病毒蛋白脂酰化及其功能
刘红,叶荣
复旦大学基础医学院,上海 200032
脂酰化(fatty acylation)是蛋白修饰的重要方式之一,这种修饰可以是静态的,也可以是动态的,修饰后蛋白的功能呈现多样性。近年来随着方法学的进步,人们对脂酰化分子机制的认识也逐渐清晰[1,2]。蛋白的脂酰化修饰主要有棕榈酰化(palmitoylation)、豆蔻酰化(myristoylation)、异戊烯化(prenylation)和糖基化磷脂酰肌醇(glycosylphosphatidylinositol, GPI)共价结合4种方式。脂酰化可发生在蛋白合成过程中或合成完成后,修饰位点可在蛋白的N端、C端或中间[3]。脂酰化的蛋白通常定位于细胞膜结构附近,这是因为与脂类共价结合导致蛋白与膜的亲和力增加,去除这些脂质基团会影响这些修饰后蛋白的生物学功能。脂质基团在蛋白上形成特定的脂质锚,随后插入膜结构的磷脂双分子层内,从而稳定蛋白与膜的结合,也有利于将蛋白导入一些特定亚细胞的膜结构域如脂筏等发挥其功能[4]。病毒在宿主细胞内的复制过程中,一些病毒蛋白发生脂酰化,这些修饰促进病毒识别及进入细胞,帮助病毒蛋白在细胞内运输及定位,从而有利于病毒复制和装配。一些重要病毒蛋白的脂酰化方式及其生物学功能的变化见图1。
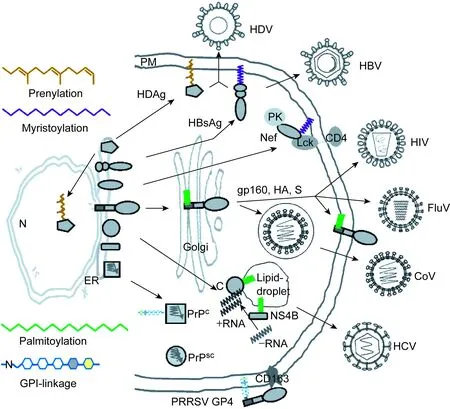
图1 重要病毒蛋白的脂酰化形式Fig.1 Diagram summary of the fatty acylation of viral proteins
1 Ⅰ型病毒膜蛋白的棕榈酰化
棕榈酰化通常发生在含天冬氨酸-组氨酸-组氨酸-半胱氨酸(Asp-His-His-Cys,DHHC)保守序列的Cys富集区(cysteine-rich domain, CRD)。含DHHC-CRD结构的蛋白广泛存在于从低等真核生物如酵母到包括人在内的哺乳动物细胞中,目前已从人细胞中分离鉴定出23种含类似结构的蛋白[5]。蛋白的棕榈酰化具有可逆性,蛋白在棕榈酰基转移酶(palmitoylacyl transferase, PAT)作用下发生棕榈酰化,转运至不同的膜结构,然后在棕榈酰蛋白硫酯酶(palmitoyl protein thioesterase, PPT)作用下去棕榈酰化,通过这种循环实现对蛋白功能的不同调控[3,6]。大部分含DHHC-CRD结构的蛋白具有PAT活性,兼具酶和底物的特性[6,7]。靠近跨膜区的Cys棕榈酰化可增强跨膜区的疏水性,使跨膜蛋白容易定位至脂筏[8]。最常发生棕榈酰化的Ⅰ型病毒膜蛋白包括流感病毒(influenza virus, FluV)的血凝素(hemagglutinin, HA)、人类免疫缺陷病毒(human immunodeficiency virus, HIV)的包膜蛋白(envelope, Env)、冠状病毒(coronavirus, CoV)的棘突蛋白(spike, S)等[9]。此外,副黏病毒F蛋白、披膜病毒包膜糖蛋白(envelope glycoprotein, gp)、杆状病毒gp64等也可发生棕榈酰化[10-12]。
流感病毒HA的棕榈酰化研究开展得最早、最广泛。其前体分子HA0由细胞蛋白酶裂解为N端HA1和C端HA2[13]。HA的棕榈酰化位点为Cys残基。甲型流感病毒有2个Cys残基位于胞质尾区(cytoplasmic tail, CT)、1个位于跨膜区(transmembrane domain, TMD);乙型流感病毒HA 的2个Cys残基均位于胞质尾区;丙型流感病毒HA-酯酶融合糖蛋白(HA-esterase fusion glycoprotein, HEF)只有1个Cys残基,位于TMD与胞质尾区之间。这些位点突变导致感染性病毒颗粒产生明显减少[14-16]。 HA棕榈酰化有助于定位脂筏,一些亚型的HA如H1、H2和H7等出现在富含鞘磷脂和胆固醇的特异性膜结构域[17,18]。脂筏是病毒成熟相关蛋白的集聚位点,也是病毒蛋白激活细胞信号转导的平台,因此脂筏中除了HA,其他流感病毒蛋白也常富集于此[19,20]。HA通常在内质网(endoplasmic reticulum, ER)发生棕榈酰化,然后装配入病毒颗粒,与基质蛋白1相互作用参与病毒的出芽过程[21]。流感病毒不能形成合胞体,表明HA介导的细胞-细胞融合受到影响,可能与HA的去脂酰化有关[22]。研究还发现,甲型流感病毒H1、H7和乙型流感病毒HA的棕榈酰化,对病毒感染过程中膜融合孔的形成和增大及水性融合通道的形成有明显促进作用[23-26]。
