基于 2-(4′-羧基苯基)咪唑-4,5-二羧酸构筑的三个镉/锌配位聚合物的合成、拓扑结构、荧光光谱及DNA作用
严世承 武大令 张敏芝 管全银 赵国良*,,
(1浙江师范大学行知学院,金华 321004)(2浙江师范大学化学与生命科学学院,金华 321004)
Coordination polymers,as a type of important materials,exhibits attractive application prospects in the fields of gas adsorption,storage and separation,adsorption of dyes,electrical conductivity,optical materials,magnetic materials,catalyzer,and so on[1-14].Those applications bring the new dawn for the porous materials science.Multifunctionalorganic ligands,especially the N-heterocyclic carboxylates,contain multi-oxygen and nitrogen atoms,and possess the ability to coordinate with metal ions in versatile ways.These building blocks lead to the formations of various coordination polymers with specific topologies and useful properties[16-17].
Imidazole-4,5-dicarboxylic acid (H3IDC),which can be partially or fully deprotonated to generate H2IDC-,HIDC2-,IDC3-at different pH values and afford various coordination modes,is favored by multitudinous research groups.According to the reported coordination polymers[18-23]constructed from H3IDC,this kinds of ligand still remain extremely widely researched.Recently,according to purposefully changing the substituent group on the 2-position of imidazole-4,5-dicarboxylicacid,excellentligandshasbeen obtained,which can be used to construct coordination polymers with rapidly changing topological structures and useful properties,such as 2-methyl-1H-imidazole-4,5-dicarboxylic acid,2-ethyl-1H-imidazole-4,5-dicarboxylic acid,2-propyl-1H-imidazole-4,5-dicarboxylic acid,2-phenyl-1H-imidazole-4,5-dicarboxylic acid,2-hydroxymethyl-1H-imidazole-4,5-dicarboxylic acid and 2-(pyridyl)-1H-imidazole-4,5-dicarboxylic acid.
Herein,taking into account the factors mentioned above,a new H3IDC derivative,2-(4′-carboxyphenyl)-1H-imidazole-4,5-dicarboxylic acid (H4CPhIDC),was purposely synthesized by condensation and oxidation reactions.Three coordination polymers of cadmium and zinc{[Cd2(CPhIDC)(bimb)]·H2O}n(1),{[Cd2(CPhIDC)(phen)2]·3H2O}n(2),{[Zn2(CPhIDC)(bpp)]·1.5H2O}n(3),(bimb=1,4-bis(imidazol-1-yl)butane,phen=1,10-phenanthroline,bpp=1,3-di(pyridin-4-yl)propane)have been synthesized by solvothermal reaction and characterized.The carboxyl containing group on the 2-position[24]of H4CPhIDC is successfully applied to construct coordination polymers.Its fully deprotonated motifs of CPhIDC4-can exhibit very flexible coordination modes(Scheme 1),and form a large diversity of supramolecular architectures.

Scheme 1 Coordination modes of CPhIDC4-for the three polymers
1 Experimental
1.1 Materials and measurements
H4CPhIDC was prepared according to literature[25-28]with some proper modification.The other reagents were of analytical grade and used without further purification.Calf thymus DNA(ct-DNA)was prepared with 0.1 mol·L-1NaCl.The concentration of ct-DNA was 200 μg·mL-1(cDNA=0.372 mmol·L-1).The ct-DNA solutions were stored at 4℃and gave a ratio of UVVis absorbance at 260 and 280 nm,A260/A280=1.8,indicating that DNA was sufficiently free of protein.The buffer solution,0.01 mol·L-1Tris-HCl(tris(hydroxymethyl)aminomethane hydrochloride,pH=7.4),was prepared with double-distilled water.
Elemental analysis was performed on C,H,N elemental analyzer,Elementar Vario ELⅢ.FTIR spectra were recorded on a Nicolet NEXUS 670 FTIR spectrophotometer using KBr discs in the range of 4 000~400 cm-1.A Mettler Toledo thermal analyzer TGA/SDTA 851ewas used to carry out the thermogravimetric analysis with a heating rate of 10℃·min-1from 30~800 ℃ in air atomsphere.Powder X-ray diffraction(PXRD)data were collected on a PW 3040/60 Focus X-ray diffractometer using Cu Kα radiation(λ=0.154 06 nm,2θ=2°~60°)at room temperature with acceleration voltage of 40 kV and current of 40 mA.Fluorescence spectra were measured at room temperature with an Edinburgh FL-FS920 TCSPC system.
