Dendritic Deep Learning for Medical Segmentation
Zhipeng Liu , Zhiming Zhang , Zhenyu Lei ,Masaaki Omura , Rong-Long Wang , and Shangce Gao ,,
Dear Editor,
This letter presents a novel segmentation approach that leverages dendritic neurons to tackle the challenges of medical imaging segmentation.In this study, we enhance the segmentation accuracy based on a SegNet variant including an encoder-decoder structure, an upsampling index, and a deep supervision method.Furthermore, we introduce a dendritic neuron-based convolutional block to enable nonlinear feature mapping, thereby further improving the effectiveness of our approach.The proposed method is evaluated on medical imaging segmentation datasets, and the experimental results demonstrate that it is superior to state-of-the-art methods in terms of performance.
Introduction: The significance of medical image segmentation in computer-assisted diagnosis and treatment planning cannot be overstated.It plays a crucial role in accurately identifying and outlining regions of interest, which aids in further analysis and medical decision-making.Nevertheless, the complexity and diversity of image content present challenges in the process of medical image segmentation [1].Particularly, precisely identifying regions of interest while minimizing errors and artifacts that may occur during image capture and processing can be particularly difficult.For example, the presence of tumors with unique textures compared to the surrounding tissue can impede their accurate identification and segmentation.Furthermore, it is worth noting that the existing simple feature mapping techniques in computer vision exhibit limitations in effectively capturing the intricate details and fine-grained structures present in medical images.This deficiency considerably impedes the accurate representation and analysis of the images complex nuances and subtle characteristics, consequently compromising the quality of segmentation outcomes.
In recent years, the utilization of deep learning techniques for medical image segmentation has gained substantial interest, driven by the availability of an increasing number of medical image datasets and remarkable advancements in methodologies.Fully convolutional networks (FCN) simplify image segmentation by converting it into a pixel-level classification task, but may suffer from challenges in capturing overall context and preserving fine details [2].Another widely used FCN architecture is U-Net, which incorporates skip connections to fuse high-level and low-level features, however, downsampling operations utilized in the network may result in information loss [3].Therefore, through the use of pooling indices derived from the corresponding encoder’s max pooling step, SegNet [4] can implement non-linear upsampling in the decoder, thus ensuring the effective preservation of spatial information in the original image and the accurate restoration of fine image details.Although it has exhibited noteworthy efficacy in diverse semantic segmentation tasks, it may suffer from information loss when dealing with non-uniform sampled images.
To address the limitations of current medical image segmentation approaches, various researchers have introduced novel mechanisms to enhance segmentation accuracy.Abraham and Khan [5] proposed a deeply supervised attention U-Net for tumor segmentation in BUS images.The model was enhanced with a multi-scaled input image pyramid to improve the quality of intermediate feature representations.Chenet al.[6] added six side-out deep supervision modules,enabling the network to learn to predict accurate segmentation masks at multiple scales.We have introduced a multi-scale approach for constructing the network, demonstrating the effectiveness of the training strategy involving deep supervision for addressing these challenges.
Larkum [7] emphasized the importance of dendritic structures in neural understanding, revealing limitations in the conventional neuron description that neglect dendrites’ computational properties.This omission has impeded progress in comprehending higher-level neural processes.Therefore, the utilization of dendritic neurons as a substitute for McCulloch-Pitts (MCP) neurons is both biologically interpretable and necessary.In alignment with this notion, the dendritic neuron model (DNM) has emerged as a prominent approach for classification and prediction tasks, leveraging its inherent nonlinear information processing capability [8].Gaoet al.[9] extend the DNM from a real-valued domain to a complex-valued domain and conduct an extensive evaluation of the resulting model, while we further extend the utilization of dendritic neurons to image segmentation.
Expanding on prior research, we propose a novel method called dendritic deep supervision neural network (DDNet), which combines biologically interpretable dendritic neurons with deep supervision during the training process.The use of dendritic neurons is expected to capture intricate details and fine-grained structures through nonlinear feature mapping, while deep supervision facilitates the learning of hierarchical representations and improves training efficiency and accuracy, thereby conferring supplementary advantages for medical image segmentation.The contributions of this study are summarized as follows:
1) Novel network architecture: We propose a novel network architecture that integrates dendritic neurons and a deep supervision mechanism into the SegNet framework, specifically designed to enhance medical image segmentation.
2) Improved feature representation: Our dendritic neuron-based feature extractor captures precise and informative features from medical images while enabling access to nonlinear feature mappings, further enhancing the performance of our approach.
3) Enhanced training effectiveness: Our approach incorporates a deep supervision mechanism and a customized loss function, optimizing the training process.These enhancements improve efficiency and effectiveness by providing additional guidance for feature acquisition and facilitating the capture of fine-grained details and hierarchical representations.
4) Superior performance: The effectiveness of our proposed method is demonstrated through its outperformance of existing networks in experiments conducted on several datasets.
