected pigs show mild symptoms or remain asymptomatic.In such scenarios,the infected pigs might continuously contaminate the environment and act as potential source of infection.Therefore,accurate serological diagnosis is essential for us to be able to monitor the presence of seropositive animals with sub-acute or chronic forms of ASF.In July 2021,the China Centre for Animal Disease Control and Prevention approved 33 ASFV antibody detection enzyme-linked immunosorbent assay (ELISA) kits for emergency use through a rigorous comparative evaluation process,aiming at the early detection of ASFV-infected pigs through regular antibody screening and providing important guidance for ASF prevention and control.However,improper sample handling and preservation may yield false-positive results in ELISA,and all positive and inconclusive samples from ELISA must be confirmed by using alternative serological tests like the indirect immunofluorescence assay (IFA),the indirect immunoperoxidase test (IPT),and the immunoblotting test (IBT) as recommended by the WOAH(Gallardoetal.2015,2019).
The serological detection methods recommended by WOAH and the current Chinese national standard for the diagnosis of ASF (GB/T 18648-2020) include ELISA and IFA.The IFA method used as a confirmatory test for ASFV antibodies was established by Sanchez-Vizcaino in 1987 (Sanchez-Vizcaino 1987),and has not been meaningfully updated in the past 35 years.It is mainly limited by the lack of suitable cells (WOAH 2019) that not only support the proliferation of ASFVs,but are also suitable for use in an IFA.At present,the cells used for IFA detection of ASFV antibodies are mainly African green monkey kidney cells (Vero cells) and monkey stable cells.To the best of our knowledge,there have been no detailed studies of IFA methods for detecting ASFV antibodies.
In this study,we established a wild boar kidney cell line (BK2258) that can be passaged stably and evaluated its susceptibility for ASFV.Then,using the BK2258 cell line,we established a new IFA method for ASFV antibody detection,and assessed its sensitivity and specificity compared with conventional iELISA.The newly established IFA method is very sensitive and specific,and can be used as a confirmatory test for ASFV antibody detection,which will benefit the clinical diagnosis and epidemiological surveillance of ASF in China.
2.Materials and methods
2.1.Viruses and cells
The genotype I virus SD/DY-I/21 and genotype II virus HLJ/HRB1/20 were isolated from the field samples and kept in our laboratory (Sunetal.2021a,b).
Two fetal kidneys were surgically excised from a wild boar fetus with a gestational age of about 80 days.The kidneys were decapsulated,the cortex carefully removed,the medulla discarded,and the remaining kidney tissue was chopped into 1-2 mm3pieces.The pieces were then placed in a 100-mL beaker and washed five times with PBS.Tissue pieces were further treated using trypsin digestion solution (0.25% trypsin,8.0 mg mL-1NaCl,0.4 mg mL-1KCl,0.06 mg mL-1Na2HPO4·H2O,0.06 mg mL-1KH2PO4,0.37 mg mL-1NaHCO3) for 10 min at 37°C.To terminate the digestion reaction,the supernatant was added to RPMI 1640 medium (containing 10% fetal bovine serum,100 U mL-1penicillin,and 100 U mL-1streptomycin).The remaining tissue blocks were digested with trypsin digestion solution in a water bath at 37°C,and the supernatant was collected by repeated stirring every 5 min until the tissue blocks were completely digested.Subsequently,the collected suspension was centrifuged at 800 r min-1for 10 min at 4°C,the supernatant was discarded,10 mL RPMI 1640 culture medium was added to the precipitation,and the suspension was then filtered through a 200-mesh stainless steel screen.The filtrate was divided and cultured in an incubator at 37°C with 5% CO2.After 72 h,the cells grew into a monolayer that was digested and dispersed with trypsin,and the cell concentration was adjusted to 2×105cells mL-1for further subculture.
2.2.Swine serum samples
To validate the established IFA method,a panel of 479 pig serum samples was analyzed.The origins of the samples were as follows:
Standard controls: One positive and one negative serum were prepared from sera of known origin as standard controls and stored in our laboratory.The standard positive serum was obtained from pigs experimentally infected with the highly virulent HLJ/18 isolate (Zhaoetal.2019),and the standard negative sera were obtained from specific pathogen free (SPF) pigs.
Serum samples obtained from pigs immunized with the seven-gene-deleted vaccine candidate (HLJ/18-7GD): 149 serum samples were collected during the development of the HLJ/18-7GD vaccine candidate (Chenetal.2020).