HIV Env被细胞蛋白酶裂解为2个亚单位:gp120和gp41。gp120与受体结合,gp41具有包括膜融合在内的多种功能[27]。gp41胞质尾区含有2个棕榈酰化位点:Cys-764和Cys-837[28,29]。其中1个位点突变成Ser,HIV-1感染性降低2倍;2个位点同时突变成Ser,HIV-1感染性则降低60倍[30,31]。病毒装配出芽过程中,要求gp160胞质区与Gag相互作用,单个棕榈酰化位点突变对Env-Gag的相互作用影响很小,双位点同时突变导致Env-Gag相互作用减弱,从而影响Env特异性装配至病毒颗粒[28,32]。然而,棕榈酰化在gp160表达、细胞内运输及细胞-细胞膜融合过程中的作用仍未完全阐明,有待进一步研究[29]。用β-甲基环糊精除去细胞的胆固醇后,HIV-1不但感染性降低,而且合胞体的形成减少[33-35]。HIV病毒颗粒的组装及出芽依赖脂筏结构,棕榈酰化的Env在细胞膜内侧锚定,但2个棕榈酰化位点缺失后,Env失去了脂筏定位能力,不能抵抗非离子去垢剂的作用[29]。
冠状病毒棘突蛋白为典型的Ⅰ型跨膜糖蛋白,以三聚体的形式发挥生物学功能,不仅决定冠状病毒的宿主特异性,还介导病毒与细胞膜融合[36]。棘突蛋白膜内区有1个含8~10个Cys的保守区域,容易发生棕榈酰基修饰[37]。严重急性呼吸综合征(severe acute respiratory syndrome,SARS)冠状病毒和鼠冠状病毒的棘突蛋白发生棕榈酰化后,棘突蛋白介导的细胞-细胞融合、病毒感染性及病毒颗粒装配受影响[38-40]。重组鼠冠状病毒表达和羟胺处理实验发现,棘突蛋白的棕榈酰化要求膜内区3个以上Cys参与,棕榈酰化有利于其定位至脂筏[41]。棕榈酰化可能为棘突蛋白提供受体结合和膜融合所必要的膜锚定力,从而增强病毒的感染性。
2 丙型肝炎病毒NS4B与核心蛋白的棕榈酰化
丙型肝炎病毒(hepatitis C virus, HCV)非结构蛋白NS4B含有261个氨基酸,是高度疏水的跨膜蛋白,参与基因组复制及病毒颗粒的组装和释放[42]。NS4B在HCV RNA复制复合体的形成过程中,C端膜内区发生棕榈酰化,用棕榈酰化抑制剂2-溴代棕榈酸盐处理后RNA复制明显降低[43]。用蛋白转运抑制剂布雷菲德菌素A(brefeldin A)破坏高尔基复合体,NS4B棕榈酰化并不受影响,表明修饰发生在蛋白进入高尔基复合体之前[43]。NS4B 的Cys-257是主要棕榈酰化位点,但HCV 基因型3、5和6的NS4B第257位不是Cys,而是Thr或Tyr[43]。所有HCV基因型NS4B末端的Cys-261均高度保守,其突变可明显减弱病毒RNA复制。但由于Cys-261也是NS3蛋白酶的识别位点,这些功能减低与棕榈酰化的关系仍无法确定。另外,Cys-257和Cys-261同时突变时,NS4B本身聚合作用及其与NS5A或其他蛋白的相互作用明显减弱[44]。
HCV核心蛋白由191个氨基酸残基组成,在含脂滴的内质网上与病毒RNA结合形成核衣壳颗粒[45]。靠近核心蛋白C端疏水部位的Cys-172是主要棕榈酰化位点,该位点突变后引起核心蛋白与脂滴相互作用减弱,导致内质网上核心蛋白减少,形成的病毒颗粒也减少[46]。用信号肽酶裂解核心蛋白使疏水尾部减少至4个氨基酸残基,可导致其与内质网结合不稳定,而棕榈酰化有利于病毒核心与内质网的稳定结合[47]。
3 HIV Nef蛋白和乙型肝炎病毒L蛋白的豆蔻酰化
发生豆蔻酰化的蛋白通常含有特异性保守序列Gly-X-X-X-Ser/Thr,且Gly后通常为1个Cys残基,修饰的豆蔻酰基团从豆蔻酰CoA获得,由N-豆蔻酰转移酶(N-myristoyltransferase, NMT)催化[48]。NMT分为NMT1和NMT2两种,包括1个催化结构域和1个N端结构域,后者可特异性结合至核糖体[49]。蛋白的N端如果集中出现碱性氨基酸残基,可作为蛋白豆蔻酰化的附加信号。这些碱性氨基酸残基与磷脂头部带负电荷的基团发生静电作用,豆蔻酰基作为“静电开关”调控该蛋白表面的正电荷,从而改变蛋白与膜的亲和力[2]。发生豆蔻酰化的病毒蛋白有多种,其中HIV-1的Nef蛋白和乙型肝炎病毒(hepatitis B virus,HBV)的L蛋白豆蔻酰化对病毒的复制和组装是必需的。
