Identification and Genetic Diversity Analysis of Chinese Mitten Crab(Eriocheir sinensis) in the Liao River Area
Wang Shi-hui, Li Chi-tao, Shang Mei, Jia Zhi-ying, Hu Xue-song, Ge Yan-long, and Shi Lian-yu
Key Laboratory of Freshwater Aquatic Biotechnology and Breeding, Ministry of Agriculture, Heilongjiang Fisheries Research Institute, Chinese Academy of Fishery Sciences, Harbin 150070, China
Introduction
Chinese mitten crab (Eriocheir sinensis) is commonly called the "river crab" in China, because it spends most of its life, except its reproductive and larval stages, in fresh water (Suiet al., 2009), and it is mainly distributed in the East China Sea, particularly the coast and coastal estuaries of the Bohai Sea. The crab has high social and economic values in China,due to its flavour and richness in proteins and vitamins(Chenet al., 2013). Due to long-term evolutionary impacts, the species have differentiated into several geographic populations in different river systems, and the three main populations inhabit the Liao River,the Yangtze River and the Oujiang River (Wang and Li, 2002). Chinese mitten crab stocks had been depleted since the 1960s and the number of it had been declined by 0.2 t in 1998, due to overfishing, dam construction and water pollution (Tanget al., 2000).Fortunately, Chinese mitten crab aquaculture has further progressed since the 1990s with advances in larviculture techniques (Zhao, 1980; Dai, 2012).Nowadays, fry produced in hatcheries are the major source of farm-raised crabs because of increasing market demand. In recent years, the yield and value of cultured crabs have steadily improved and the crab production has increased from 232 000 t in 2000 to 812 103 t in 2016 (Bureau of Fsheries Fishery Management, 2017). These showed that Chinese mitten crab production had been an important component of the freshwater aquaculture industry in China.
The Yangtze River population, exhibiting the highest growth rate and the best flavour, was considered the best option for establishing artificial cultures. So mongers sold Oujiang crab fry to the Liao River Estuary as the Yangtze crab fry and transported the Liao River crabs to the Yangtze River, because the natural stocks in the Yangtze River Estuary had been seriously depleted since the 1960s (Zhou, 2009),this led to the mixture of three natural geographic crab populations (Xu and Lin, 1997). Moreover,the introduced Japanese mitten crab led to the crab germplasm much more complex. Currently, the progeny of Chinese mitten crab is growing slowly,leading to smaller individuals of lower economic value. So constructing the systematic breeding selection process is essential to increase the growth rate and size of Chinese mitten crab and to rescue the germplasm of this species (Chen, 2007; Wang, 2007).Successful systematic breeding selection depends on fully studying the population structure and genetic diversity of Chinese mitten crab (Zhuet al., 2008).
As widely used molecular markers, microsatellites exhibit high polymorphism, strong individual specificity and codominant inheritance characters, and they have been extensively applied to detect germplasm resources and developed sustainable fishing practices(Baker, 1994). Recently, genetic diversity studies of Chinese mitten crab populations have made great progresses by using the microsatellite marker technique (Suiet al., 2009). The genetic diversity of crabs in four areas (Jiangsu, Anhui and Liaoning provinces and Tianjin City) was determined using microsatellite markers, and three populations (Jiangsu and Anhui provinces and Tianjin City) clustered together whereas the Liao River crab populations were isolated (Maet al., 2007). Although the Liao River crab and the Yangtze River crab are not clustered together, its divergence is weak (Changet al., 2008).By analyzing the genetic diversity of selected cultured Chinese mitten crab stocks, another study found that they are not obviously differentiated from the natural stocks of the Yangtze River population (Liet al.,2010). Finally, three cultured Chinese mitten crab populations (Xinghua, Xuancheng and Panjin cities)are found to have high genetic diversity and low differentiation based on microsatellite markers (Xionget al., 2012).
Crabs originating from the Yangtze River basin are regarded as the most valuable, due to multiple attributes including better growth patterns, larger size and better flavour (Liet al., 2000). For example, Chinese mitten crab resources should be protected (Gu and Zhao, 2001), and the genetic variability in the Yangtze River crabs is studied by using amplified fragment length polymorphism (AFLP) markers (Huanget al.,2008). In contrast, the Liao River crab has not been given sufficient attention due to small size. So there is an urgent need for in-depth analyse of the germplasm resources of this population. So, firstly the crabs between the Liao River and the Yangtze crabs were identified; secondly, 13 highly polymorphic microsatellite loci were selected to analyze the germplasm resources; finally, the genetic diversity,differentiation and structure of Chinese mitten crab were researched.
Materials and Methods
Sample collection and geographical information
Healthy adult Chinese mitten crabs (weighing ♀ 97.02±9.70 g, ♂123.05±27.46 g) were collected from two sites; one was the rice fields of four towns, Hujia(HJ), Chenjia (CJ), Baqiangzi (BQZ) and Xinli (XL),distributed on both sides of the Liao River, and the other site was in Zouqu County of Jiangsu Province,south of the Yangtze River (Fig. 1). A total of 120 Liao River and 37 Yangtze River crabs were collected; each sub-population of the Liao River crabs was sampled by collecting 30 individuals, 15 females and 15 males.All the specimens were transported to the laboratory,and muscle tissues were collected from the legs and preserved at -80℃.