1.2 Synthesis of the polymers
1.2.1 Synthesis of{[Cd2(CPhIDC)(bimb)]·H2O}n(1)
A mixture of H4CPhIDC (0.140 g,0.5 mmol),bimb(0.147 g,0.75 mmol),CdCl2·2.5H2O (0.170 g,0.75 mmol),and H2O/EtOH (15 mL,4∶1,V/V)with the pH value of 8 adjusted by 0.5 mol·L-1NaOH was sealed in a 20 mL Teflon-lined stainless steel vessel and heated at 160℃for 3 d.After the mixture was cooled to room temperature at a rate of 10 ℃·h-1,colorless crystals suitable for single-crystal analysis and physical measurements were obtained,washed with distilled water,and dried in air.Yield:41%(based on CdCl2·2.5H2O).Anal.Calcd.for C44H40N12O13Cd4(%):C 37.46,H 2.86,N 11.92;Found(%):C 37.71,H 2.83,N 11.98.IR(KBr,cm-1):3 538(m),2 943(w),2 860(w),1 588(s),1 542(s),1 525(s),1 440(s),1 405(s),1 375(m),1350(m),1 293(s),1 229(s),1 113(s),1 097(s),941(s),862(s),823(s),789(s),785(s),652(s),570(m),499(m),462(s).
1.2.2 Synthesis of{[Cd2(CPhIDC)(phen)2]·3H2O}n(2)
The synthesis method is similar to 1,where the metal source is Cd(OH)2,the auxiliary ligand is changed to phen,the pH value is not adjusted,the solvents are H2O/i-PrOH/acetone(15 mL,2∶1∶1,V/V).Yield:36%(based on Cd(OH)2).Anal.Calcd.for C36H26N6O9Cd2(%):C 47.44,H 2.88,N 9.22;Found:C 47.28,H 2.83,N 9.29.IR(KBr,cm-1):3 499(m),1 603(s),1 588(s),1 542(s),1 514(s),1 429(s),1 392(s),1 279(m),1 223(w),1 145(m),1 013(w),970(m),859(s),842(s),792(m),727(s),547(m),506(m),457(m).
1.2.3 Synthesis of{[Zn2(CPhIDC)(bpp)]·1.5H2O}n(3)
The synthesis method is roughly the same as 1,where the metal source is changed to zinc acetate,the auxiliary ligand is changed to bpp,the pH value is adjusted to close to 6.5,the solvents are H2O/EtOH(15 mL,3∶2,V/V).Yield:44%(based on Zn(CH3COO)2).Anal.Calcd.for C25H21N4O7.5Zn2(%):C 47.79,H 3.37,N 8.92;Found(%):C 47.72,H 3.25,N 9.17.IR(KBr,cm-1):3 480(m),2 936(w),2 874(w),1 608(s),1 590(s),1 550(s),1 430(s),1 403(s),1 381(s),1 352(m),1 271(s),1 224(m),1 175(m),1 122(s),1 068(m),1 028(s),872(m),830(s),813(s),796(s),789(s),749(s),727(s),664(w),634(w),574(w),525(m),489(m),463(s).
1.3 X-ray diffraction analysis
The single crystals of the polymers with approximate dimensions were mounted on a Bruker Smart ApexⅡCCD diffractometer.The diffraction data were collected using a graphite monochromated Mo Kα radiation(λ=0.071 073 nm)at 296(2)K.Absorption corrections were applied using SADABS[29].The structure was solved by using the SHELXS-97[30]program package and refined with the full-matrix least-squares technique based on F2using the SHELXL-97[31]program package.Remaining hydrogen atoms were added in calculated positons and refined as riding atoms with a common fixed isotropic thermal parameter.Hydrogen atoms on water molecules were located in a difference Fourier map and included in the subsequent refinement using restrains (dO-H=0.085 nm,dH…H=0.130 nm)with Uiso(H)=0.15Ueq(O).Other hydrogen atoms were added theoretically.The detail about the crystal data is summarized in Table 1.Selected bond distances and bond angles are given in Table S1 to Table S3(Supporting information).
CCDC:969814,1;937738,2;937741,3.
1.4 DNA binding
1.0 mL of 200 μg·mL-1DNA solution,1.0 mL of 200 μg·mL-1EB solution and 2.0 mL of Tris-HCl buffer solution with pH=7.40 were added to a 10 mL colorimetrictubeand allowed tostand atroom temperature for 2 h.Then,a solution of 0.50 or 0.10 mmol·L-1compound was added to the mixed solutionand diluted to a scale with distilled water.After 4 h at room temperature,the fluorescence spectra of the composite system in 520~700 nm range are recorded by exciting at 251 nm.