HIV-1 Nef是相对分子质量为27 000的磷酸化蛋白,早期被认为是负调控因子,与细胞的膜结构结合从而抑制病毒基因组表达[50]。后期研究发现Nef具有增强病毒复制的能力,用特异性抑制剂阻断其N端结构域与核糖体的结合可抑制HIV-1颗粒的产生[51]。Nef豆蔻酰化由NMT2催化,其N端Thr通过静电作用首先结合至双层脂质膜一侧表面,再与肉豆蔻酸一起进入膜内。此时Nef的构象并没有变化,而NMT的构象发生明显变化[51]。Nef发生豆蔻酰化时,其N端区域也需Arg和Lys等碱性氨基酸残基及至少1个豆蔻酰化特异性基序,其中Lys被认为是Nef进入脂筏的关键因素[52]。然而,相对于细胞蛋白,Nef豆蔻酰化的作用较弱,表明豆蔻酰化不是Nef与细胞膜结合的主要因素[53]。Nef豆蔻酰化也可影响细胞内蛋白功能来增强病毒复制(如下调细胞表面CD4,病毒出芽时与CD4结合受阻),从而影响感染性病毒颗粒的形成[54]。Nef还可下调细胞表面主要组织相容性复合体(major histocompatibility complex,MHC)Ⅰ类分子,促进HIV-1免疫逃避,介导细胞内信号转导及T细胞活化,增加病毒颗粒产生[55]。
另外,豆蔻酰化的Nef可优先激活巨噬细胞。与肿瘤坏死因子α(tumor necrosis factor α,TNF-α)相似,可溶性Nef激活丝裂原活化蛋白激酶(mitogen-activated protein kinase,MAPK)/核因子κB(nuclear factor κB,NF-κB)信号途径,刺激产生炎性细胞因子和趋化因子,如白细胞介素1β(interleukin 1β,IL-1β)、IL-6、TNF-α、巨噬细胞炎性蛋白1α(macrophage inflammatory protein 1α,MIP-1α)、MIP-1β等,激活巨噬细胞[56]。G2A突变后,Nef无豆蔻酰化,不能激活MAPK/NF-κB信号途径,导致CD163表达及巨噬细胞活化降低[56,57]。Nef激活巨噬细胞后形成纳米管,并与MHCⅡ、TGN蛋白及SH3蛋白等结合,诱导抗炎因子产生。Nef进入B细胞,使其分化因子增加,从而干扰生发中心因子的产生,与HIV-1导致的淋巴瘤发生有关[58]。
HBV外膜蛋白由S、M和L组成。L比M多了preS1序列,含21~47个氨基酸残基。HBV外膜蛋白也参与包装丁型肝炎病毒(hepatitis D virus, HDV),preS1序列是与肝细胞受体结合位点的可能区域[59,60]。最近发现牛磺胆酸钠共转运肽(sodium taurocholate cotransporting polypeptide, NTCP)作为HBV和HDV的功能性受体,能特异性结合该区域[61]。早期研究发现,preS1豆蔻酰化对病毒出芽无明显影响,将豆蔻酰化信号人工缺失,仍能产生与野毒株形状无明显差别的病毒颗粒[62]。后来才发现,preS1 N端的Gly突变使结合至宿主细胞膜上的L蛋白减少,导致病毒丧失感染性,但Gly豆蔻酰化并不是病毒装配所必需的,提示preS1的豆蔻酰化是产生有感染性HBV的决定性因素[63]。HBV与细胞膜的结合主要位于preS1的第9~18位氨基酸残基,第28~48位氨基酸残基具有辅助病毒结合的功能,豆蔻酰化发生在N端Gly残基[63,64]。preS1豆蔻酰化后构象发生改变,蛋白结合至细胞膜,从而影响宿主细胞的脂质双分子层或表面成分,促使病毒结合和定位至宿主细胞上,病毒颗粒与HBV受体的亲和性明显增加[64]。最近有研究发现,豆蔻酰化信号缺失的蛋白也能促进中和抗体产生,为HBV疫苗研制提供了新思路[65]。
此外,乳头瘤病毒VP2、小RNA病毒VP4、呼肠孤病毒μ1蛋白也常发生豆蔻酰化[66-68]。猪繁殖与呼吸综合征病毒(porcine reproductive and respiratory syndrome virus, PRRSV)、沙粒病毒及蛙病毒的一些结构和非结构蛋白也具有不同程度的豆蔻酰化[69-71]。这些蛋白豆蔻酰化主要影响病毒进入细胞的过程和病毒颗粒的装配。
4 HDV L蛋白的异戊烯化