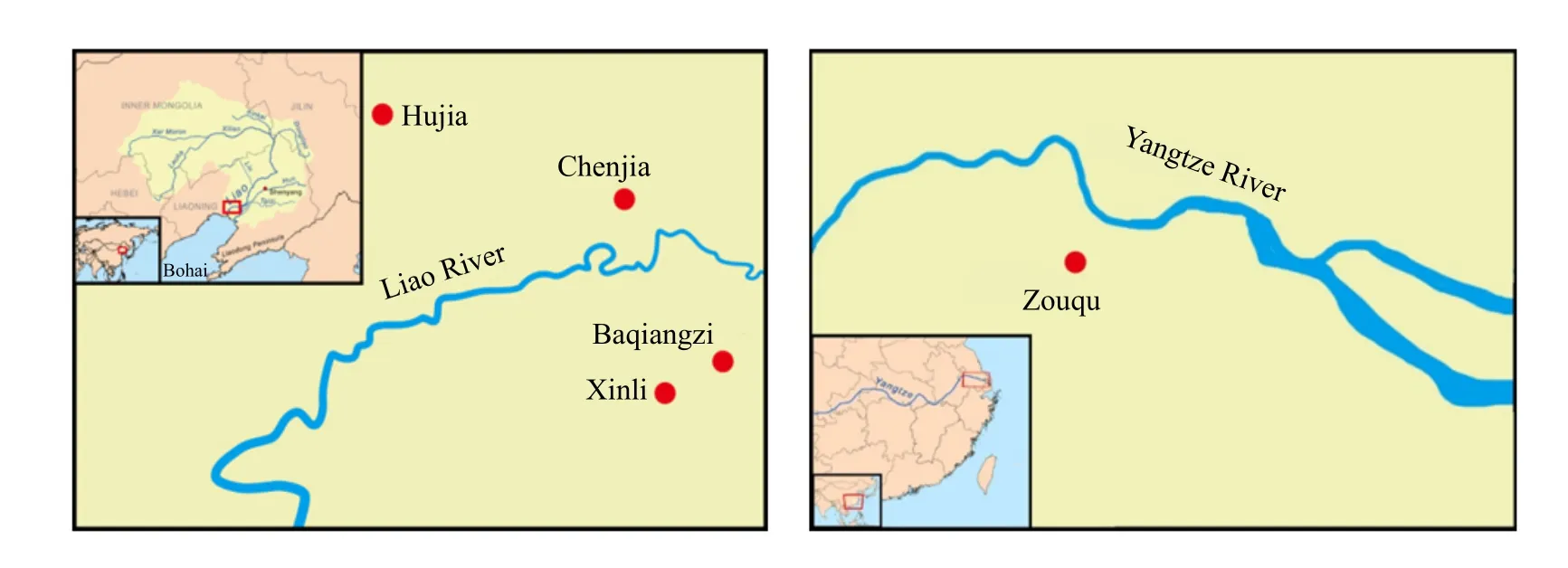
Fig. 1 Sampling location of Chinese mitten crab of the Liao River and the Yangtze River population (E. sinensis)
DNA extraction
Firstly, put the muscle into a mortar containing liquid nitrogen and grind. Then, DNA from each sample was extracted separately using a MagPure Tissue DNA KF Kit (Guangzhou Megiddo Biological Technology Co.,Ltd.). The integrity of DNA was detected by agarose gel electrophoresis, and the concentration and purity were determined using a UV spectrophotometer (Bio-Rad GelDoc XR). DNA was stored at -80℃ and diluted to 50 µg · mL-1.
SSR primers and PCR amplification
Two DNA samples from each town were used to screen 55 SSR primers as reported by Xionget al. (2012),Maoet al. (2009) and Maet al(2007). To identify the germplasm resources of the Liao River crabs, a pair of primers reported by Zhou and Gao (1999) was used. All the primer information is shown in Table 1.PCR instrument (Biometra) was used to amplify PCR with a total volume of 20 µL containing 10 µL 2×ESTaqmixture, 1 µL gene-specific primers, 1 µL DNA template and 7 µL RNase-free water (Beijing ComWin Biotech Co., Ltd). PCR amplification (25 cycles) conditions were as the followings: initial denaturation for 2 min at 94℃ followed by a locusspecific number of cycles of 30 s at 94℃, 30 s at optimal annealing temperature and 30 s at 72℃ with a final extension step of 10 min at 72℃. PCR product was stored at -20℃ before electrophoresis.
Polyacrylamide gel electrophoresis
PCR-amplified product of each polymorphism was analyzed by 8% non-denaturing polyacrylamide gel electrophoresis with silver staining as the followings:pre-electrophoresis for 10 min followed by loading PCR product on the gel and electrophoresis for 6 h at 150 V. After electrophoresis, the gel was first marked and washed in ddH2O for 5 min followed by staining with 0.1% AgNO3solution for 10 min, rinsed twice in ddH2O, and then finally colourized in a developing solution (2% NaOH solution, 2 mL formaldehyde) for 10 min. A picture was taken for analysis using a Nikon camera (Quet al., 2004).