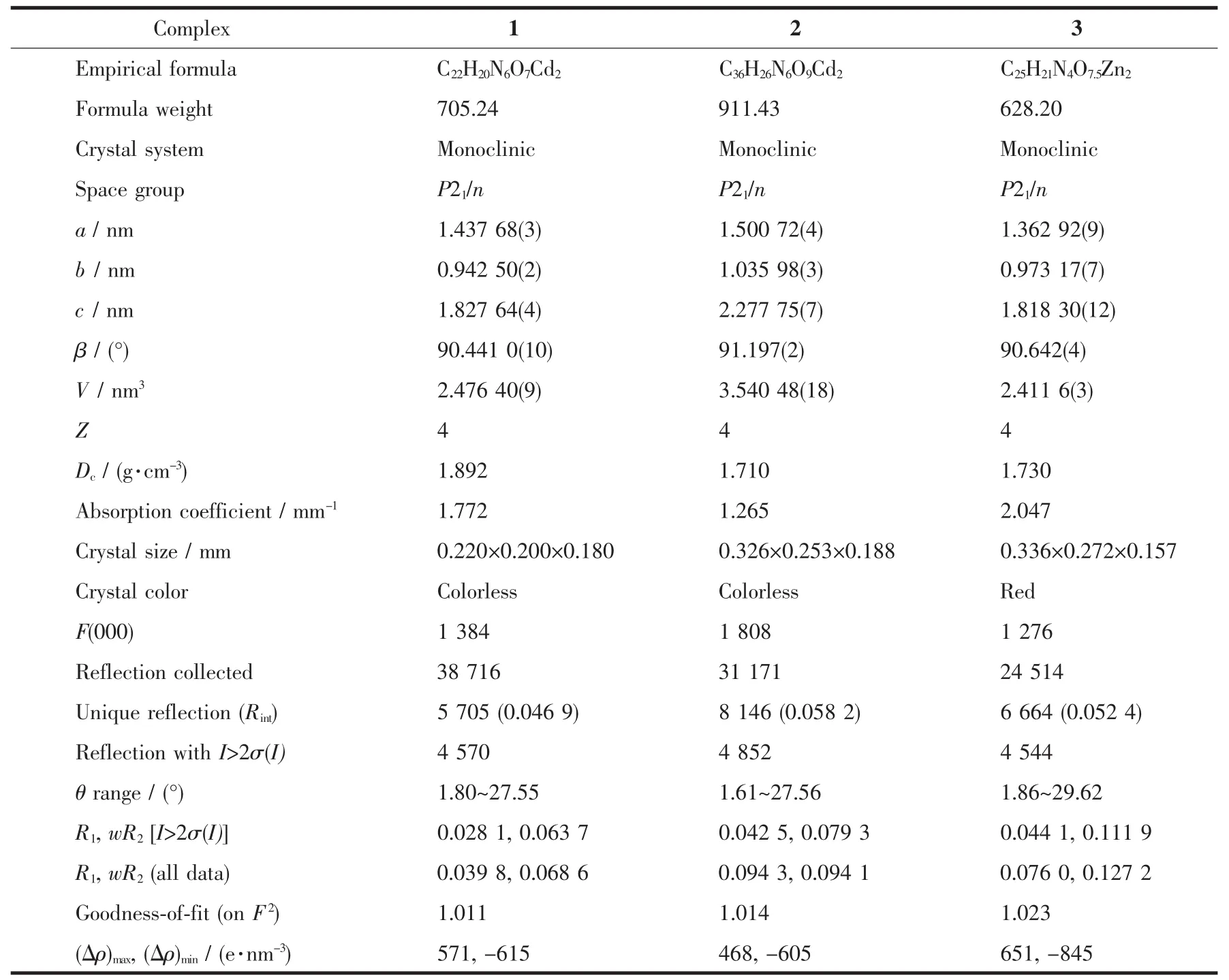
Table 1 Crystal data and structure refinement parameters for polymers 1~3
2 Results and discussion
2.1 Crystal structures of the complexes
2.1.1 Crystal structure of{[Cd2(CPhIDC)(bimb)]·H2O}n(1)
Single-crystal analysis shows polymer 1 crystallizes in the monoclinic system with space group P21/n and the asymmetric unit cell contains two Cd2+ions,one CPhIDC4-anion,one 1,4-bis(imidazol-1-yl)butane molecule and one lattice water molecule.As shown in Fig.1a,the Cd(2)adopts distorted octahedral geometry with a six-coordinated mode by four oxygen atoms(O5,O1#3,O2#3,O5#4,dCd-O=0.223 6(2)~0.251 9(3)nm)and one nitrogen atom(N2,dCd-N=0.227 4(3)nm)from three CPhIDC4-ligands and one nitrogen atom(N3,dCd-N=0.225 2(3)nm)from one bimb molecule.The Cd(1)adopts distorted trigonal bipyramid coordinated with three oxygen atoms(O3,O6,O4#1,dCd-O=0.221 0(2)~0.236 3(2)nm),one nitrogen atom(N1#1,dCd-N=0.221 2(3)nm)from two CPhIDC4-ligands and one nitrogen atom(N6#2,dCd-N=0.223 3(3)nm)from one bimb molecule.From Cd-O,Cd-N bond distances and O3-Cd1-N6#2,O3-Cd1-O4#1,N6#2-Cd1-O4#1 bond angles for 1,we could know that the five atoms are practically in the same plane while the O1 and N3 atoms are at the side of it.Thus the two Cd2+ions are bridged by the ends of one bimb molecule respectively.The CPhIDC4-ligand adopt conformations with μ5-κO∶κ2O′,N∶κ2O″,N′∶κ2O‴,O″″∶κO″‴.The two Cd2+ions are bridged by the ends of one bimb molecule respectively.The CPhIDC4-ligand adopts conformations with μ5-κO ∶κ2O′,N ∶κ2O″,N′∶κ2O‴ ,O″″∶κO″‴coordination fashion(Scheme 1a)to connect Cd(1)and Cd(2)ions.The selected distances and bond angles fall in the normal regions which are comparable to the values reported in literatures[32].For 1,the Cd2+ion is bridged by the imidazole ring to form a 2D plane ellipsoid lattice(0.785 6(4)nm×0.922 3(3)nm)(Fig.1b).The 2D surface adopts staggered conformations connected with bimb molecules and CPhIDC4-anions to build up a 3D[Cd2(CPhIDC)(bimb)]framework(Fig.1c).Both bimb molecules reside in the tunnels and the extensive hydrogen bonds (Table S4)contribute themselves to stabilize the crystal structure.

