Transcription of tcdA and tcdB and toxins expressionof Clostridium difficile between Chinese highisolates and European epidemic hyper-virulent strain
LI Xian-ping,LI Wen-ge,WANG Jing,LU Jin-xing
(State Key Laboratory for Infectious Disease Prevention and Control, National Institute for Communicable DiseaseControl and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, China)
Clostridiumdifficile(C.difficile) is a Gram-positive, spore-forming, obligately anaerobic bacterium. During the latest decade, the reports of cases ofClostridiumdifficileinfection (CDI) from hospital had a significant increase due to emergence of epidemic hyper-virulentC.difficile. Researches concerningC.difficileandClostridiumdifficileassociated disease (CDAD) have been paid more and more attention.C.difficilecan cause a wide spectrum of diseases, ranging from mild self-limiting diarrhea to pseudomembranous colitis and toxic megacolon[1]. AndC.difficilecan also cause diarrhea of pigs and cattle and others animals. Therefore, it is a common pathogen found in human and animals[2].
Two types ofC.difficilewere classified according to the toxins produced by bacteria: one type is non-toxigenicC.difficiletoxin typing A-B-. There is no pathogenicity locus (PaLoc) domain, and the responding gene location is replaced by 127 nucleotides in A-B-strain. This kind ofC.difficilefails to produce any toxin; the other is toxigenicC.difficilewhich contains PaLoc domain. PaLoc domain contains 19.6 k nucleotides, which responsible for toxin genestcdA,tcdBand regulatory genestcdR,tcdCandtcdE. ThetcdAgene contains 8 133 nucleotides, which encodes 308 kDa toxin A, andtcdBgene contains 7 101 nucleotides, encodes 270 kDa toxin B. ToxigenicC.difficileis also classified two toxin typing, A-B+and A+B+, according to the deletion oftcdAgene. Figure 1 showed there was a deletion of 1.8 k nucleotides in 3’ region intcdAgene of A-B+strain, some deletion is even 6 k nucleotides. One A-B+toxin typingC.difficilestrain was isolated from China-Japanese Friendship Hospital in 2008 and the entire genome was sequenced, and the strain was named BJ08. This was the first whole genome sequencing strain of A-B+toxin typingC.difficilein China and even in Asia. In this study, we compared the differences of growth rates and levels oftcdAandtcdBmRNA and productions of toxin B between strain BJ08 whose strain toxin typing is A-B+and highly isolated in China and strain UK1 which strain toxin typing is A-B+and is European epidemic hyper-virulent strain. Our study is useful for understanding the growth and toxin synthesis in strain BJ08 and for prevention and control of possible outbreaks of CDI caused by strain BJ08 in China.

Fig.1StructureschemeofC.difficilePaLocdomainresponsibleforcodingtoxins
1A represents structure scheme of strain BJ08;
1B represents structure scheme of UK1.
Letters above rectangles represent coding genes.
The numbers in rectangles represent the nucleotide numbers of coding genes.
The black arrows indicate the direction of transcription.
Materials and methods
Experimental strains
Strain BJ08 was isolated from China-Japanese Friendship Hospital and sequenced the entire genome; strain UK1 is European epidemic hyper-virulent strain which genotype is BI/NAP1/027 and isolated from the United Kingdom, and it is a kindly present from Dr. Feng Han-ping in Tufts University.