Field samples: 300 field sera were collected from Heilongjiang and Liaoning provinces,and Inner Mongolia Autonomous Regions,China during routine epidemiological surveillance.
Serum samples obtained from pigs infected with low virulent ASFVs: 12 serum samples were collected on days 7,9,and 11 from four pigs infected with the lowvirulent HLJ/HRB1/20 strain,and 84 serum samples were collected from days 2 to 13 after inoculation with the lowvirulent SD/DY-I/21 strain.
Negative serum samples: 30 serum samples were collected from SPF pigs.
Polyclonal sera positive for other important swine pathogens: Positive swine serum to Classical swine fever virus (CSFV),Porcine reproductive and respiratory syndrome virus (PRRSV),Porcine circovirus type 2 (PCV2),Porcine pseudorabies virus (PRV),Foot-and-Mouth disease virus type O (FMDV/O),and Porcine epidemic diarrhea virus (PEDV) were kindly provided by laboratories inthe Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (CAAS).
2.3.ASFV replication dynamics in BK2258 cells
To determine the replication dynamics of ASFV,wild boar kidney cells (BK2258) were infected with ASFV SD/DYI/21 (MOI=0.1).Briefly,BK2258 cells were incubated with virus in 24-well plates for 1 h in a 5% CO2incubator at 37°C,culture medium was then discarded and the cells were washed three times with PBS (HyClone,USA).Subsequently,DMEM culture medium containing 2% fetal bovine serum was added and the cells were incubated in a 5% CO2incubator at 37°C to observe cytopathic effect(CPE).Supernatants were collected every 24 h until 192 h after inoculation.The p72 gene in the supernatants was detected by qPCR and the viral replication curve was determined.
2.4.Quantitative PCR
ASFV genomic DNA from cell supernatants was extracted using QIAamp®DNA Mini Kits (Qiagen,Germany).Quantitative PCR (qPCR) was performed using an ABI QuantStudio5 (ABI,USA) as described elsewhere (Kingetal.2003).
2.5.Establishment of an lFA for ASFV antibodies detection
Initially,BK2258 cells (up to the F15 generation) were infected with ASFV SD/DY-I/21.Then,the ASFV infection dose was determined by 10-fold serial dilution.Infected BK2258 cells were grown in 96-well plates and incubated at 37°C with 5% CO2.After 48 h,the culture medium was discarded and the cells were washed twice with PBS.The infected cells were then fixed with 4% paraformaldehyde(Solarbio,China) for 30 min and permeabilized with 0.25% Triton-X100 for 15 min at room temperature.Swine serum samples diluted at 1:100 were then added in duplicate and the cells were incubated at 37°C for 1 h.Positive and negative control standard sera for ASFV,diluted at 1:100,were included in duplicate.After being washed with PBS,the cells were incubated with a 1:200 dilution of FITC-labelled rabbit anti-pig IgG and incubated at 37°C for 45 min.Finally,the cells were visualized by using a fluorescence microscope;results were considered positive if specific green fluorescence was detected and negative if no such fluorescence was observed.
2.6.Optimization of the immunofluorescent assay
To determine the optimal dose for virus infection,ASFV SD/DY-I/21 was serially diluted 10-fold and BK2258 cells were infected with virus at 10-3to 10-6dilutions (4 wells for each dilution) at 37°C and 5% CO2for 48 h before IFA assay.Similarly,to optimize the dilution of the primary and secondary antibodies,ASFV standard positive and negative serum samples were diluted 2-fold (1:1,024),and FITC-labelled rabbit anti-pig IgG (Sigma,Germany)was serially diluted from 1:100 to 1:1,600.Likewise,the optimal incubation time were evaluated at 37°C for 30,45,and 60 min.After the above conditions were optimized,fluorescence microscopy was used to observe the wells that corresponded to clustered,bright,specific green fluorescence.
2.7.Analysis of the detection limit and sensitivity of the lFA
To determine the detection limit of the IFA,a strong positive serum was serially diluted 2-fold and assessed with the IFA.As a comparison,the diluted serum samples were also tested in a commercial ELISA kit (Beijing Jinnuo Biotechnology Co.,Ltd.,China).To determine the sensitivity of the IFA,four and seven pigs were intramuscularly infected with the low virulent genotype II virus HLJ/HRB1/20 and the genotype I virus SD/DYI/21 strain at a dose of 106.0TCID50,respectively.Blood was collected at 7,9,and 11 days post-infection (dpi)from pigs infected with HLJ/HRB1/20 and from 2 to 13 dpi from pigs infected with SD/DY-I/21.The serum samples were then evaluated in the IFA and commercial ELISA kit,respectively.