Fig.1 (a)Ball-and-stick structural view of 1;(b)2D plane structure for 1 viewed along a axis;(c)3D framework for 1 viewed along b axis;(d)(3,4,5)-topological connected for 1
In the 3D[Cd2(CPhIDC)(bimb)]frameworks,from the topological point of view,CPhIDC4-anions ligand are each bonded to five Cdギions(three Cd(1)and two Cd(2)),while Cd(1)ions are each coordinated to five atoms(O1,O2,O3,O3#2,N2)from three CPhIDC4-anions in a κO∶κ2O′,N∶κO″coordination fashion and one nitrogen atom (N3)from bimb molecule in κN coordination fashion.Cd(2)ions are each coordinated to four atoms (O6,O4,O5,N1)from two CPhIDC4-ligand anions in a κ2O,O′∶κ2O″,N and one atom from one bimb ligand in κN coordination fashion.From topological point of view the 3D framework can be simplified in some underlying net,where Cd(1)and Cd(2)atoms,CPhIDC4-anions and bridge bimb ligands are presented by 4-coordinated,3-coordinated,5-coordinated nodes,respectively.
Viewed along a axis for 1 without bimb and benzeneedge,respectively (Fig.1d),topology of the 3,4,5-coordinated trinodalunderlying netcan be described with point symbol(5·6·7)(4·52·6·74·82)(4·52·6·7).The structure of 1 is completely different from the reported(3,4,5)-connected frameworks with(6·8·10)(6·82)(63)4(64·102)(64·84·102),(63)2(66)(68·82),(4·62)2(43·67)2(44·62),(42·6)(44·62)(44·63·83),(5·6·7)(54·6·8)(54·63·83),(4·6·8)2(4·82)(4·64·85)(42·62·82),(4·62)(42·6)(42·84)(43·6·86)(42·65·83),(4·6·8)2(42·62·82)(42·65·83)2,and(4·62)2(4·67·82)2(65·10)topologies[33-34],which presents a new trinodal (3,4,5)-connected 3D network topology.
2.1.2 Crystal structure of{Cd2(CPhIDC)(phen)2]·3H2O}n(2)

Fig.2 (a)Ball-and-stick structural view of 2;(b)3D stacking structure for 2 with hydrogen bond and π…π from phen molecules;(c)Single-screw structure of 2 and the double-helix structure for DNA;(d)(4,4)-topological connected for 2
Single-crystal analysis shows polymer 2 crystallizes in the monoclinic system with space group P21/n and the asymmetric unit cell contains two Cd2+ions,one CPhIDC4-anion,two 1,10-phenanthroline molecules and three lattice waters.As shown in Fig.2a,polymer 2 is a double-core structure bridged by the CPhIDC4-anions and adopt six-coordinated with two Cd2+ions(Cd1 and Cd2).The Cd1 is connected with three oxygen atoms(O3#1,O4,O6,dCd-O=0.222 0(3)~0.235 1(3)nm), one nitrogen atom(N1#1,dCd-N=0.223 0(3)nm)from two CPhIDC4-ligands and two nitrogen atoms(N5,N6,dCd-N=0.236 9(4)~0.238 8(4)nm)from one 1,10-phenanthroline molecule to form a distorted octahedralgeometry.Cd2 ispractically identical to Cd1,but the six-coordinated atoms form a different severe flattening octahedral geometry.Cd-O distances,Cd-N distances and bond angles for 2(Table S2)fall in the normal regions which are comparable to the values reported in literatures[35].The CPhIDC4-anions adopt conformations with each bridging four Cd2+ions in μ4-κ2O,O′∶κ2N,O″∶κ2N′,O‴∶κ2O″″,O″‴ coordination fashion (Scheme 1b)to form a ternary-chelate ring,two five-chelate rings and a seven-chelate ring.
Viewed along c axis without phen molecular,the Cd2+ion is bridged by the CPhIDC4-to form a 2D plane grid.Viewed along b axis only,the Cd2+ions bridging by ligands to form a bent long chain with phen molecules connected in the two sides of it.Meanwhile,the two adjacent chains stack with the function of π…π from phen molecules to form a 3D network (Fig.2b).In addition,the hydrogen bonds(O2B-H2C…O3W#1)connected by the oxygen atom from free water molecules and hydrogen bond (O2WH2A…O3,O2B-H2C…O3W#1,O2B-H2D…O1#2,O3W-H3C…O2),which is shown in Table S5,connected by oxygen atom from carboxylic acid and free water and thus contribute themselves to stabilize the crystal structure.