Data analysis
All the bands on the electrophoretic gel were analyzed using Gel Pro software (Media Cybernetics), and "tag data format conversion" software was used to convert the positions of the bands into genotypes for each sample. The number of alleles (Na), effective number of alleles (Ne), observed heterozygosity (H0), expected heterozygosity (He), genetic similarity coefficient(I), genetic distance (Ds) and coefficient of genetic differentiation (FST) were calculated using Popgen1.32 software (Nei, 1978).

Table 1 Identification and characterization of microsatellites of E. sinensis
PIC_CALC 0.6 software was applied to determine the polymorphism information content (PIC); whenPIC>0.5, the tested site was a highly polymorphic locus (Botsteinet al., 1980). Nei'sDsof the four subpopulations was calculated using Popgen1.32 software(Francis Yeh), and an unweighted pair group method with arithmetic mean (UPGMA) phylogenetic tree was constructed using MEGA 5.0 software (Tamuraet al.,2011).
Results
Identification of the Liao River crabs
The Liao River crab identification results are shown in Fig. 2; the primer was effective at detecting both the Liao River and the Yangtze River crabs. In the Liao River population, several bands were amplified by PCR reaction, but only one or no band was detected in the Yangtze River population. Thus, the Liao River crabs were not heavily mixed, so there was a potential for cultivate purebred breeding selection.
Genetic diversity of the four crab sub-populations and microsatellite polymorphism
The genetic diversity results of the four Liao River Chinese mitten crab sub-populations are shown in Table 2.
A total of 106 alleles (Na) were detected with an average of 7.6154, 7.6154, 7.1538 and 7.6154 alleles per locus among the four sub-populations,andNeranged from 4.7883 to 5.2963. HJ sub-population had the highestNerelative to the three remaining populations. The averageH0of HJ, CJ,BQZ and XL sub-populations was 0.6145, 0.6112,0.5768 and 0.5707, respectively, andHewas 0.7996,0.7853, 0.7744 and 0.7959 for the sub-populations.PICranged from 0.7264 to 0.7588, and all the four sub-populations showed high polymorphism(PIC>0.5) according the standard of Botsteinet al(1980).