3.Results
3.1.Establishment of a cell line using wild boar fetus kidney cells
The IPKM cell line was established by using primary kidney macrophages from a 6-day old pig (Masujinetal.2021),and the cell line ZMAC-4 was generated from the lungs of a porcine fetus (Portugaletal.2020).Here,to establish a cell line,the kidneys from a wild boar fetus at 80 days of gestation were fully minced and digested with trypsin (Fig.1-A).The primary kidney cells (F1),when were observed under an inverted microscope,comprised different morphological cells,including long spindle forming fibroblasts,oval renal epithelial cells,and small tissue masses (Fig.1-B).The cells were subcloned using limiting dilution,and the oval renal epithelial cells were selected for expansion and passage.The morphology of the cells was stable over several passages,with the cells remaining tightly packed in oval shapes (Fig.1-B).The stable cell line was named as BK2258.

Fig.1 Characterization of the wild boar fetus kidney cells.A,a wild boar fetus at a gestational age of approximately 80 days.B,morphological observation of BK2258 cells at different passages.Oval renal epithelial cells are indicated by the solid red arrow and long spindles forms of fibrocytes are shown by the open red arrow.C,cytopathic effect (CPE) in BK2258 cells infected with ASFV SD/DY-I/21 at different timepoints.D,the in vitro replication kinetics of ASFV SD/DY-I/21 in BK2258 cells.
3.2.Susceptibility of BK2258 cells to ASFVs
To evaluate the susceptibility of BK2258 cells to ASFVs,the cells were infected with ASFV SD/DY-I/21 at an MOI of 0.1.Cell morphological changes were observed at different times post-infection.At 24 h post-infection (hpi),no obvious morphological changes were observed,a few cells gathered into small clusters at 48 hpi,and the size of the clusters increased at 72 hpi.Viable cells developed plaques at 96 hpi as a result of aggregation,shrinkage,and lysis,and the size of the plaques expanded further at 120 hpi (Fig.1-C).The cells were completely exfoliated at 144 hpi.Cell supernatants were collected for viral DNA detection at different times post-infection using qPCR.The results showed that ASFV SD/DY-I/21 efficiently replicated in BK2258 cells with viral DNA copy numbers as high as 5.35×109copies mL-1(Fig.1-D).
3.3.Establishment of lFA for ASFV antibody detection
Based on the efficient replication of ASFV SD/DY-I/21 in BK2258 cells,an IFA was established to detect ASFV antibodies using FITC-conjugated anti-pig IgG.We observed that ASFV-infected cells reacted with ASFVpositive serum,forming specific green fluorescent foci,but did not react with ASFV-negative serum,which suggests that ASFV SD/DY-I/21 and BK2258 cells are suitable to establish an IFA for ASFV antibody detection.Then,we optimized the reaction conditions for the IFA for ASFV antibody detection.
3.4.Determination of optimal dose and time of virus infection for the lFA
To determine the optimal infection dose of ASFV for the IFA,BK2258 cells were infected with different doses of ASFV SD/DY-I/21 ranging from 10-3to 10-6dilutions of virus stock (107.8TCID50mL-1),respectively.The results revealed that all four virus dilutions (10-3to 10-6)showed specific green fluorescence in response to ASFVpositive serum,but not negative serum.Cells infected with the 10-4dilution of ASFV reacted with the positive serum to produce clustered,bright,and specific green fluorescence,and the number of fluorescent spots was moderate and easy to assess (Fig.2-A);therefore,10-4dilution (MOI=0.001) of SD/DY-I/21 was selected as the optimal infection dose.