Strain growth curves, colony count and samples preparation
Strains BJ08 and UK1 were inoculated on pre-reduced CCFA agar plate in an anaerobic jar for 48 hours, respectively. Anaerobic conditions provided by the anaerobic jar (MarkII, Holland) were 80% N2, 10% H2, 10% CO2at 37 ℃. After cultivation, one colony was inoculated in 5 mL pre-reduced brain heart infusion (BHI) broth for 24 hours. Ten microliters was then added to another 5 mL pre-reduced BHI broth and incubated for 12 hours. An inoculum of 3×106mL-1was incubated in 250 mL BHI with 180 r/m shake culture at 37 ℃ anaerobic environment. Bacteria growth were measured by reading the optical density (OD) at 600 nm and counting plated cells at 0, 3, 6, 9, 12, 15, 18, 21, 24, 27 and 30 hours, respectively. At each time point, 1 mL culture was taken for measure by Cary 50Bio ultraviolet-visible spectrophotometry (Varian, USA) at OD600 nm, and serial 10-fold dilutions of the culture were plated on pre-reduced BHI blood agar and incubated for 48 hours for colony count. For each time point, samples were taken as follows: (i) 1 mL for absorbance at OD600 nm; (ii) 3-10 mL for RNA extraction according to the number of bacteria; (iii) 3-10 mL for toxin A/B ELISA test according to the number of bacteria. Samples (ii) were centrifuged for 10 min at 5 000 g, and then 0.5 mL supernatant was transferred to new 1.5 mL tubes for toxin A/B ELISA. The remaining pellet was sonicated in Ultrasonic Processor (Bandelin, Germany) for 5 min, centrifuged for 10 min at 5 000 g and the supernatant was transferred to a new 1.5 mL tube. Both supernatants were stored at -20 ℃ until toxins analysis. Samples (iii) were centrifuged for 10 min at 5 000 g and the supernatant was discarded. The pellets were stored at -80 ℃ until RNA extraction[3].
Real-time PCR reaction
The pellets were taken from -80 ℃ refrigerator, washed twice with PBS and the volume of samples was adjusted to ensure each sample contain 1×107C.difficileaccording to colony count. Total RNA were extracted from 107C.difficilecells using TianGen total RNA extraction kit (Tiangen biotech Co., Ltd., China). The extraction total RNA was stored at -80 ℃ for reverse transcription. A portion of RNA samples were used for reverse transcription reaction using TianGen first strand synthesis kit (Tiangen biotech Co., Ltd., China). cDNA synthesis was performed as prescribed in kit.
For quantification of mRNAs oftcdAandtcdB, real-time PCR was performed using ABI7500 Fast Real-Time PCR systerm (ABI, USA) and TianGen SuperReal Premix Plus (SYBR Green) (Tiangen biotech Co., Ltd., China), with 16 S as an internal control. Primers sequences, which were designed using primer express 3.0 according to the conserved region oftcdAandtcdB, were listed in Table 1. A total 20 μL reaction system was mixed per reaction as followed: 10 μL 2×SuperReal PreMix Plus; 0.5 μL primer (20 μM); 2 μL cDNA template; 0.4 μL 50×ROX and 6.6 μL sterile water. After initial 15 s denaturation at 95 ℃, PCR was running for 40 cycles: 10 s/95 ℃ denaturation and 32 s/60 ℃ elongation. To verify the specificity of amplification, a melting curve analysis was done after amplification.
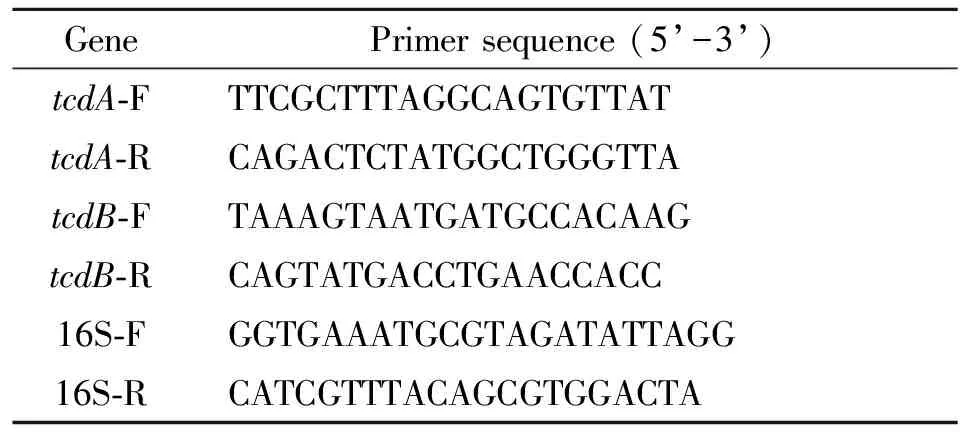
Tab.1 Primer sequences
Detection of toxin A/B in supernatants and cells by ELISA
The extracellular and intracellular supernatants were taken from -20 ℃ refrigerator, and volume of sample was adjusted to ensure each sample contain toxins of 1×107C.difficileaccording to colony count. The 50 μL supernatant was used for detection of toxin A/B by ELISA according to A/B ELISA kit (HCB, Canada). After adding to stop buffer, the OD value was measured at 450 nm by BioTek microplate reader for ELISA (BioTek, USA).