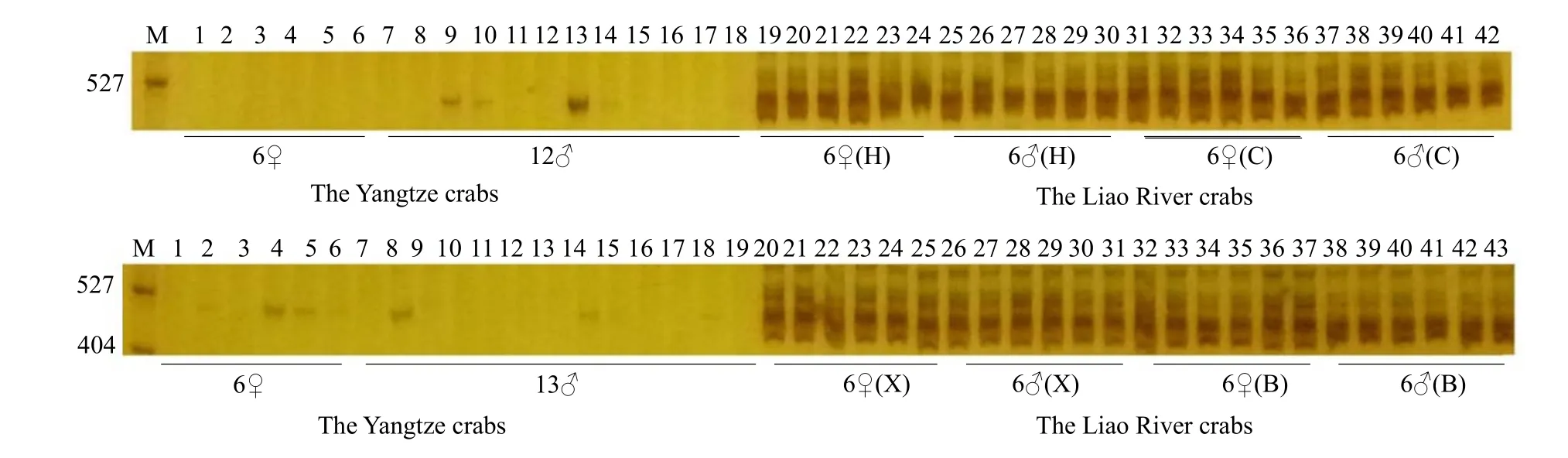
Fig. 2 Identification results of the Liao River crab germplasm resources
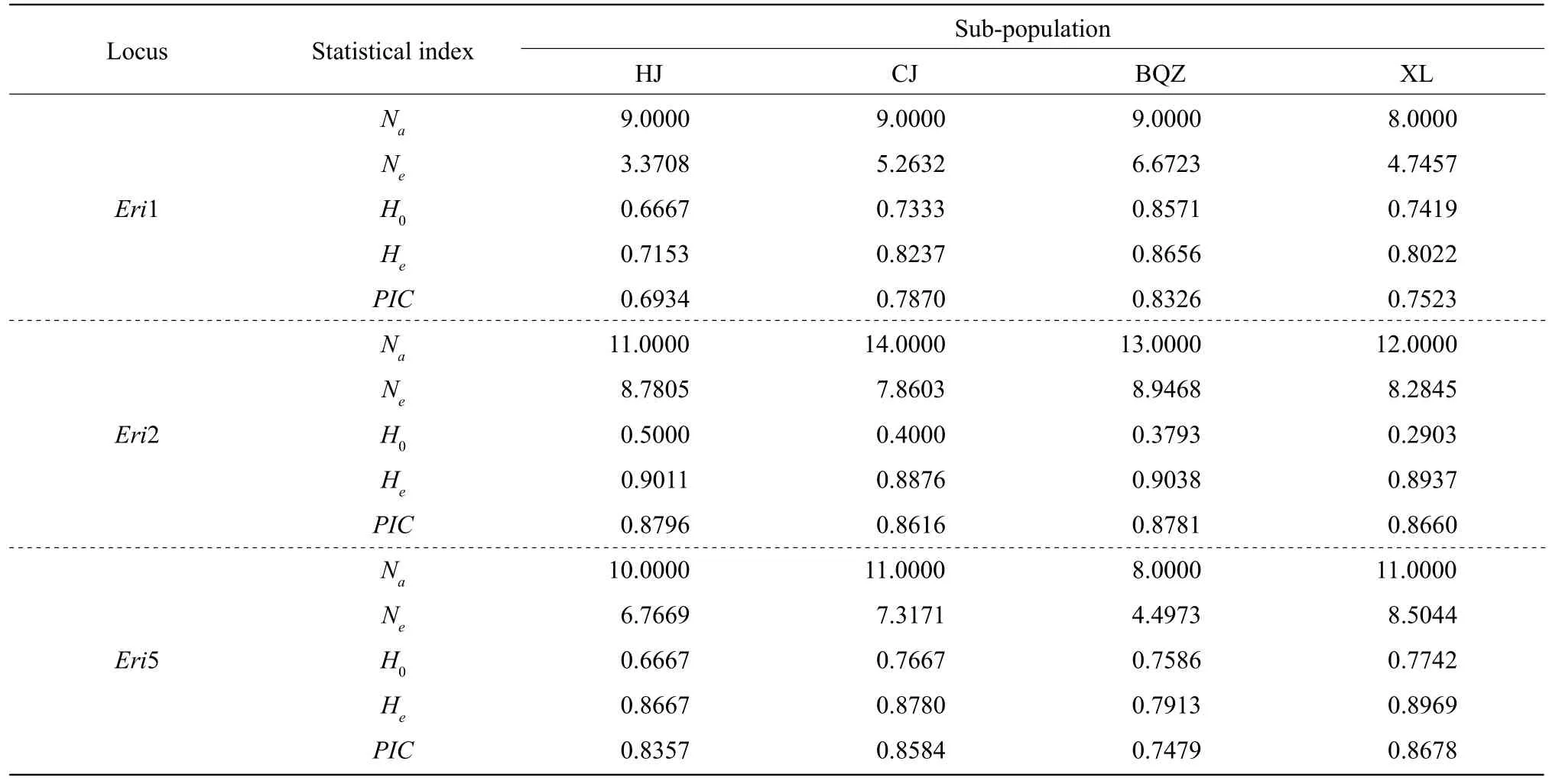
Table 2 Polymorphism of the four sub-populations of the Liao River Chinese mitten crab (E. sinensis) at 13 microsatellite loci
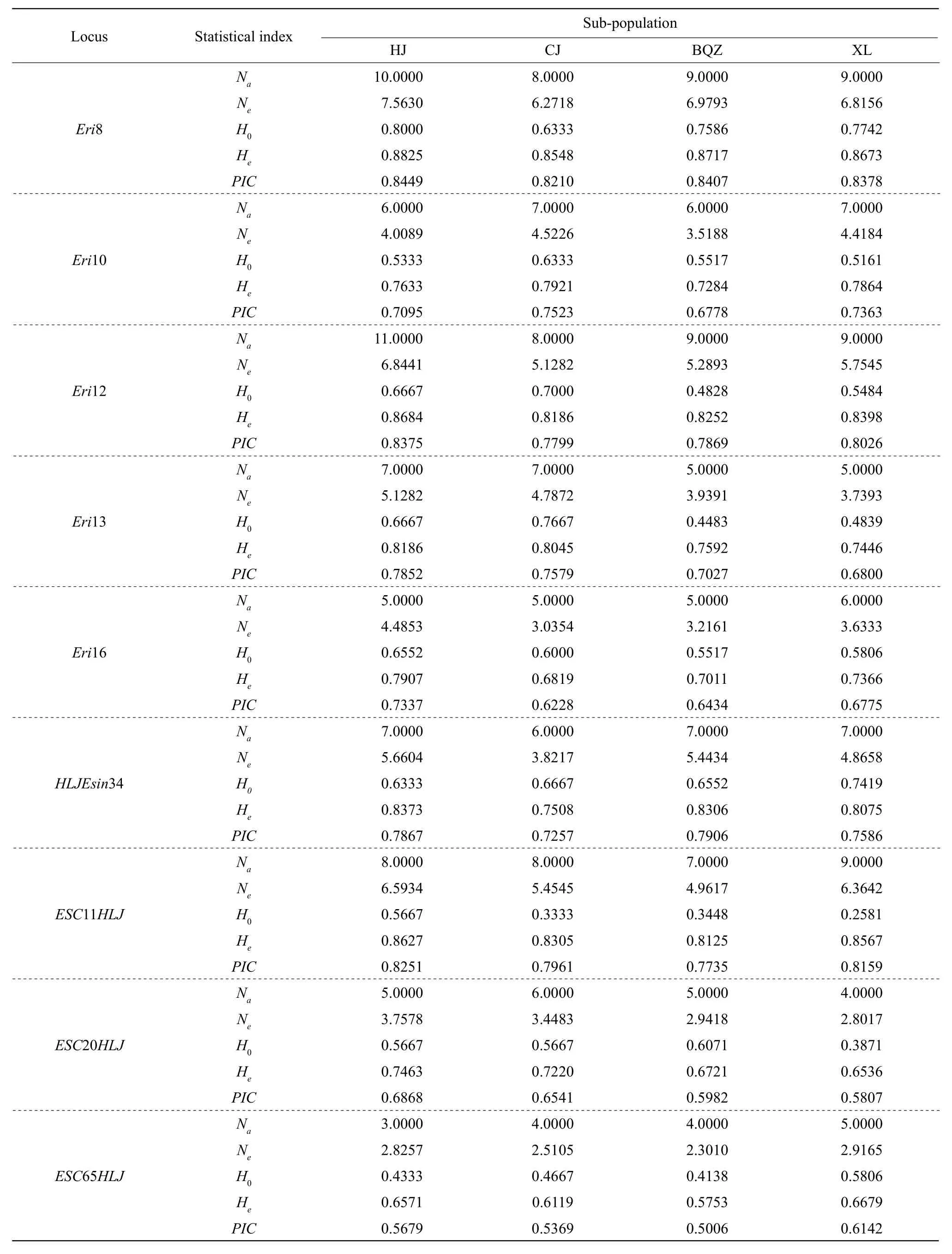
Continued
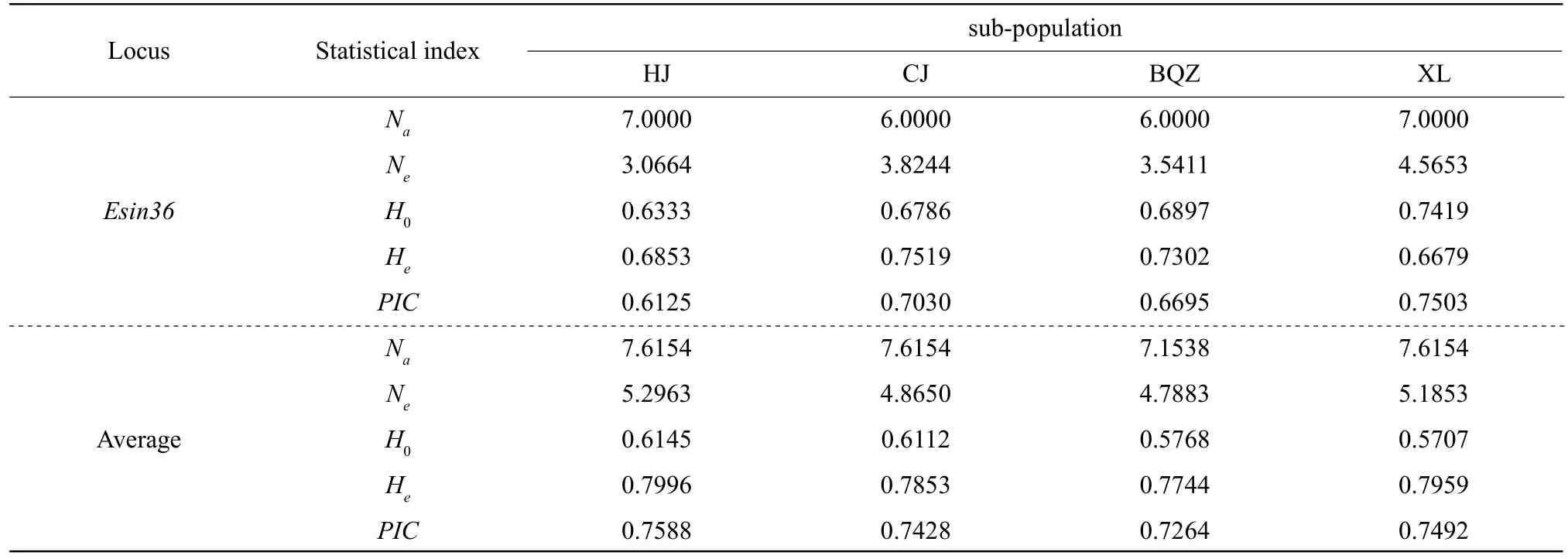
Continued
The genetic diversity of the Liao River population(all the four sub-populations combined) is shown in Table 3. The meanNaper locus was 8.9231, with a range from 5 to 15, andNeranged from 2.9777 to 9.9516 with an average of 5.7468. The averageH0was 0.5931 with a range of 0.3750 to 0.7479, and the averageHeper locus was 0.8064 with a range of 0.6669 to 0.9033.PICranged from 0.6028 to 0.8913 with an average of 0.7753.FSTvalues ranged from 0.0175 to 0.0696 with a mean of 0.0342. These data showed that the Liao River population had a low level of differentiation. The results of the above analyses indicated that the Liao River population of Chinese mitten crab had high genetic diversity.