1978年,异戊烯化作为参与酵母杂交的一个主要因素被首次证明,随后发现哺乳动物细胞蛋白和HDV L蛋白也存在这种修饰[72-74]。异戊烯化包括15-C的法尼酰基化(farnesylation)和20-C的双牻牛儿酰基化(geranylgeranylation),通常发生在蛋白C端特异性序列CAAX(C,半胱氨酸;A,脂肪族氨基酸;X,任意氨基酸)的Cys残基,而X残基的性质决定发生法尼酰基化还是双牻牛儿酰基化[75]。异戊烯基通过硫酯键连接至Cys残基的过程由异戊烯基转移酶催化,通常不可逆。蛋白的异戊烯化对其膜结合功能至关重要,如阻断Ras蛋白法尼酰基化可逆转Ras转化细胞的功能[76]。此外,异戊烯化也可介导蛋白-蛋白间相互作用,并在蛋白运输中发挥作用[77]。
HDV包膜与HBV相同,核心部分由HDV RNA和HDV抗原(HDV antigen,HDAg)组成,其中大HDAg(Large HDAg, L-HDAg)C端包含异戊烯化位点CXXX(C,半胱氨酸;X,任意氨基酸),由法尼酰基转移酶和双牻牛儿酰基转移酶Ⅰ催化,另一些能被双牻牛儿酰基转移酶Ⅱ催化,其识别的序列不同[78]。HDV颗粒的装配需HBV表面抗原(HBV surface antigen,HBsAg)与L-HDAg相互作用,异戊烯修饰的L-HDAg易运输至内质网膜上,同时L-HDAg的异戊烯化也反馈抑制HDV RNA的合成,促使HDV RNA与L-HDAg一起装配成HDV颗粒[79]。根据HDAg异戊烯化设计抑制剂,可干扰HDV组装,目前已用于临床治疗慢性丁型肝炎[80,81]。
5 朊蛋白与GPI共价结合
GPI与蛋白质共价结合是一种间接脂酰化修饰,修饰的蛋白通常附着于细胞膜的外侧,参与膜蛋白转运、信息传递、细胞黏附和补体调节等作用。哺乳动物有150多种蛋白以此种方式与膜结合,包括水解酶、受体、黏附分子、补体抑制因子和功能不详的表面抗原等[82]。GPI由糖基化的磷脂和乙醇胺残基组成,排列复杂,被其修饰的蛋白以糖基-磷脂酰-肌醇的顺序锚定于细胞膜外侧。如果用特异性磷脂酶C处理细胞表面,GPI锚定的蛋白被释放出来[83]。GPI锚定的蛋白通常在N端有信号序列,在C端有疏水序列[84]。最早证明发生GPI修饰的病毒蛋白为登革病毒NS1[85]。最近发现,PRRSV GP4 C端的GPI结合位点突变后,其不能进入脂筏,从而不能与受体CD163结合,导致病毒丧失感染性[86]。
朊病毒(prion)感染导致脑神经元退化,其主要成分是朊蛋白(prion protein,PrP),由正常细胞编码的PrP前体蛋白称为细胞型朊蛋白(cellular prion protein,PrPc),不致病,PrPc的异构体羊痒疫朊蛋白(scrapie prion protein,PrPsc)为其致病形式[87]。PrPc是一种糖基化膜蛋白,包括N端信号肽序列、中间高度保守疏水区和C端GPI锚定区。PrPc在核糖体合成后被转运至粗面内质网和高尔基复合体内进行糖基化,然后在高尔基复合体上与GPI饱和酰基及脂筏内饱和鞘脂类结合,最后转运定位至细胞膜[88]。成熟的朊蛋白通常以与脂筏结合的形式存在,但未糖基化的PrPc前体在内质网也能与脂筏结合,这种结合能促进蛋白的折叠[89]。PrPsc中GPI锚的生物学作用很难用实验证实,因为PrPsc的聚合形式保护GPI不被磷脂酶C消化。人们首次证明GPI锚定与PrPsc致病性的关系,是用缺失特定GPI锚定元件的转基因小鼠产生大量PrPsc蛋白,但发现这些PrPsc蛋白并未引起动物产生与羊痒疫相似的临床症状[90]。GPI锚通过改变膜结构及胞质内磷脂变化,致使PrPc交联或PrPsc聚集,引起突触发生变性,导致海绵样病变。磷脂酶的消化会导致PrPc的交联形式消失[91]。因为朊病毒引起突触变性取决于其GPI锚组成,提示可针对GPI锚建立治疗朊病毒感染疾病的新策略。
6 结语
脂酰化作为一种常见的蛋白修饰方式,参与细胞内多种蛋白的膜相关功能的发挥;但长期以来由于方法上的限制,该研究进展缓慢。近年来,新的标记和标记方法的出现,脂酰化研究越来越受到重视,其中脂酰化蛋白组学的开展为鉴定被修饰蛋白及参与修饰反应的酶具有重要推进作用。病毒感染过程中合成蛋白的膜结合能力对病毒的复制与致病性至关重要,尤其是一些包膜病毒的装配。近年来,PrPsc和HDAg脂酰化修饰研究为相关药物开发提供了新靶点和新思路。同样,一些病毒蛋白有明确的结构和功能特点,也是研究脂酰化的良好模型,可加快脂酰化机制的阐明。
[1] Hannoush RN, Sun J. The chemical toolbox for monitoring protein fatty acylation and prenylation [J]. Nat Chem Biol, 2010, 6(7): 498-506.