Proposed method: As illustrated in Fig.1, the comprehensive framework comprises two fundamental components: deep supervision SegNet (DSegNet) and DNM modules.The DSegNet module employs deep supervision by utilizing the feature results from both deep and shallow layers of the SegNet variant.The final feature maps generated by the last layer of DSegNet are further processed by the DNM module to perform nonlinear feature mapping, resulting in the desired segmentation outcomes.These structures work in tandem to achieve improved medical image segmentation performance.
DSegNet: Within the proposed framework, the DSegNet module effectively captures multi-scale information for more accurate segmentation outcomes.To improve the SegNet architecture, we introduce an additional layer for deep supervision and feature enrichment.By incorporating multiple deep supervision signals (DS1,DS2,DS3,DS4, andDS5) in Fig.1 from intermediate layers and upsampled feature maps, we refine segmentation results at various scales, effectively utilizing both local and global contextual information.This deep supervision mechanism enhances the model’s capacity to handle images with irregular sampling patterns and accurately restore intricate image details.Furthermore, SegNet’s pooling indices in the decoder path preserve spatial information during upsampling.This feature ensures the faithful restoration of fine image details and robust handling of unevenly sampled images.
DNM: To further improve the effectiveness of the network, we introduce the DNM module, which leverages the unique computational properties of dendritic neurons.In Fig.1, the DsegNet output is processed by the DNM module through the synapse layer, dendritic layer, membrane layer, and soma layer.According to the final feature maps generated by the last layer of SegNet, the DNM module performs nonlinear feature mapping, enhancing the model’s ability to capture intricate details and fine-grained structures in medical images.This nonlinear mapping facilitates the extraction of more discriminative and informative features, contributing to improved segmentation accuracy.Fig.2 visually illustrates the distinct segmentation results obtained by different methods, where sub-figures located in the top three rows represent distinct segmentation for DatasetB,while the rest of the figure is the illustration of Polyp.The green and red curves represent the contours of the masks, while the white pixels indicate the predicted areas.This visual representation highlights the accurate prediction and delineation capabilities of our method for the segmented regions.

Fig.2.Segmentation results of different methods.
In contrast to prior methods, such as the utilization of proposed neurons high-order coverage function neural network (HCFNN)instead of fully connected layers [10], our method focuses on optimizing convolutions and incorporates additional preprocessing steps to enhance the utilization of the dendritic layer.Specifically, to optimize the utilization of the dendritic layer, we perform several dimension transformations on the input feature.Next, we apply normalization to the input feature, ensuring that the data falls within a suitable range for optimal processing.Following, we replicate the input feature vectors along the channel dimension to match the number of dendritic branches to achieve the desired effect of feature reuse.These modifications enable the proposed neurons to serve as replacements for neuron feature maps, expanding their role beyond simple feature extraction in fully connected layers.In addition, the learnable parameters, includingk,wij, andqi j, are randomly initialized within the range of ( 0,1).
whereZi jrepresents the output of thej-th dendritic branch for thei-th element of the inputx.The operation involves element-wise multiplication between the weightswijand the inputx, followed by subtracting the thresholdqi j.The rectified linear unit (ReLU) activation function is then applied, which clips negative values to zero, introducing non-linearity and facilitating the preservation of informative signals while suppressing noise and irrelevant information.The amplification of the resulting signal is controlled by the parameterk.Equation (1) captures the essential information integration and processing within the dendritic layer.
Then the dendritic layer receives signals from the previous synaptic layer and performs a summation operation for each dendritic branch.In this context, thej-th dendritic branch aggregatesNinput signalsZi j, resulting in the computation:
Subsequently, the membrane layer accumulates the signals from all dendritic branches through another summation operation.This layer combines the outputs ofMdendritic branches to generate a collective representation denoted as.Yrepresents the overall input integration from the dendritic layer.
Finally, the soma layer processes the output of the membrane layer using a sigmoid activation function.This function determines the firing behavior of the neuron, and the final output predictionPis given by
whereksandqsare additional learnable parameters, which are both initially randomly set within the range of (0,1).During the training phase, we employ the Adam optimizer to optimize the learnable parameters.
Loss function: A combined loss function is proposed in our framework, comprising a binary cross-entropy (BCE) loss from the final DNM module and a depth-supervised focal loss for upsampling layers
where B measures dissimilarity between predicted and ground truth labels, aiding in representation acquisition.F incorporates intermediate supervision signals, leveraging parametersαandγto handle class imbalance and prioritize challenging samples (α =0.8, γ=2).In the loss function,ndenotes the pixel count,TiandPirepresent true labels and prediction results, respectively, andλdenotes the deep supervision loss coefficient.
Experiment: This section presents experimental evaluations on three distinct datasets using four-fold cross-validation.Additionally,we performed ablation experiments and parameter analysis to enhance the evaluation of our proposed approach.The datasets employed in our experiments encompass the DatasetB (primarily designed for tumor segmentation tasks), the STU dataset (comprising a smaller tumor dataset comprising a mere 42 images), and an extensively utilized dataset for Polyp segmentation tasks.