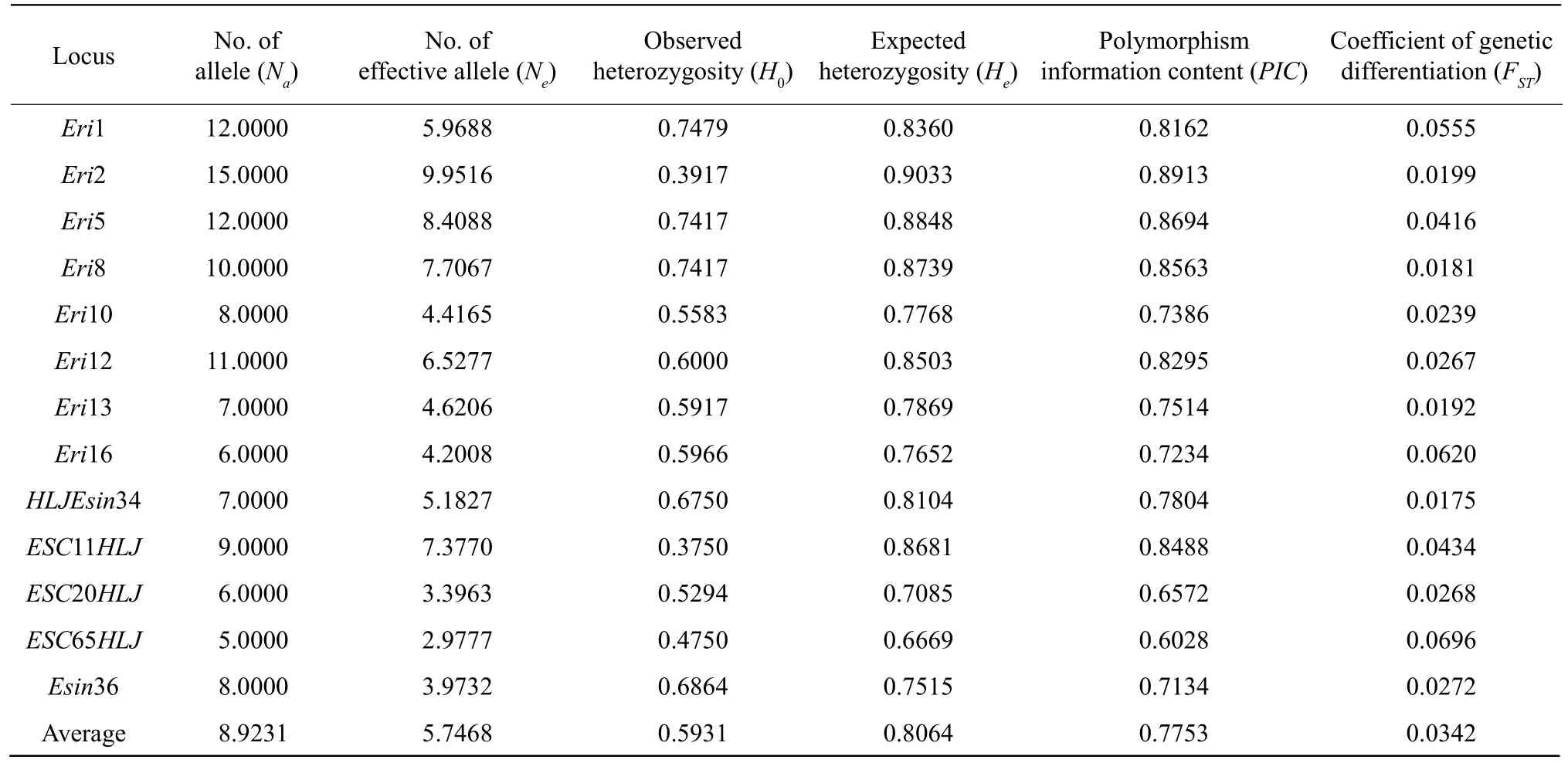
Table 3 Polymorphism of the Liao River Chinese mitten crab (E. sinensis) at 13 microsatellite loci
Genetic differentiation among the four subpopulations
BQZ and XL sub-populations had the greatestDs(0.2628) and the smallestI(0.7689). Meanwhile, HJ and XL sub-populations had the smallestDs(0.1162)and the largestI(0.8903) (Table 4).
Based on UPGMA phylogenetic tree, HJ, XL and CJ sub-populations belonged to the same cluster, while BQZ sub-population was isolated (Fig. 3).