Interestingly,comparing the space filling figure for 2 with the structure of DNA,the single-screw structure of 2 is quite similar to the double-helix structure for DNA (Fig.2c).The inner hydrogen bond from the spiral chain and the phen molecules reside in the tunnel contribute the framework to stabilize the spiral chain structure.From the topological point of view,each CPhIDC4-ligand,linked to four Cd2+ions(two Cd1 and two Cd2),represents a 4-connected node while each Cdギion connects to two CPhIDC4-anions as an edge of underlying net.Thus the chelating effect in 2 leads to 4-coordinated 2D underlying net with point symbol(44)(62)and vertex symbol(4·4·4·4)(Fig.2d).
2.1.3 Crystal structure of{[Zn2(CPhIDC)(bpp)]·1.5H2O}n(3)
Single-crystal analysis shows polymer 3 crystallizes in the monoclinic system with space group P21/n and the asymmetric unit cell contains two Zn2+ions(Zn(1)and Zn(2)),one CPhIDC4-anion,one bpp molecule,one and a half lattice water.As shown in Fig.3a,Zn(1)is five-coordinated by three oxygen atoms(O2#1,O3#2,O3,dZn-O=0.199 4(2)~0.213 7(2)nm)and one nitrogen atom(N2,dZn-N=0.210 8(2)nm)from three imidazole carboxylic acid ligands and one nitrogen atom(N3,dZn-N=0.205 3(3)nm)from one bpp molecule.Zn(1)is only 0.000 03 nm away from the O2#1,O3 and N3 plane,whereas the N2,O3#2 are at the side of the plane with the N2-Zn1-O3#2 bond angle of 149.28(8)°,which deviates from the straight line about 30.72°and thus the six atoms form a distorted trigonal bipyramid geometry.Zn(2)adopts the five-coordinated mode to form a distorted trigonal bipyramid geometry by three oxygen atoms(O6,O4,O5#4,dZn-O=0.203 5(3)~0.209 1(2)nm)and one nitrogen atom(N1#4,dZn-N=0.209 8(3)nm)from one CPhIDC4-ligand and one nitrogen atom(N4#3,dZn-N=0.208 9(3)nm)from one bpp molecule.The bond angles of O6-Zn2-N4#3,O6-Zn2-O5#4,N4#3-Zn2-O5#4 are 117.31(13)°,117.69(11)°,124.90(13)°,respectively,and the sum of the three is 359.90°,which shows that O6,N4#3,O5#4 and Zn(2)are almost in the same plane,the bond angle(N1#4-Zn2-O4)is 169.54(10)°.In addition,two Zn2+ions are linked by the two sides of the bpp molecule.The ligand adopts conformations with μ5-κO∶κ2O′,N∶κ2O″,N′∶κ2O‴,O″″∶κO″‴ coordination fashion (Scheme 1c)to connect Zn1 and Zn2 to form a five-member chelate ring and a seven-member chelate ring,respectively.The imidazole carboxylic acid ligand in the 3 adopt single dentate and bidentate chelate coordination mode connected to the metal ions.The Zn-O distances and the Zn-N distances and bond angles fall in the normal regions which are comparable to the values reported in literatures[36].
Viewed along c axis without bpp,the Zn2+ion is bridged by the imidazole ring to form a 2D-grid sheet structure(Fig.3b).The 2D-grid sheet staggered conby the CPhIDC4-anions, bpp molecules expands to 3D Zn2(CPhIDC)(bpp)framework(Fig.3c).The3D Zn2(CPhIDC)(bpp)framework is found to be stabilized by the bpp padding molecules and the abundant hydrogen bonds(Table S6).