As shown in Table 1, we compare DDNet with classical and stateof-the-art networks, among which RRCNet utilizes a deep supervision technique and employs a refinement residual convolutional network for breast tumor segmentation, MBSnet is a multi-branch medical image segmentation network that captures local and global information [11].To ensure fairness, all comparative experiments are conducted with the optimal configuration of parameters and loss functions using the same experimental setup.
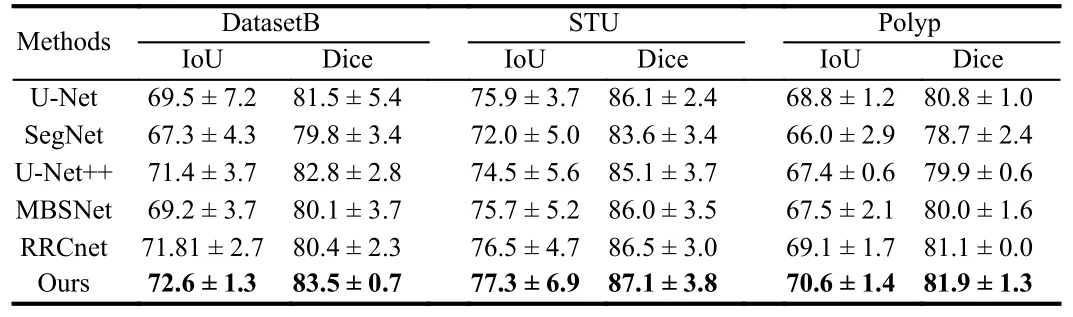
Table 1.Comparison of Results
Moreover, the results of the parameter discussion are presented in Tables 2 and 3.The optimal result is achieved when settingλto 0.1 andMto 10 in the framework.It is worth noting that we conduct separate parameter discussions on both DatasetB and the STU dataset,and the effectiveness of the chosen parameters is confirmed in each case.Additionally, we configure the epoch to 100, the batch size to 8,and the learning rate to 4E-5.Table 4 displays the outcomes of the ablation experiments performed to assess the efficacy of our proposed methodology.The objective of these experiments is to evaluate the influence of incorporating deep supervision (DS) and deep supervision with a dendritic neuron model (DS+DNM) on the performance of the SegNet baseline.Intersection over union (IoU), Dice coefficient, F1 score, and recall are adopted as evaluation metrics to quantify the enhancements achieved by integrating these components.

Table 2.Discussion on the Parameters M and λ in DatasetB
Conclusion: This investigation explores the issue of segmentation in medical images.In order to obtain better segmentation outcomes,we propose a novel network that integrates biologically interpretable dendritic neurons and employs deep supervision during training to enhance the model’s efficacy.To evaluate the effectiveness of our proposed methodology, we conducted comparative trials on datasets STU, Polyp, and DatasetB.The experiments demonstrate the superiority of our proposed approach.

Table 3.Discussion on the Parameters M and λ in STU
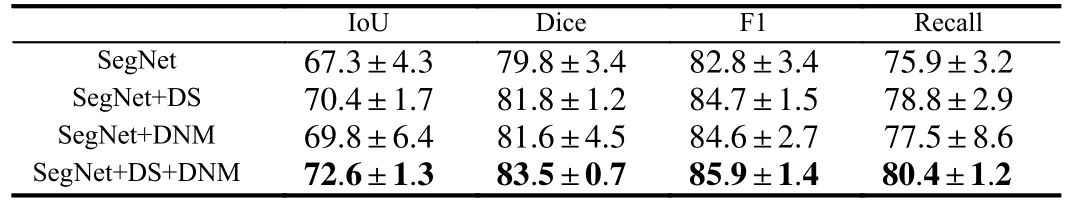
Table 4.Ablation Experiments on DatasetB
Acknowledgments: This work was partially supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI(JP22H03643), Japan Science and Technology Agency (JST) Support for Pioneering Research Initiated by the Next Generation(SPRING) (JPMJSP2145) and JST through the Establishment of University Fellowships Towards the Creation of Science Technology Innovation (JPMJFS2115).
 IEEE/CAA Journal of Automatica Sinica2024年3期
IEEE/CAA Journal of Automatica Sinica2024年3期
- IEEE/CAA Journal of Automatica Sinica的其它文章
- A Dual Closed-Loop Digital Twin Construction Method for Optimizing the Copper Disc Casting Process
- Adaptive Optimal Output Regulation of Interconnected Singularly Perturbed Systems With Application to Power Systems
- Sequential Inverse Optimal Control of Discrete-Time Systems
- More Than Lightening: A Self-Supervised Low-Light Image Enhancement Method Capable for Multiple Degradations
- Set-Membership Filtering Approach to Dynamic Event-Triggered Fault Estimation for a Class of Nonlinear Time-Varying Complex Networks
- Dynamic Event-Triggered Consensus Control for Input Constrained Multi-Agent Systems With a Designable Minimum Inter-Event Time