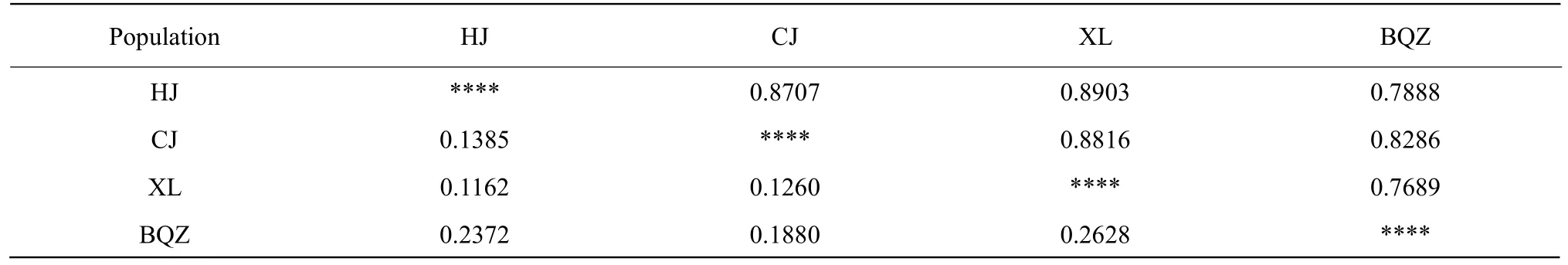
Table 4 Genetic distance (Ds, below diagonal) and similarity (S, above diagonal) of the four sub-populations of the Liao River Chinese mitten crab (E. sinensis)
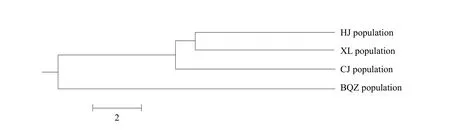
Fig. 3 UPGMA phylogenetic tree of the four sub-populations of the Liao River Chinese mitten crab (E. sinensis) based on Nei's genetic distance
Discussion
Identification analysis of the Liao River crabs
Chinese mitten crab is an important germplasm resource that is widespread in China; the three main populations are found in the Liao River, the Yangtze River and the Oujiang River (Wang and Li, 2002). It is difficult to distinguish among these populations,due to the limitations of current microsatellite maker methods, but several genetic differences have been detected by random amplification of polymorphic DNA (RAPD). In this study, one primer (HX01-400)was used to distinguish the Liao River from the Yangtze River crabs, demonstrating that the primer was effective at differentiating the samples, which was consistent with previous studies (Zhou, 2009).Zhou and Gao (1999) first developed the means to distinguish the Liao River from the Yangtze River crab. They collected the Liao River crabs in Panjin City and the Yangtze River crabs in Jiangsu Province and then, successfully identified the Yangtze River crabs mixed with the Liao River crabs or the Oujiang crabs from several farms. However, this study was the first to show that the Liao River crab did not mix with the Yangtze River crab. The reason might be that the Yangtze River crab was susceptible to cold temperatures. Perhaps, the Yangtze River crab was not suitable for the survival overwinter. Therefore, the resource conservation of the Liao River crabs should be strengthened and this had laid a solid foundation for further research on the breeding of the Liao River population.
Genetic diversity analysis of the Liao River population
Biodiversity included genetic diversity, species diversity and ecosystem diversity, and genetic diversity were the basis for the evolution of life and species differentiation. The origin of species diversity, variation and evolution could be revealed by studying genetic diversity (Yinet al., 2008);Na,Ne,H0,He,I,Ds,FSTand other indicators were the major factors involved in the analysis of genetic diversity.
In this study, out of 55 pairs of population specific primers for the microsatellite loci, 13 were used to analyze the genetic diversity of the Liao River crab population; the meanNawas 8.9231,Newas 5.7468,H0was 0.5931,Hewas 0.8064, andPICwas 0.7753.These data showed that Chinese mitten crabs from the local Liao River population showed high genetic diversity, which was consistent with the results of previous studies (Hänfling and Weetman, 2003; Suiet al., 2009; Changet al., 2008). Changet al. (2008)collected samples from the four sites in Jiangsu,Anhui and Liaoning provinces and Tianjin City, and they demonstrated thatNawas nine atESC11HLJ,eight atESC20HLJand eight atESC65HLJ. Our analysis showed thatNawas nine atESC11HLJ, six atESC20HLJJ and five atESC65HLJin the Liao River population, suggesting that the germplasm resources of the Liao River crab populations were abundant. Liuet al. (2015) analyzed the genetic variation among wild and cultured populations of Chinese mitten crab from the Yangtze River, the Huanghe and the Liao River basins using microsatellite markers and found that crabs from different rivers showed abundant genetic diversities. Compared with the Yangtze River population of Chinese mitten crab, cultured crabs from the Liao River population showed richer diversities,which agreed with the results from prior studies. Xionget al. (2012) analyzed the genetic diversity of cultured populations of Chinese mitten crab using microsatellite markers and proposed thatNevalues of the cultured population in Liaoning Province were 4.51 atEri1, 6.82 atEri2 and 5.49 atEri5, but the results showed thatNevalues of the Liao River population were 5.9688 atEri1, 9.9516 atEri2 and 8.4088 atEri5. These results were not consistent with those of previous studies, because a greater number of samples were collected for this study, so the results were more representative of the Liao River Chinese mitten crab population. Although the results were not consistent,the conclusions were the same in that Chinese mitten crab breeding populations had high levels of genetic diversities, low levels of genetic differentiation and potential value for breeding. However, no previous reports analyzed the genetic diversity of the Liao River Chinese mitten crab population. The research on the genetic diversity of the Liao River crab population increased the understanding of biodiversity and contributed to the protection of the crab population and its local environment.