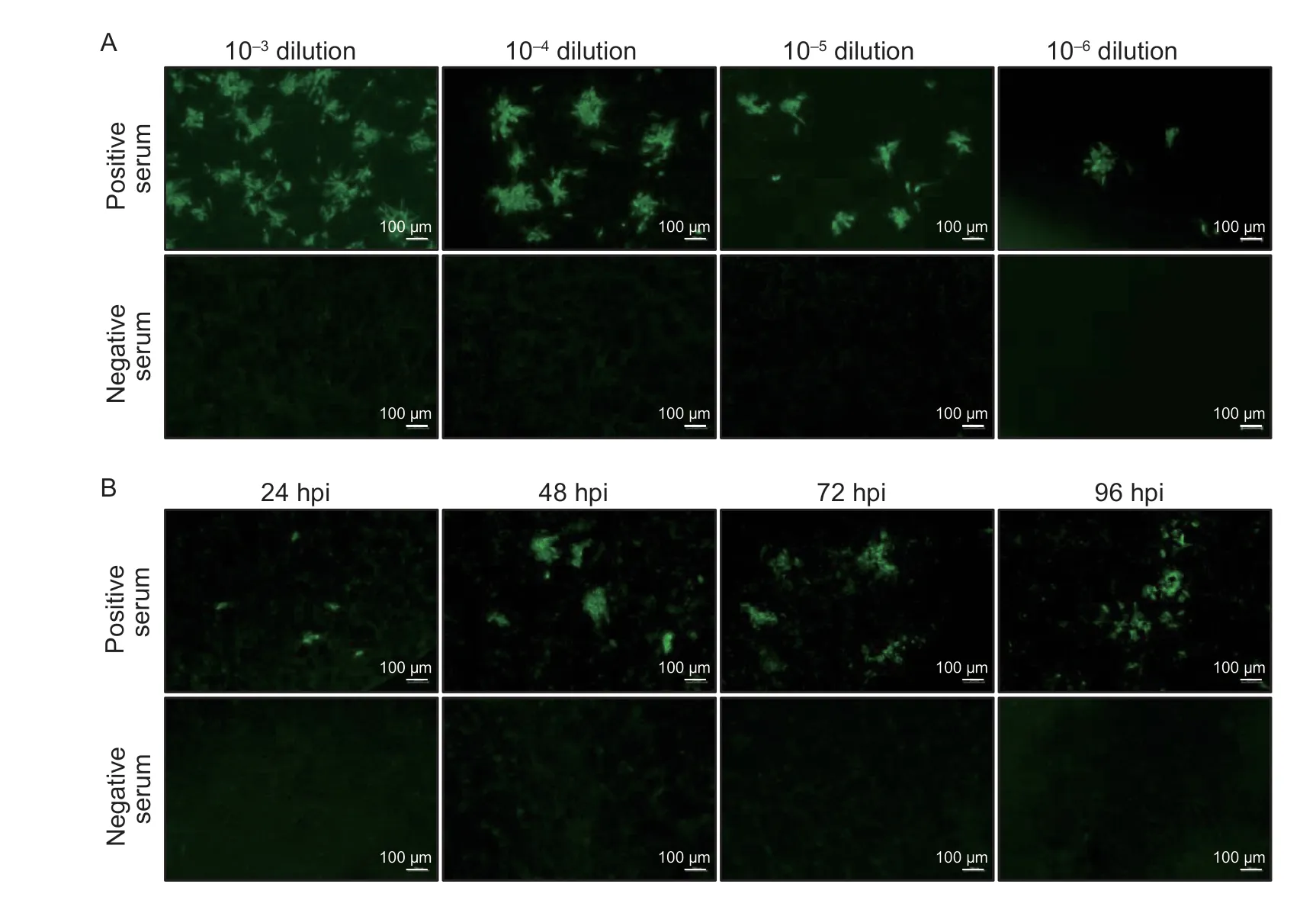
Fig.2 Determination of the optimal virus infection dose and incubation time after African swine fever virus (ASFV) infection.A,determination of the optimal virus infection dose.BK2258 cells were infected with ASFV SD/DY-I/21 at a 10-3 to 10-6 dilution,and then were tested against standard ASFV positive and negative pig sera.B,optimal incubation time after ASFV infection to perform the immunofluorescence assay (IFA).IFA was performed using ASFV positive and negative sera at different hours post-infection (hpi).
To determine the optimal infection time,BK2258 cells were infected with ASFV SD/DY-I/21 and cultured in the incubator for different times.ASFV-infected cells were detected based on a response to positive serum using the IFA.The results showed that the fluorescence was small at 24 hpi;however,at 48 hpi,the fluorescence was clustered,bright,specific,and easy to observe.The fluorescence was diffuse at 72 and 96 hpi (Fig.2-B).Therefore,48 hpi was deemed the optimal time of ASFV infection for the IFA.
3.5.Determination of the optimal serum dilution
To determine the optimal serum dilution for the IFA,a positive serum was serially diluted from 1:2 to 1:1,024 and then tested using ASFV-infected BK2258 cells.Specific green fluorescence was observed in the ASFVinfected cells reacting with all the 10 different dilutions of positive serum,whereas green fluorescence was not detected using the negative serum,except for some background colors at the 1:2 dilution (Fig.3-A).The green fluorescence was brightest using positive serum at 1:16 dilution with much weaker background.Hence,1:16 dilution was chosen as the optimal dilution of porcine serum.Furthermore,we selected three weakly positive sera confirmed by iELISA for the IFA testing,and found that all three sera were positive for the IFA,indicating that 1:16 dilution of the serum was feasible and reliable.