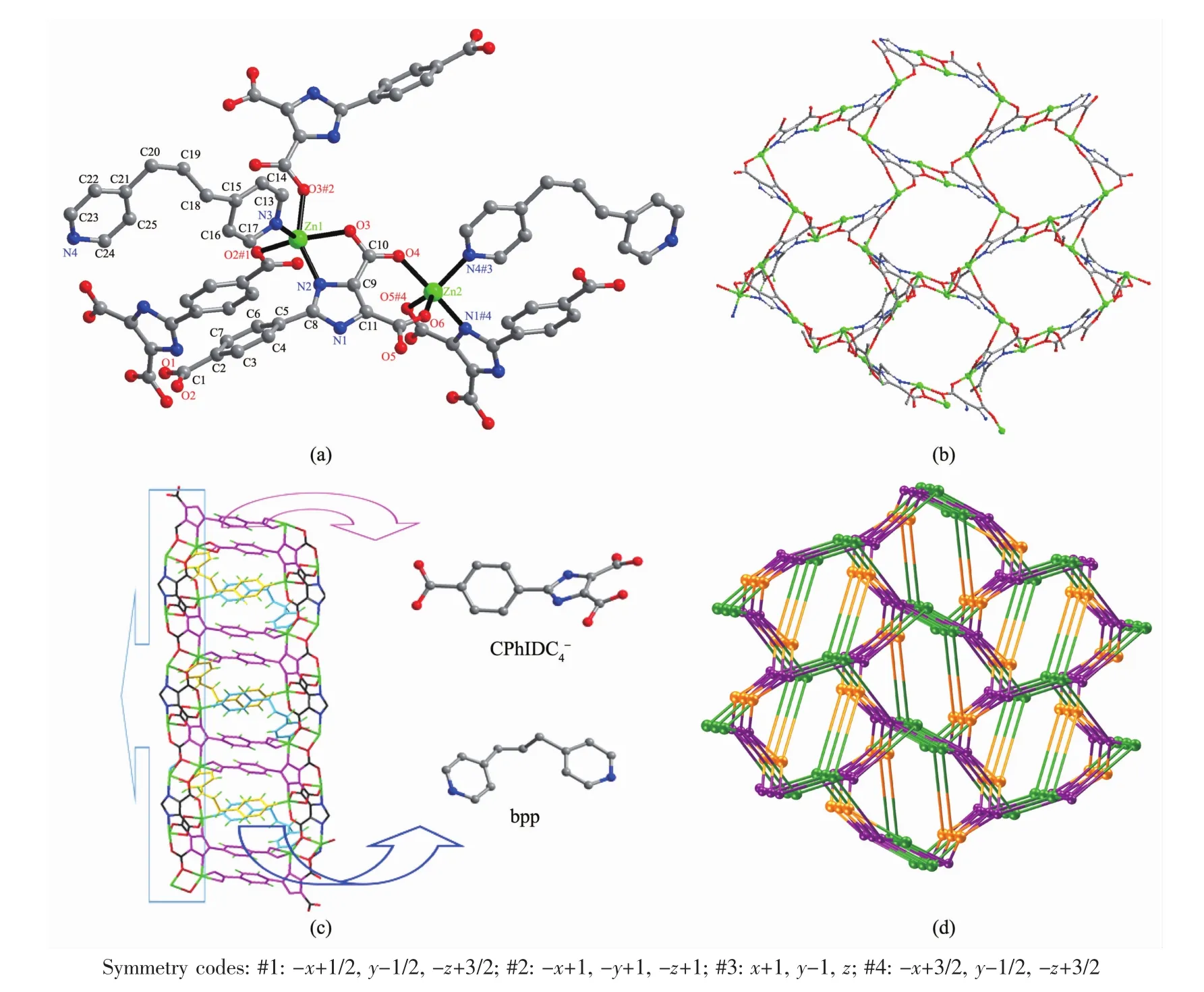
Fig.3 (a)Ball-and-stick structural view of 3;(b)2D plane structure for 3 viewed along a axis;(c)3D framework for 3 viewed along b axis;(d)(3,4,5)-topological connected for 3
As the 3D topological connected for 3 in Fig.3d,the CPhIDC4-anions are each bonded to four Znギions(two Zn1 and two Zn2).The Zn(1)ions are each coordinated to six atoms(O3#2,O2#1,O1#1,O3,N2,N3)from three CPhIDC4-anions in a κO∶κ2O′,N∶κ2O″,O‴ coordination fashion and one bpp molecule in κN coordination fashion.The Zn(2)ions are each coordinated to four atoms(O6,O4,O5,N1)from two CPhIDC4-ligand anions in κ2O,O′∶κ2O″,N coordination fashion and one nitrogen atom from one bpp ligand in κN coordination fashion.Therefore,each Zn1 ion,Zn2 ion and CPhIDC4-anion can now be viewed as 4-connected nodes,3-connected nodes and 5-connected nodes,respectively and leads to trinodal net with point symbol(4·52·6·72)(5·6·7)(4·52·6·74·82),which is same as 1(Fig.3d).
2.2 Analysis of FTIR spectra and PXRD
A broad absorption peak at 3 435 cm-1in the free ligand can be assigned to the stretching vibration of phenolic hydroxyl group νO-Hin the carboxyl group.The absorption peak at 3 012~3 109 cm-1is the stretching vibration peak of νN-Hon the imidazole ring.The stretching vibration peak of the carbonyl group νC=Oappears at 1 716 cm-1,and the stretching vibration peak of νC=Nin imidazole ring is located at 1 614 cm-1.The infrared spectra of polymers 1~3 are similar,indicating that they have similar coordination modes.The broad peaks appear from 3 403 to 3 538 cm-1in the three polymers due to the O-H stretching vibration ofwater.Thecharacteristic absorption peaksof carboxyl groups at 1 716 cm-1disappear in polymers.The asymmetric stretching vibration νas(-COO-)peaks appear at 1 542~1 550 cm-1,and symmetrical stretching vibration peaks νs(-COO-)appear at 1 428~1 440 cm-1and 1 375~1 395 cm-1,indicating that the carboxyl group of ligand are in the form of bidentate chelating,bridging and monodentate coordination[19,37].νC=Nstretching vibration of imidazole ring in ligand has redshift from 1 614 to 1 588~1 590 cm-1.Two vibration peaks at 2 943 cm-1(1),2 936 cm-1(3)and 2 860 cm-1(1),2 874 cm-1(3)are C-H stretching vibration of-CH2-group in auxiliary ligands bimp and bpp.However,polymer 2 don′t have these two peaks.Another two absorption peaks at 499~506 cm-1and 457~463 cm-1can be attributed to the stretching vibration of νM-Oand νM-N.All of above are consistent with the results of single crystals structure analysis.