[2] Triffo SB, Huang HH, Smith AW, Chou ET, Groves JT. Monitoring lipid anchor organization in cell membranes by PIE-FCCS [J]. J Am Chem Soc, 2012, 134(26): 10833-10842.
[3] Tom CT, Martin BR. Fat chance! Getting a grip on a slippery modification [J]. ACS Chem Biol, 2013, 8(1): 46-57.
[4] Steinhauer J, Treisman JE. Lipid-modified morphogens: functions of fats [J]. Curr Opin Genet Dev, 2009, 19(4): 308-314.
[5] Korycka J1,ach A, Heger E, Bogusawska DM, Wolny M, Toporkiewicz M, Augoff K, Korzeniewski J, Sikorski AF.Human DHHC proteins: a spotlight on the hidden player of palmitoylation [J]. Eur J Cell Biol, 2012, 91(2): 107-117.
[6] Guan XM, Fierke CA. Understanding protein palmitoylation: Biological significance and enzymology [J]. Sci China Chem, 2011, 54(12): 1888-1897.
[7] Linder ME, Jennings BC. Mechanism and function of DHHC S-acyltransferases [J]. Biochem Soc Trans, 2013, 41(1): 29-34.
[8] Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, Waldmann H. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins [J]. Cell, 2010, 141(3): 458-471.
[9] Veit M. Palmitoylation of virus proteins [J]. Biol Cell, 2012, 104(9): 493-515.
[10] Branigan PJ, Day ND, Liu C, Gutshall LL, Melero JA, Sarisky RT, Del Vecchio AM.The cytoplasmic domain of the F protein of human respiratory syncytial virus is not required for cell fusion [J]. J Gen Virol, 2006, 87(Pt 2): 395-398.
[11] Smit JM, Bittman R, Wilschut J. Deacylation of the transmembrane domains of Sindbis virus envelope glycoproteins E1 and E2 does not affect low-pH-induced viral membrane fusion activity [J]. FEBS Lett, 2001, 498(1): 57-61.
[12] Zhang SX, Han Y, Blissard GW. Palmitoylation of the Autographa californica multicapsid nucleopolyhedrovirus envelope glycoprotein GP64: mapping, functional studies, and lipid rafts [J]. J Virol, 2003, 77(11): 6265-6273.
[13] Luo M. Influenza virus entry [J]. Adv Exp Med Biol, 2012, 726: 201-221.
[14] Zurcher T, Luo G, Palese P. Mutations at palmitylation sites of the influenza virus hemagglutinin affect virus formation [J]. J Virol, 1994, 68(9): 5748-5754.
[15] Ujike M, Nakajima K, Nobusawa E. Influence of acylation sites of influenza B virus hemagglutinin on fusion pore formation and dilation [J]. J Virol, 2004, 78(21): 11536-11543.
[16] Kordyukova LV, Serebryakova MV, Baratova L A, Veit M. S acylation of the hemagglutinin of influenza viruses: mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine [J]. J Virol, 2008, 82(18): 9288-9292.
[17] Zhang J, Pekosz A, Lamb RA. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins [J]. J Virol, 2000, 74(10): 4634-4644.
[18] Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane [J]. J Biol Chem, 1999, 274(4): 2038-2044.
[19] Barman S, Ali A, Hui EK, Adhikary L, Nayak DP. Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses [J]. Virus Res, 2001, 77(1): 61-69.
[20] Takeda M, Leser GP, Russell CJ, Lamb RA. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion [J]. Proc Natl Acad Sci USA, 2003, 100(25): 14610-14617.