Fig.3 Determination of the optimal dilutions of primary and secondary antibody.A,determination of the optimal dilution of primary antibody.Standard positive (left panel) and negative (right panel) sera were serially diluted and tested with the immunofluorescence assay (IFA).B,optimal dilution of secondary antibody.FITC-labelled rabbit anti-pig IgG was serially diluted and tested using standard ASFV positive (left panel) and negative (right panel) sera.The dilution ratio is presented in each image.
3.6.Determination of the optimal secondary antibody dilution
To select the appropriate secondary antibody dilution,different dilutions of a purchased FITC-labelled rabbit antipig IgG (Sigma,F1638) were tested.When the antibody was diluted to 1:100,specific green fluorescence with high brightness was observed in ASFV-infected cells reacting with the positive serum (Fig.3-B).However,as the dilution ratio of the secondary antibody was increased,the intensity of the specific green fluorescence in the ASFV-infected wells decreased substantially (Fig.3-B).Negative serum samples at various dilutions showed no reaction with the ASFV-infected cells (Fig.3-B).Consequently,1:100 was chosen as the optimal dilution of this secondary antibody.
3.7.Determination of the optimal incubation time for pig serum with the secondary antibody
To determine the best incubation time for pig serum and the secondary antibody with ASFV-infected cells,different incubation times (30,45,and 60 min) were tested for fluorescence intensity at a fixed temperature (37°C),respectively.Longer incubation of pig serum or the secondary antibody with ASFV-infected cells did not affect the green fluorescence intensity for the positive sera,and did not increase the background for the negative sera (Fig.4-A).Therefore,to save time and maintain the stability of the fluorescence,a 30-min incubation at 37°C for pig serum and the secondary antibody was selected as the optimal incubation time.
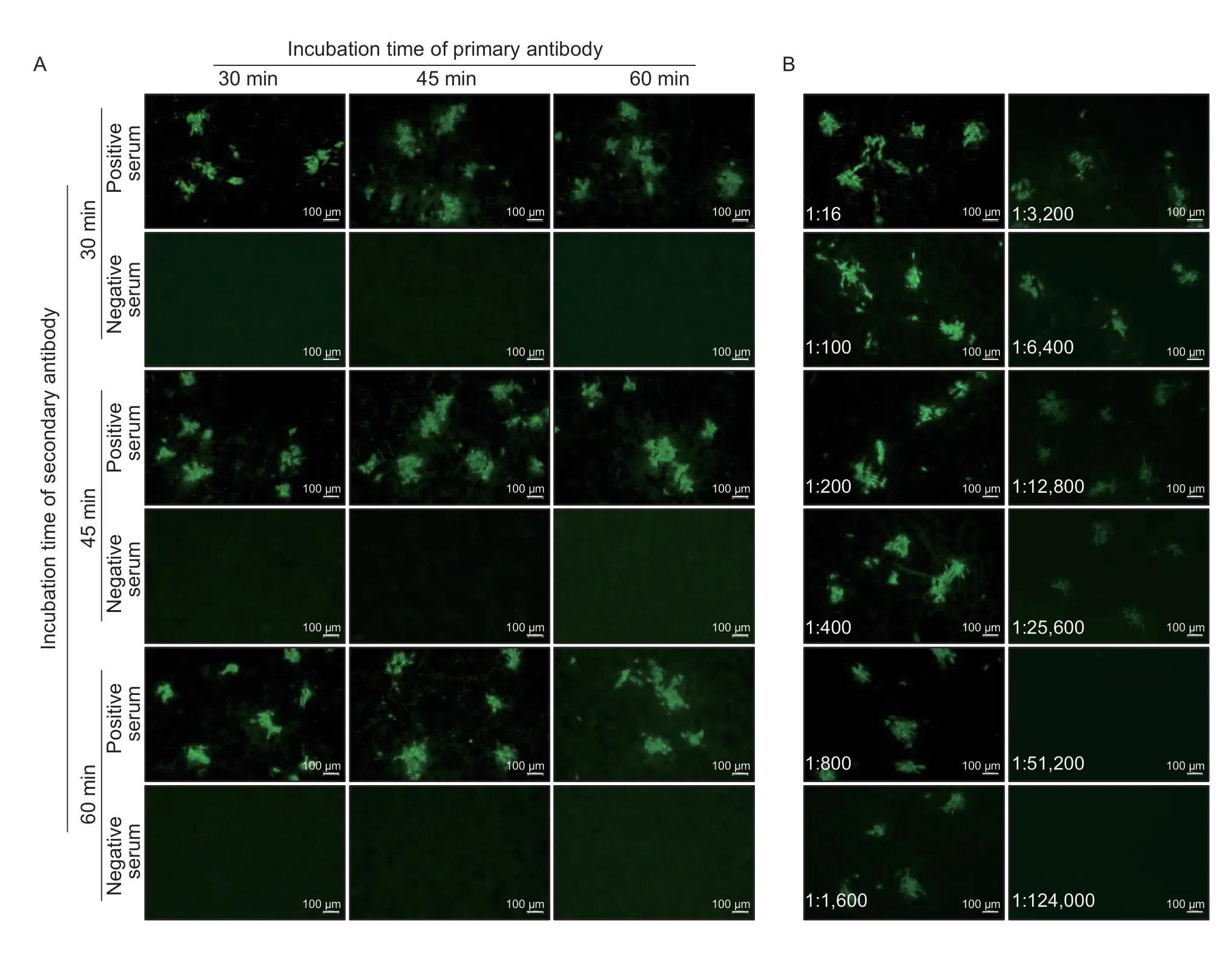
Fig.4 Determination of the optimal incubation time for the antibodies and end-point antibody titer of the immunofluorescence assay(IFA).A,determination of the optimal incubation time for the primary antibody (pig sera) and secondary antibody (FITC-labelled rabbit anti-pig IgG).B,end-point antibody titer determination for the new IFA method.A strong positive serum sample was tested in serial dilution,and found to be positive in the IFA up to the 1:25,600 dilution.The dilution ratio is presented in each image.