Results
Structure scheme of C. difficile PaLoc domain
There was a 1.8 k nucleotides deletion intcdAgene of BJ08 strain and an 18 nucleotides deletion intcdCgene of UK1 strain. The other genes such astcdB,tcdRandtcdEof PaLoc domain was almost the same in length between strain BJ08 and UK1. Because of high variability inC.difficilegene[4], there definitely were existing disproportionately nonsense mutations in the 5 genes of PaLoc domain.
Growth curve of B strain J08 and UK1
Figure 2 showed the similar tendency of growth curve between strain BJ08 and UK1. Both exponential phases started at 3 hours and reached stationary phase at 15 hours which lasted forapproximately 6 hours. After 21 hours, strains reached decline phase because of increase of amount ofC.difficileand of metabolic waste and toxins in culture medium, and OD values decreased significantly. The highest OD value of UK1 was significantly lower than that of BJ08. Both OD values began to rise significantly at 3 hours, but growth rate of BJ08 was faster than that of UK1. The growth rate of UK1 was significantly lower between 9 and 15 hours. While BJ08 kept higher growth rate from 3 to 15 hours. After 21 hours, both strain BJ08 and UK1 reached decline phase. And after 24 hours, the OD value of UK1 decreased significantly, while the OD value of BJ08 decreased slightly.

Fig.2ComparisonofgrowthcurvesofstrainBJ08andUK1indifferentculturedtimes
“*” representsP< 0.05, which OD value of strain BJ08 compares to that of UK1.
Transcription level of tcdA and tcdB mRNA in strain BJ08 and UK1
Though a 1.8 k nucleotides deletion existed in strain BJ08tcdA3’terminal, the transcription oftcdAmRNA was detected. To make data fromtcdAandtcdBmRNA transcription more comparable, the sample was adjusted to ensure each sample contain 107C.difficileaccording to the colony count. Figure 3A showed UK1tcdAmRNA was detected at 12 hours, while BJ08tcdAmRNA at 15 hours. The level of BJ08tcdAmRNA transcription was always lower than that of UK1 at each time point. BothtcdAmRNAs reached their peaks at 18 hours and kept higher transcription level till 30 hours. In short, the transcription level of UK1tcdAmRNA was earlier and higher than that of strain BJ08. Figure 3B showedtcdBmRNA of both strain were detected at 15 hours, and reached the highest level at 18 hours and kept higher transcription level till 30 hours. There was no significant difference in transcription level oftcdBmRNA in all time between strain UK1 and BJ08, except that of UK1 was higher than of BJ08 at 18 hours.

Fig.3ExpressionsoftcdAandtcdBmRNAindifferentculturedtimesbetweenstrainBJ08andUK1
3A shows the expressions oftcdAbetween BJ08 and UK1;
3B shows the expressions oftcdBbetween BJ08 and UK1.
“*” representsP< 0.05, the comparison oftcdAandtcdBmRNA between strain BJ08 and UK1.