The simulated and experimental PXRD patterns of coordination polymers 1~3 are given in Fig.S1~S3.The results suggest that the crystal structures are truly representative of the bulk materials.The differences in intensity are due to the preferred orientation of the powder samples.
2.3 Luminescent properties
There are few reports about the strong luminescent properties in imidazole-4,5-dicarboxylic acid.However,2-(4′-carboxyphenyl)-1H-imidazole-4,5-dicar-rboxylic acid (H4CPhIDC)has strong luminescent property at room temperature.
As illustrated in Fig.4,the solid-state luminescence spectra at room temperature for H4CPhIDC ligand,polymers 1,2 and 3 are observed to have their main emission at 524,527,526 and 524 nm(609 nm)(λex=467 nm),respectively.The imidazole ligand,1 and 2 can emit a certain intensity green luminescent while 3 can not only emit green but also emit orange luminescent.The green luminescent for 1,2 and 3 are from imidazole ligand and stronger than the free ligand,which can be ascribed to the luminescent of H4CPhIDC ligand sensitized by metalions.In addition,we can presume that the emission for 3 is neither metal-to-ligand charge transfer (MLCT)nor ligand-to-metal transfer(LMCT)in nature,because the Znギion is difficult to be oxidized or reduced due to its d10configuration.Thus,emissions observed at 609 nm for 3 may be assigned the band to an intra-ligandfluorescent emission of bpp anxiliary ligand[38].
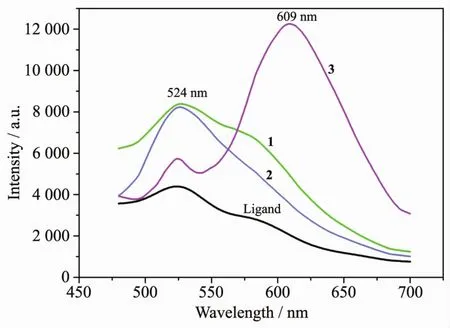
Fig.4 Fluorescence spectra for the H4CPhIDC ligand and polymers 1~3
2.4 DNA binding
The interaction of ligand and polymers with calf thymus DNA(CT-DNA)was studied by an EB fluorescent probe.Fig.5 shows the emission spectra of EB bonded to DNA with compounds or not.As the increasing concenof the compounds,the emission intensity at 592 nm of EB-DNA system changed in different degrees.According to the classical Stern-Volmer equation[39]:I0/I=1+Ksqr,where I0and I represent the fluorescence intensities in the absence or presence of the compounds,respectively;r is the concentration ratio of the compounds to DNA;Ksqis a linear Stern-Volmer quenching constant,the Ksqvalue was obtained as the slope of I0/I versus r linear plot.
From the inset in Fig.5,the Ksqvalue were 16.53,0.88,21.77 and 1.07 for H4CPhIDC ligand,polymers 1,2 and 3.It suggested that the interaction of the ligand with DNA are strong and can release more free EB molecules from EB-DNA,because of the present of benzene and imidazole rings.Especially,polymer 2 has the strongest interaction with the DNA,which is attributed to not only big planar molecules phen but also its similar double-helix structure with DNA(Fig.2c).Thus,its molecules are more likely to enter the double helix structure of the DNA molecules.While the other two polymers are weaker than that of H4CPhIDC ligand,which could be ascribed to that the planarity of molecules of 1 and 3 is not as good as H4CPhIDC ligand and 2.

Fig.5 Emission spectra of EB-DNA system in the absence and presence of the H4CPhIDC ligand(a),polymers 1(b),2(c)and 3(d)
3 Conclusions
H4CPhIDC was purposely synthesized by condensation and oxidation reactions and successfully applied to constructing three novel coordination polymers{[Cd2(CPhIDC)(bimb)]·H2O}n(1),{[Cd2(CPhIDC)(phen)2]·3H2O}n(2),{[Zn2(CPhIDC)(bpp)]·1.5H2O}n(3).Complexes 1 and 3 exhibit analogous 3D[Cd2(CPhIDC)(bimb)]and 3D[Zn2(CPhIDC)(bpp)]frameworks with(5·6·7)(4·52·6·72)(4·52·6·74·82)topology,but the metal ions and auxiliary ligands are different.Complex 2 is 2D wave-like fishing net structure with 44·62topology.Moreover,the luminescent properties shows that polymer 3 can emit green and orange luminescence,while the imidazole ligand and the other two polymers can emit a certain intensity green luminescence.In addition,the interaction of the ligands with DNA is strong and could release more free EB molecules from EB-DNA,because of the present of benzene and imidazole rings.Among the four compounds,the polymer 2 has the strongest interaction with the DNA due to the addition of the big planar molecules phen and the particularity of its structure.This class of materials provides a new impetus to the construction ofnovelmultifunctionalcoordination polymers materials.