3.8.Analysis of the detection limit and sensitivity of the lFA
To determine the detection limit and sensitivity of the IFA,one strong ASFV positive serum was selected and evaluated with serial 2-fold dilutions side by side by using the IFA and a commercial iELISA kit.The results showed that the selected serum was still positive for the IFA at a 1:25,600 dilution (Fig.4-B) and positive for the ELISA at a 1:6,400 dilution (Table 1).Thus,the end point titers of the IFA and iELISA were 1:25,600 and 1:6,400,respectively,suggesting that the IFA was more sensitive.
We previously evaluated the pathogenicity of genotype I virus SD/DY-I/21 and genotype II virus HLJ/HRB1/20 in pigs (Sunetal.2021a,b).Here,to assess the sensitivity of the IFA,serum samples collected from seven pigs infected with SD/DY-I/21 and four pigs infected with HLJ/HRB1/20 at the indicated dpi were tested in the IFA and in a commercial iELISA kit.Our newly established IFA detected ASFV antibodies in one pig infected with SD/DYI/21 as early as 6 dpi and in all seven pigs from 7 dpi,but the iELISA kit only started to detect ASFV antibodies from 7 dpi (Table 2).For the serum samples from pigs infected with HLJ/HRB1/20,the IFA detected ASFV antibody from all four pigs from 7 dpi,but just one pig was positive on 9 dpi using the iELISA kit (Table 2).These results suggest that the IFA is more sensitive than the commercial iELISA kit for ASFV antibody detection.
3.9.Specificity test for the lFA
To confirm its specificity,the newly established IFA was used to detect 18 polyclonal swine sera against other important pig pathogens,namely,CSFV,PRRSV,PCV2,PRV,FMDV/O,and PEDV.Three positive serum samples against each pathogen did not react with the ASFVinfected cells (Fig.5),indicating that the established IFA was highly specific for ASFV antibodies.
3.10.Validation of the developed lFA using clinical samples
To further validate the IFA,a total of 479 pig serum samples were tested,including 149 serum samples from pigs immunized with the HLJ/18-7GD vaccine candidate,300 clinical serum samples collected from pig farms,and30 serum samples from SPF pigs.As expected,all serum samples from immunized pigs were found to be ASFV seropositive,and all clinical sera and serum samples from SPF pigs were ASFV seronegative.