[21] Veit M, Schmidt MF. Timing of palmitoylation of influenza virus hemagglutinin [J]. FEBS Lett, 1993, 336(2): 243-247.
[22] Naeve CW, Williams D. Fatty acids on the A/Japan/305/57 influenza virus hemagglutinin have a role in membrane fusion [J]. EMBO J, 1990, 9(12): 3857-3866.
[23] Wagner R, Herwig A, Azzouz N, Klenk HD. Acylation-mediated membrane anchoring of avian influenza virus hemagglutinin is essential for fusion pore formation and virus infectivity [J]. J Virol, 2005, 79(10): 6449-6458.
[24] Armstrong RT, Kushnir AS, White JM. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition [J]. J Cell Biol, 2000, 151(2): 425-437.
[25] Kozerski C1, Ponimaskin E, Schroth-Diez B, Schmidt MF, Herrmann A. Modification of the cytoplasmic domain of influenza virus hemagglutinin affects enlargement of the fusion pore [J]. J Virol, 2000, 74(16): 7529-7537.
[26] Sakai T, Ohuchi R, Ohuchi M. Fatty acids on the A/USSR/77 influenza virus hemagglutinin facilitate the transition from hemifusion to fusion pore formation [J]. J Virol, 2002, 76(9): 4603-4611.
[27] Checkley MA, Luttge BG, Freed EO. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation [J]. J Mol Biol, 2011, 410(4): 582-608.
[28] Yang C, Spies CP, Compans RW. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated [J]. Proc Natl Acad Sci USA, 1995, 92(21): 9871-9875.
[29] Rousso I, Mixon MB, Chen BK, Kim PS. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity [J]. Proc Natl Acad Sci USA, 2000, 97(25): 13523-13525.
[30] Bhattacharya J, Peters PJ, Clapham PR. Human immuno-deficiency virus type 1 envelope glycoproteins that lack cytoplasmic domain cysteines: impact on association with membrane lipid rafts and incorporation onto budding virus particles [J]. J Virol, 2004, 78(10): 5500-5506.
[31] Chan WE, Lin HH, Chen SS. Wild-type-like viral replication potential of human immunodeficiency virus type 1 envelope mutants lacking palmitoylation signals [J]. J Virol, 2005, 79(13): 8374-8387.
[32] Yu X, Yuan X, McLane MF, Lee TH, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions [J]. J Virol, 1993, 67(1): 213-221.
[33] Liao Z, Cimakasky LM, Hampton R, Nguyen DH, Hildreth JE. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1 [J]. AIDS Res Hum Retroviruses, 2001, 17(11): 1009-1019.
[34] Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release [J]. Proc Natl Acad Sci USA, 2001, 98(24): 13925-13930.
[35] Viard M, Parolini I, Sargiacomo M, Fecchi K, Ramoni C, Ablan S, Ruscetti FW, Wang JM, Blumenthal R. Role of cholesterol in human immunodeficiency virus type 1 envelope protein-mediated fusion with host cells [J]. J Virol, 2002, 76(22): 11584-11595.
[36] Masters PS. The molecular biology of coronaviruses [J]. Adv Virus Res, 2006, 66: 193-292.
[37] Shulla A, Gallagher T. Role of spike protein endodomains in regulating coronavirus entry [J]. J Biol Chem, 2009, 284(47): 32725-32734.
[38] Petit CM, Chouljenko VN, Iyer A, Colgrove R, Farzan M, Knipe DM, Kousoulas KG. Palmitoylation of the cysteine-rich endodomain of the SARS-coronavirus spike glycoprotein is important for spike-mediated cell fusion [J]. Virology, 2007, 360(2): 264-274.
[39] McBride CE, Machamer CE. Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein [J]. Virology, 2010, 405(1): 139-148.
[40] Thorp EB, Boscarino JA, Logan HL, Goletz JT, Gallagher TM. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity [J]. J Virol, 2006, 80(3): 1280-1289.
[41] Yang J, Lv J, Wang Y, Gao S, Yao Q, Qu D, Ye R. Replication of murine coronavirus requires multiple cysteines in the endodomain of spike protein [J]. Virology, 2012, 427(2): 98-106.
[42] Li S, Yu X, Guo Y, Kong L. Interaction networks of hepatitis C virus NS4B: implications for antiviral therapy [J]. Cell Microbiol, 2012, 14(7): 994-1002.
[43] Yu G Y, Lee KJ, Gao L, Lai MM. Palmitoylation and polymerization of hepatitis C virus NS4B protein [J]. J Virol, 2006, 80(12): 6013-6023.
[44] Gouttenoire J, Penin F, Moradpour D. Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory [J]. Rev Med Virol, 2010, 20(2): 117-129.
[45] Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production [J]. Nat Cell Biol, 2007, 9(9): 1089-1097.