Supporting information is available at http://www.wjhxxb.cn
[1]Weston M H,Colón Y J,Bae Y S,et al.J.Mater.Chem.A,2014,2(2):299-302
[2]Kumar K V,Preuss K,Titirici M,et al.Chem.Rev.,2017,117(3):1796-1825
[3]Lin J Y S.Science,2016,353(6295):121-122
[4]Rodenas T,Luz I,Prieto G,et al.Nat.Mater.,2015,14(1):48-55
[5]Haque E,Lo V,Minett A,et al.J.Mater.Chem.A,2014,2(1):193-203
[6]Sun L,Park S S,Sheberla D,et al.J.Am.Chem.Soc.,2016,138(44):14772-14782
[7]Sakamoto R,Iwashima T,Kogel J F,et al.J.Am.Chem.Soc.,2016,138(17):5666-5677
[8]Medishetty R,Nalla V,Nemec L,et al.Adv.Mater.,2017,29(17):1605637
[9]Ricco R,Malfatti L,Takahashi M,et al.J.Mater.Chem.A,2013,1(42):13033-13045
[10]WANG Li-Ping(王丽苹),WANG Gong-Ying(王公应).Journal of Molecular Catalysis(China)(分子催化),2015,29(3):275-287
[11]CHEN Xiao-Qian(陈晓倩).Thesis for the Master of Minnan Normal University(闽南师范大学硕士论文).2015.
[12]HAO Li-Min(郝 丽 敏).Thesis for the Master of Chang′an University(长安大学硕士论文).2015.
[13]Hermes S,Schröter M K,Schmid R,et al.Angew.Chem.Int.Ed.,2005,44(38):6237-6241
[14]Bradshaw D,Garai A,Huo J.Chem.Soc.Rev.,2012,41(6):2344-2381
[15]Tanabe K K,Cohen S M.Inorg.Chem.,2010,49(14):6766-6774
[16]Miao Y R,Su Z,Suslick K S.J.Am.Chem.Soc.,2017,139(13):4667-4670
[17]Stock N,Biswas S.Chem.Rev.,2012,112(2):933-969
[18]Arstad B,Fjellvg H,Kongshaug K O,et al.Adsorption,2008,14(6):755-762
[19]Janiak C,Vieth J K.New J.Chem.,2010,34(11):2366-2388[20]Henninger S K,Habib H A,Janiak C.J.Am.Chem.Soc.,2009,131(8):2776-2777
[21]Torrisi A,Bell R G,Mellot-Draznieks C.Cryst.Growth Des.,2010,10(7):2839-2841
[22]Li K,Olson D H,Lee J Y,et al.Adv.Funct.Mater.,2008,18(15):2205-2214
[23]Lee C Y,Farha O K,Hong B J,et al.J.Am.Chem.Soc.,2011,133(40):15858-15861
[24]Wang W,Niu X,Gao Y,et al.Cryst.Growth Des.,2010,10(9):4050-4059
[25]Sharghi H,Asemani O,Khalifeh R.Synth.Commun.,2008,38(6):1128-1136
[26]Coppola G M.Synth.Commun.,2008,38(20):3500-3507
[27]Wang F Q,Zheng X J,Wan Y H,et al.Inorg.Chem.,2007,46(8):2956-2958
[28]Tan C,Wang Q.Inorg.Chem.,2011,50(8):2953-2956
[29]Sheldrick G M.SADABS,Program for Empirical Absorption Correction of Area Detector Data,University of Göttingen,Germany,1997.
[30]Sheldrick G M.SHELXS-97,Program for Crystal Structure Solution,University of Göttingen,Germany,1997.
[31]Sheldrick G M.SHELXL-97,Program for Crystal Structure Refinement,University of Göttingen,Germany,1997.
[32]ZHAN Pei-Ying(战佩英).Chinese J.Inorg.Chem.(无机化学学报),2014,30(7):1629-1634
[33]Xue M,Zhu G,Ding H,et al.Cryst.Growth Des.,2009,9(3):1481-1488
[34]Han Z B,Zhang G X.CrystEngComm,2010,12(2):348-351
[35]DU Fang-Yuan(杜芳园),LI Shi-Kun(李士坤),LIN Qiu-Yue(林秋月),et al.Chinese J.Inorg.Chem.(无机化学学报),2015,31(4):813-823
[36]Tao J,Shi J X,Tong M L,et al.Inorg.Chem.,2001,40(24):6328-6330
[37]Nakamoto K,Translated by HUANG De-Ru(黄德如),WANG Ren-Qing(汪仁庆).Infrared and Raman Spestra of Inorganic and Coordination Compounds(无机和配位化合物的红外和拉曼光谱).Bejing:Chemistry Industry Press,1986.
[38]He K H,Li Y W,Chen Y Q,et al.Cryst.Growth Des.,2012,12(6):2730-2735
[39]Lakowicz J R,Weber G.Biochemistry,1973,12(21):4161-4170