4.Discussion
For over a century,ASF has been a major constraint to pig production.The impact of the disease is particularly felt in China,which accounts for more than half of the world’s pig population.Since neither a vaccine nor treatment is available against ASFV,control and preventive measures rely on strict biosecurity management,and early and accurate diagnosis (Ouraetal.2013;Gallardoetal.2015;Tesfagaberetal.2021;Tsegayetal.2022;Dingetal.2023).Antibody detection is particularly relevant for ASF diagnosis,as the presence of anti-ASFV antibodies is a good indicator of current or previous infection (Gallardoetal.2019;WOAH 2019).Moreover,low virulent ASFVs lead to persistent and chronic disease,and the infected pigs remain asymptomatic but shed low levels of virus that can be missed by viral nucleic acid detection methods.Chronically infected pigs maintain the virus for a long time,remain ASFV seropositive,and play an important role in the epidemiological cycle of ASF (Cubillosetal.2013;Sánchez-Vizcaínoetal.2015).In these situations,ASF can be diagnosed by specific ASFV antibody detection(Cubillosetal.2013).ASFV antibodies appear as early as 7-9 days in pigs infected with low virulent ASFVs and can persist for several months or even years (Reisetal.2007;Sánchez-Vizcaínoetal.2015).The serological testsfor ASFV antibody detection approved by WOAH include ELISA,immunoblotting test (IB),immunofluorescent assay,and indirect immuno-peroxidase test (IPT) (Sanchez-Vizcaino 1987;Pastoretal.1989;Gallardoetal.2015;Bergeronetal.2017).Among these,the IFA is considered a gold standard serological assay for confirming the diagnosis of uncertain samples detected by ELISA.
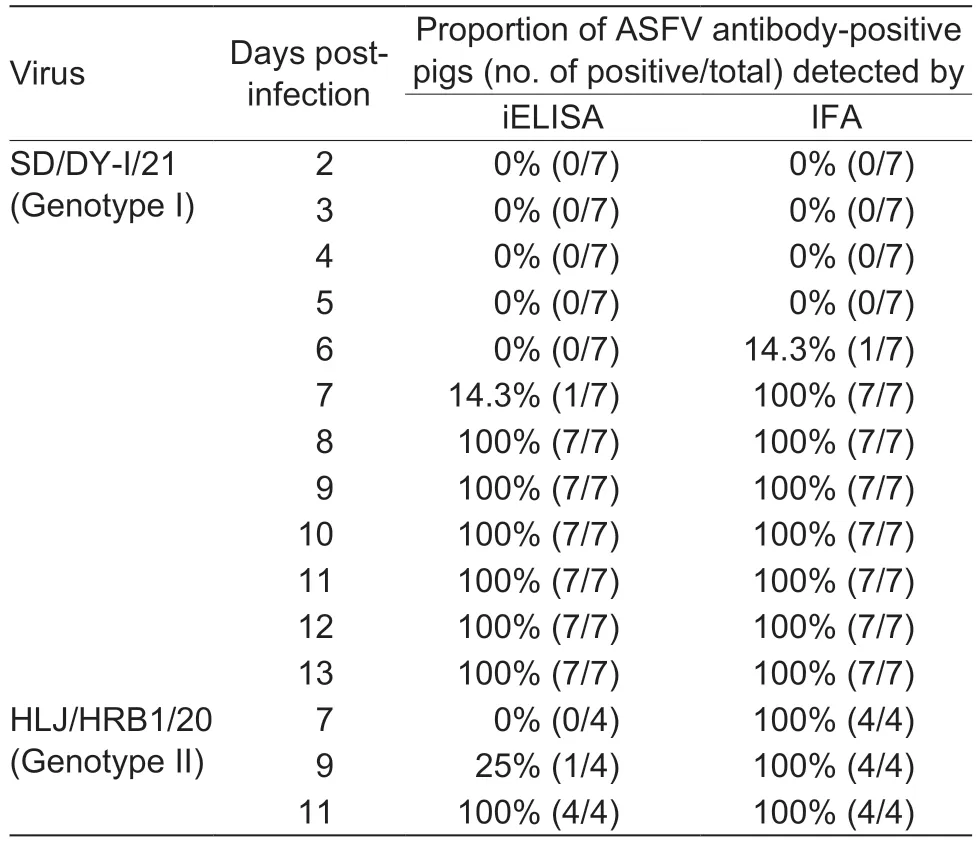
Table 2 Comparison of iELISA and the new immunofluorescence assay (IFA) for detecting African swine fever virus (ASFV)antibodies of pigs infected with low virulent ASFVs at different timepoints post-infection
The principle of IFA is based on the detection of ASFV antibodies that bind to fixed cultured Vero or monkey stable monolayer cell lines infected with celladapted ASFV (WOAH 2019).The CPE of these cells infected with ASFV was mainly characterized by rounded shrinkage of a single cell or the aggregation of several cells (Hurtadoetal.2010;de Leónetal.2013).Therefore,we prepared the wild boar cell line BK2258 that stably supports ASFV replication and forms stable CPE characterized by cell aggregation,shrinkage,lysis,and plaques.When using this cell line to establish the IFA for ASFV antibody detection,it reacted with ASFV-positive serum and formed bright clusters of fluorescence,but no fluorescence with negative serum.