[46] Majeau N, Fromentin R, Savard C, Duval M, Tremblay MJ, Leclerc D. Palmitoylation of hepatitis C virus core protein is important for virion production [J]. J Biol Chem, 2009, 284(49): 33915-33925.
[47] Okamoto K, Mori Y, Komoda Y, Okamoto T, Okochi M, Takeda M, Suzuki T, Moriishi K, Matsuura Y. Intramembrane processing by signal peptide peptidase regulates the membrane localization of hepatitis C virus core protein and viral propagation [J]. J Virol, 2008, 82(17):8349-8361.
[48] Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins [J]. Biochim Biophys Acta, 1999, 1451(1): 1-16.
[49] Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation [J]. J Biol Chem, 2001, 276(43): 39501-39504.
[50] Harris M. From negative factor to a critical role in virus pathogenesis: the changing fortunes of Nef [J]. J Gen Virol, 1996, 77(Pt 10): 2379-2392.
[51] Morgan CR, Miglionico BV, Engen JR. Effects of HIV-1 Nef on human N-myristoyltransferase 1 [J]. Biochemistry, 2011, 50(16): 3394-3403.
[52] Giese SI, Woerz I, Homann S, Tibroni N, Geyer M, Fackler OT. Specific and distinct determinants mediate membrane binding and lipid raft incorporation of HIV-1(SF2) Nef [J]. Virology, 2006, 355(2): 175-191.
[53] Bentham M, Mazaleyrat S, Harris M. Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein [J]. J Gen Virol, 2006, 87(Pt 3): 563-571.
[54] Foster JL, Garcia JV. HIV-1 Nef: at the crossroads [J]. Retrovirology, 2008, 5: 84.
[55] Hoffmann S, Jonas E, König S, Preusser-Kunze A, Willbold D. Nef protein of human immunodeficiency virus type 1 binds its own myristoylated N-terminus [J]. Biol Chem, 2007, 388(2): 181-183.
[56] Chihara T, Hashimoto M, Osman A, Hiyoshi-Yoshidomi Y, Suzu I, Chutiwitoonchai N, Hiyoshi M, Okada S, Suzu S. HIV-1 proteins preferentially activate anti-inflammatory M2-type macrophages [J]. J Immunol, 2012, 188(8): 3620-3627.
[57] Lai RP, Yan J, Heeney J, McClure MO, Göttlinger H, Luban J, Pizzato M. Nef decreases HIV-1 sensitivity to neutralizing antibodies that target the membrane-proximal external region of TMgp41 [J]. PLoS Pathog, 2011, 7(12): e1002442.
[58] Lamers SL, Fogel GB, Huysentruyt LC, McGrath MS.HIV-1 nef protein visits B-cells via macrophage nanotubes: a mechanism for AIDS-related lymphoma pathogenesis [J] ? Curr HIV Res, 2010, 8(8): 638-640.
[59] Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain [J]. J Virol, 1999, 73(3): 2052-2057.
[60] Hong HJ, Ryu CJ, Hur H, Kim S, Oh HK, Oh MS, Park SY. In vivo neutralization of hepatitis B virus infection by an anti-preS1 humanized antibody in chimpanzees [J]. Virology, 2004, 318(1): 134-141.
[61] Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus [J]. Elife, 2012, 1: e00049.
[62] Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly [J]. Proc Natl Acad Sci USA, 1991, 88(3): 1059-1063.
[63] Engelke M, Mills K, Seitz S, Simon P, Gripon P, Schnölzer M, Urban S. Characterization of a hepatitis B and hepatitis delta virus receptor binding site [J]. Hepatology, 2006, 43(4): 750-760.
[64] Schulze A, Schieck A, Ni Y, Mier W, Urban S. Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction [J]. J Virol, 2010, 84(4): 1989-2000.
[65] Niedre-Otomere B, Bogdanova A, Bruvere R, Ose V, Gerlich WH, Pumpens P, Glebe D, Kozlovska T. Posttranslational modifications and secretion efficiency of immunogenic hepatitis B virus L protein deletion variants [J]. Virol J, 2013, 10: 63.
[66] Krauzewicz N, Streuli CH, Stuart-Smith N, Jones MD, Wallace S, Griffin BE. Myristylated polyomavirus VP2: role in the life cycle of the virus [J]. J Virol, 1990, 64(9): 4414-4420.
[67] Chow M, Newman JF, Filman D, Hogle JM, Rowlands DJ, Brown F. Myristylation of picornavirus capsid protein VP4 and its structural significance [J]. Nature, 1987, 327(6122): 482-486.
[68] Nibert ML, Schiff LA, Fields BN. Mammalian reoviruses contain a myristoylated structural protein [J]. J Virol, 1991, 65(4): 1960-1967.