ToxinsmeasurementinsupernatantsandcellsofstrainBJ08andUK1
Toxin A was neither detected in supernatants nor in cells in strain BJ08, although thetcdAmRNA of BJ08 was detected. As shown in Figure 4A, toxin A was detected in cells at 21 hours, and not until 27 hours was toxin A in supernatants detected in strain UK1. This may be limited by sensitivity of ELISA kit which detection lower limit was 0.5 pg/mL. The level of toxin A was approximately 6 times in cells than that in supernatant at 27 hours, and approximately 3.5 times at 30 hours. Figure 4B showed both toxin B in supernatants were detected at 27 hours, and the toxin B content increased significantly with incubation time. There was no significant difference in the level of toxin B between strain BJ08 and UK1 at 27 or 30 hours. Intracellular toxin B of strain BJ08 and UK1 were showed in Figure 4C. Both began to synthetize toxin B at 21 hours. The level of toxin B increased significantly with incubation time extension. There was no significant difference in level of toxin B between strain BJ08 and UK1 at each time point, from 21 to 30 hours.
Discussion
The outbreaks of CDI caused by hyper-virulent BI/NAP1/027 strain led to serious threats in health and heavy burdens in economy in western countries[5]. Data from the United States CDC show that theC.difficileinfection results in 14 000 deaths and costs $1.1 billion in the United States annually. Up to now, there is no report of CDI outbreak and isolation of hyper-virulent BI/NAP1/027 strain in mainland China. According to the limited data and our previous studies, we found ChineseC.difficileisolates have their own characteristics. One of important characteristic is that the isolating rate of A-B+typing is higher than that in the western countries, up to 23.2%-48%[6-8], while the isolating rate was only 3% in Canada[9]. Higher isolating rate of A-B+typeC.difficilein China aroused our research interest.

Fig.4ExpressionsoftoxinAandtoxinBinsupernatantsandcellsbetweenstrainBJ08andUK1
4A shows the production of toxin A in supernatants and cells of UK1;
4B shows the production of toxin B in supernatants between BJ08 and UK1;
4C shows the production of toxin B in cells between BJ08 and UK1.
“*” representsP<0.05, the comparison of intracellular and extracellular toxin A in strain UK1.
Growth curves showed the OD value of UK1 was significantly lower than that of BJ08 at the same period. That was, the growth rate of UK1 was significantly lower than that of BJ08. Does faster growth rate mean that strain BJ08 is better than UK1 in environment adoption or in pathogenicity? And whether does toxin A affect the growth rate of strain UK1? Future experiments should be performed to verify the hypothesis. No toxin A, buttcdAmRNA was detected in BJ08 strain. There were two transcription units in the PaLoc domain, the genes oftcdA,B,RandEwere in one transcription unit, andtcdCgene was in an independent transcription unit. AlthoughtcdAmRNA could not translate into toxin A in BJ08, it is probably that thetcdAmRNA expresses short peptides which involves in the virulence regulation.
In our study, we improved experimental methods of Gerber M, et al[3]. Total RNA of 107colonies was extracted at each time point. Because of the same number of bacteria, the results were comparable. The level oftcdAmRNA in strain UK1 was significantly higher than that in BJ08 strain, the reason probably lies in that strain UK1 expressed toxin A, while BJ08 does not. The level oftcdBmRNA had no significant difference between strain BJ08 and UK1 at each time point, except at 18 hours, which of UK1 was slightly higher than that of BJ08. It is thought the level of mRNA is equivalent to level of gene expression in prokaryocyte. The results showed there was no significant difference in the expression level oftcdBbetween strain BJ08 and UK1, and there was also no significant difference in intracellular and extracellular toxin B content between strain BJ08 and UK1 at each time point. The result was also confirmed by ELISA. The level of toxins increased with incubation time either in cells or in supernatant, the reasons may beC.difficilesynthesize more toxins at recession phase, which is consistent with previous study[10].
Our experiments showed that strain BJ08 fails to synthesize and secret toxin A, but toxin B produced by strain BJ08 is as much as by strain UK1. It does not mean strain BJ08 is lower in virulence and harmfulness than strain UK1 although BJ08 can’t synthesize toxin A. It is reported that the A-B+strain has higher drug resistant rate which reached 85% than A+B+strain[11]. Tracy D. Wilkins’s paper says toxin B produced by A-B+strain has more virulence than that produced by A+B+strain, because substrate specificity spectrum of toxin B from A-B+strain broadened[13-14]. And increasing number of cases of CDI outbreaks caused by A-B+strains has been reported recently. Therefore, we will compare the differences of the two strains in drug resistance and animal experiments in future experiment, and to provide the theoretical support and technical reserve for prevention and control of possible CDI outbreak caused by A-B+strain which is highly isolated in China.