The WOAH recommends a high dose of IFA virus inoculation with an MOI of 0.025-0.05 (WOAH 2019).In our study,the dose of virus inoculation was MOI of 0.001.This lower infection dose allowed a longer culture time after infection (48 h),which was conducive to the effective replication of the virus and the formation of cytological lesions.The virus used to develop the IFA was the low virulent genotype I ASFV SD/DY-I/21,and serum samples collected from pigs experimentally infected with genotype I and II ASFVs were tested.The results showed that all pigs inoculated with genotype I and II viruses were ASFV antibody-positive from 7 dpi,indicating that the new IFA method can be used for the detection of both genotype I and II ASFV antibodies.
The established IFA and a commercial iELISA kit were also used to simultaneously monitor antibody dynamics in pigs infected with genotype I and II ASFVs.According to the IFA results,one pig tested positive early on 6 dpi and the remaining 10 pigs on 7 dpi,whereas the iELISA results showed that 5 pigs tested positive at the earliest on 8 dpi and the remaining 6 pigs on 11 dpi,which indicates that the IFA could detect ASFV antibodies earlier than the iELISA.
The IFA was further evaluated using swine sera obtained from pigs immunized with the ASFV HLJ/18-7GD vaccine candidate (Chenetal.2020),SPF pigs,and field samples.All 149 serum samples from the HLJ/18-7GD-inoculated pigs were ASFV antibody positive,and the 30 serum samples from the SPF pigs were ASFV antibody negative,which further demonstrates the high specificity of the IFA.In addition,all 300 epidemiological surveillance field samples were negative,which might indicate that those pig farms are free of ASFV infection.
The present study established a new IFA with excellent sensitivity and specificity for the detection of ASFV antibodies.The newly developed IFA offers a promising approach for a rapid ASFV serological diagnosis,laying the foundation for further development of a diagnostic kit.The assay is easy to use and practicable and could be a useful tool to improve the quality of ASFV antibody detection kits.It could also be used as a gold standard and confirmatory test for inconclusive results with other ASFV serological assays.
5.Conclusion
IFA is recommended as one of the gold standard serological methods for detecting ASFV antibodies by the WOAH.This study established BK2258 cells supporting efficient ASFV replication and developed a sensitive and specific IFA for detecting ASFV antibodies.The newly established IFA was more sensitive and could detect ASFV antibodies 1-4 days earlier than the commercial iELISA.The IFA showed high specificity and had not cross-reaction with other important porcine pathogen antibodies.This IFA may play an important role in confirmed diagnosis and evaluation of different ASFV antibody detection methods.
Acknowledgements
This work was supported by the National Key R&D Program of China (2019YFE0107300 and 2021YFD1800101),the Applied Technology Research and Development Project of Heilongjiang Province,China(GA19B301),and the Central Public-interest Scientific Institution Basal Research Fund,China (1610302022003).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures involving animals were carried out in strict accordance with recommendation in the guide for the care and use of laboratory animals of the Ministry of Science and Technology of the People’s Republic of China.The protocol was approved by the committee on the ethics of animal experiments of the Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (CAAS).Experiments involving live ASFVs were carried out in a biosafety level 3 (P3) facility in HVRI and were approved by the biosafety committee of HVRI.
 Journal of Integrative Agriculture2024年1期
Journal of Integrative Agriculture2024年1期
- Journal of Integrative Agriculture的其它文章
- Advances in DNA methylation and its role in cytoplasmic male sterility in higher plants
- Assessment of molecular markers and marker-assisted selection for drought tolerance in barley (Hordeum vulgare L.)
- 3D genome organization and its study in livestock breeding
- SUPER WOMAN 2 (SPW2) maintains organ identity in spikelets by inhibiting the expression of floral homeotic genes OsMADS3,OsMADS58,OsMADS13,and DROOPING LEAF
- Genetic dissection and validation of a major QTL for grain weight on chromosome 3B in bread wheat (Triticum aestivum L.)
- Coupling of reduced inorganic fertilizer with plant-based organic fertilizer as a promising fertilizer management strategy for colored rice in tropical regions