[69] Du Y, Zuckermann FA, Yoo D. Myristoylation of the small envelope protein of porcine reproductive and respiratory syndrome virus is non-essential for virus infectivity but promotes its growth [J]. Virus Res, 2010, 147(2): 294-299.
[70] Capul AA, Perez M, Burke E, Kunz S, Buchmeier MJ, de la Torre JC. Arenavirus Z-glycoprotein association requires Z myristoylation but not functional RING or late domains [J]. J Virol, 2007, 81(17): 9451-9460.
[71] Whitley DS, Yu K, Sample RC, Sinning A, Henegar J, Norcross E, Chinchar VG. Frog virus 3 ORF 53R, a putative myristoylated membrane protein, is essential for virus replication in vitro [J]. Virology, 2010, 405(2): 448-456.
[72] Anderegg RJ, Betz R, Carr SA, Crabb JW, Duntze W. Structure of Saccharomyces cerevisiae mating hormone a-factor. Identification of S-farnesyl cysteine as a structural component [J]. J Biol Chem, 1988, 263(34): 18236-18240.
[73] Maltese WA. Posttranslational modification of proteins by isoprenoids in mammalian cells [J]. FASEB J, 1990, 4(15): 3319-3328.
[74] Glenn JS, Watson JA, Havel CM, White JM. Identification of a prenylation site in delta virus large antigen [J]. Science, 1992, 256(5061): 1331-1333.
[75] Gao J, Liao J, Yang GY. CAAX-box protein, prenylation process and carcinogenesis [J]. Am J Transl Res, 2009, 1(3): 312-325.
[76] Berndt N, Sebti SM. Measurement of protein farnesylation and geranylgeranylation in vitro, in cultured cells and in biopsies, and the effects of prenyl transferase inhibitors [J]. Nat Protoc, 2011, 6(11): 1775-1791.
[77] Pfeffer S, Aivazian D. Targeting RabGTPases to distinct membrane compartments [J]. Nat Rev Mol Cell Biol, 2004, 5(11): 886-896.
[78] Glenn J S. Prenylation of HDAg and antiviral drug development [J]. Curr Top Microbiol Immunol, 2006, 307: 133-149.
[79] Huang WH, Chen CW, Wu HL, Chen PJ.Post-translational modification of delta antigen of hepatitis D virus [J]. Curr Top Microbiol Immunol, 2006, 307: 91-112.
[80] Yurdaydin C, Idilman R, Bozkaya H, Bozdayi AM. Natural history and treatment of chronic delta hepatitis [J]. J Viral Hepat, 2010, 17(11): 749-756.
[81] Yurdaydin C. Treatment of chronic delta hepatitis [J]. Semin Liver Dis, 2012, 32(3): 237-244.
[82] Fujita M, Kinoshita T. GPI-anchor remodeling: potential functions of GPI-anchors in intracellular trafficking and membrane dynamics [J]. Biochim Biophys Acta, 2012, 1821(8): 1050-1058.
[83] Tsai YH, Liu X, Seeberger PH. Chemical biology of glycosylphosphatidylinositol anchors [J]. Angew Chem Int Ed Engl, 2012, 51(46): 11438-11456.
[84] Orlean P, Menon AK. Thematic review series: lipid post-translational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids [J]. J Lipid Res, 2007, 48(5): 993-1011.
[85] Jacobs MG, Robinson PJ, Bletchly C, Mackenzie JM, Young PR. Dengue virus nonstructural protein 1 is expressed in a glycosyl-phosphatidylinositol-linked form that is capable of signal transduction [J]. FASEB J, 2000, 14(11): 1603-1610.
[86] Du Y, Pattnaik AK, Song C, Yoo D, Li G. Glycosyl-phosphatidylinositol (GPI)-anchored membrane association of the porcine reproductive and respiratory syndrome virus GP4 glycoprotein and its co-localization with CD163 in lipid rafts [J]. Virology, 2012, 424(1): 18-32.
[87] Ma J. The role of cofactors in prion propagation and infectivity [J]. PLoS Pathog, 2012, 8(4): e1002589.
[88] Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, Anderson RG, Taraboulos A, Prusiner SB. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains [J]. Proc Natl Acad Sci USA, 1996, 93(25): 14945-14949.
[89] Sarnataro D, Campana V, Paladino S, Stornaiuolo M, Nitsch L, Zurzolo C. PrPCassociation with lipid rafts in the early secretory pathway stabilizes its cellular conformation [J]. Mol Biol Cell, 2004, 15(9): 4031-4042.
[90] Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, Priola S, Caughey B, Masliah E, Oldstone M. Anchorless prion protein results in infectious amyloid disease without clinical scrapie [J]. Science, 2005, 308(5727): 1435-1439.
[91] Bate C, Williams A. Neurodegeneration induced by clustering of sialylated glycosylphosphatidylinositols of prion proteins [J]. J Biol Chem, 2012, 287(11): 7935-7944.