[1]Voth DE, Ballard JD.Clostridiumdifficiletoxins: mechanism of action and role in disease[J]. Clin Microbiol Rev, 2005, 18(2): 247-263.
[2]Cheng Y, Lu JX, Yan SK, et al. Primary study on the gene typing, molecular characteristics of virulence and resistance associated gene of 12Clostridiumdifficileclinical isolates in China[J]. Chin J Zoonoses, 2009, 25(05): 401-405. (in English)
[3]Gerber M, Walch C, Löffler B, et al. Effect of sub-MIC concentrations of metronidazole vancomycin, clindamycin and linezolid on toxin gene transcription and production inClostridiumdifficile[J]. J Med Microbiol, 2008, 57: 776-783. DOI: 10.1099/jmm.0.47739-0
[4]Sebaihia M, Wren BW, Mullany P, et al. The multidrug-resistant human pathogenClostridiumdifficilehas a highly mobile, mosaic genome[J]. Nat Gene, 2006, 38(7): 779-786.
[5]McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene-variant strain ofClostridiumdifficile[J]. N Engl J Med, 2005, 353(23): 2433-2441.
[6]Hawkey PM, Marriott C, Liu WE, et al. Molecular epidemiology ofClostridiumdifficileinfection in a major Chinese hospital: an underrecognized problem in Asia? [J]. J Clin Microbiol, 2013, 51(10): 3308-3313. DOI: 10.1128/JCM.00587-13
[7]Huang H, Wu S, Wang M, et al.Clostridiumdifficileinfections in a Shanghai hospital: antimicrobial resistance, toxin profiles and ribotypes[J]. Int J Antimicrob Agents, 2009, 33(4): 339-342. DOI: 10.1016/j.ijantimicag.2008.09.022
[8]Cheng Y, Du P, Chen C, et al. Toxin A-negative, toxin B-positiveClostridiumdifficileinfection diagnosed by polymerase chain reaction[J]. Infect Control Hosp Epidemiol, 2011, 32(5): 520-522. DOI: 10.1086/659959
[9]Martin H, Willey B, Low DE, et al. Characterization ofClostridiumdifficilestrains isolated from patients in Ontario, Canada, from 2004 to 2006[J]. J Clin Microbiol, 2008, 46(9): 2999-3004. DOI: 10.1128/ JCM.02437-07
[10]Hundsberger T, Braun V, Weidmann M, et al. Transcription analysis of the genes tcdA-E of the pathogenicity locus ofClostridiumdifficile[J]. Eur J Biochem, 1997, 244(3): 735-742.
[11]van den Berg RJ, Claas EC, Oyib DH, et al. Characterization of toxin A-negative, toxin B-positiveClostridiumdifficileisolates from outbreaks in different countries by amplified fragment length polymorphism and PCR ribotyping[J]. J Clin Microbiol, 2004, 42(3): 1035-1041.
[12]Wilkins TD, Lyerly DM.Clostridiumdifficiletesting: after 20 years, still challenging[J]. J Clin Microbiol, 2003, 41(2): 531-534.
[13]Drudy D, Harnedy N, Fanning S, et al. Isolation and characterization of toxin A-negative, toxin B-positiveClostridiumdifficilein Dublin, Ireland[J]. Clin Microbiol Infect, 2007, 13(3): 298-304.
[14]Shin BM, Kuak EY, Yoo SJ, et al. Emerging toxin A-B+ variant strain ofClostridiumdifficileresponsible for pseudomembranous colitis at a tertiary care hospital in Korea[J]. Diagn Microbiol Infect Dis, 2008, 60(4): 333-337.