Genetic structure of the Liao River population
The degree of genetic differentiation was shown byFST. AnFSTvalue between 0 and 0.05 indicated low genetic differentiation; between 0.05 and 0.15 indicated medium differentiation; and greater than 0.15 meant high genetic differentiation (Balloux and Lugon-Moulin, 2002). In this study, the averageFSTof the Liao River Chinese mitten crab population was 0.0342 with a range from 0.0175 to 0.0696, indicating low genetic differentiation. This observation was consistent with the results of research on native populations (Gao and Zhou, 1998; Wang and Yu, 1995). Suiet al. (2009)collected a total of 352 mitten crabs from nine locations in six river systems and showed that differentiation in the Liao River population was much lower than that in the Hepu population. Xionget al. (2012)analyzed the genetic diversity of cultured populations of Chinese mitten crab using microsatellite markers,and they proposed that both the Yangtze River and the Liao River populations had low differentiation.
Unlike the morphological discrimination and isozymic analyses, microsatellite markers were very sensitive to polymorphisms and provided more genetic information, such asDsand population structure. In this study, genetic structure analysis showed thatDswas the largest, 0.2628 andIwas the smallest between BQZ and XL sub-populations, andDswas the smallest,0.1162, andIwas the largest between HJ and XL subpopulations. Additionally, cluster analysis showed that HJ, XL and CJ sub-populations were in the same cluster, while BQZ sub-population was isolated.Therefore, it was suggested that HJ and XL subpopulations should form the basic breeding population,while BQZ sub-population should form a separate breeding population to prevent germplasm recession and maintain species diversity. Then, family selection could be applied.
Conclusions
In summary, the results had identified and analyzed the genetic diversity of the Liao River Chinese mitten crab, calculated Nei'sDsand constructed an UPGMA phylogenetic tree, and revealed the germplasm resources of this population. This study provided important basic information for crab germplasm evaluation, crab breeding and the development of molecular markers. The favourable gene characteristics of the Liao River should be explored in greater details bycombining traditional breeding techniques with marker-assisted breeding technology, and the development of fine seed for breeding Chinese mitten crab should be pursued. Finally, germplasm resources should be protected to promote the stable breeding of healthy Chinese mitten crabs.
Baker J S F. 1994.A global protocol for determining genetic distances among domestic livestock breeds. In: Proceedings of the 5th World Congress on Genetics Applied to Livestock Production.Canada. pp. 501-508.
Balloux F, Lugon-Moulin N. 2002. The estimation of population differentiation with microsatellite markers.Molecular Ecology,11(2): 155-165.
Botstein D, White R L, Skolnick M,et al. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms.American Journal of Human Genetics, 32(3):314-331.
Bureau of Fisheries Fishery Management. 2017.China fisheries statistical yearbook. Chinese Agricultural Press, Beijing. pp. 30.
Chang Y M, Liang L Q, Ma H T,et al. 2008. Microsatellite analysis of genetic diversity and population structure of Chinese mitten crab(Eriocheirs inensis).Journal of Genetics and Genomics, 35(3):171-176.
Chen K, Li E C, Yu Na,et al. 2013. Biochemical composition and nutritional value analysis of Chinese mitten crab,Eriocheir sinensis,grown in pond.Global Advanced Research Journal of Agricultural Science, 2(6): 164-173.
Chen W J. 2007.Research on technique of healthy aquaculture of river crab in large scale. Agricultural University of Nanjing, Nanjing. pp. 1.
Dai H X. 2012.The research of water purification and dissolved oxygen change rule of crab eco-culture ponds. Shanghai Ocean University, Shanghai. pp. 2-3.
Gao Z Q, Zhou K Y. 1998. Genetic variation of the Chinese mittenhanded crab (Eriocheir sinensis) populations detected by RAPD analysis.Chinese Biodiversity, 6(3): 186-190.
Gu X M, Zhao F S. 2001. Resources and culturing situation of Chinese mitten crab (Eriocheir sinensis) and species character conservation.Journal of Lake Sciences, 13(3): 267-271.
Hänfling B, Weetman D. 2003. Characterization of microsatellite loci for the Chinese mitten crab,Eriocheir sinensis.Molecular Ecology Notes, 3(1): 15-17.
Huang L, Wang C H, Li S F. 2008. Preliminary study on genetic variability in Chinese mitten crab (Eriocheir sinensis) from the Yangtze river by AFLP marker.Journal of Shanghai Fisheries University, 17(4): 385-389.
Li X H, Xu Z Q, Pan J L,et al. 2010. Genetic diversity of selected stock of Chinese mitten crab,Eriocheir sinensis.Journal of Fishery Sciences of China, 17(2): 236-242.
Li Y S, Li S F, Xu G Y,et al. 2000. Comparison of growth performance of Chinese mitten crab (Eriocheir sinensis) in pen culture from the Yangtze and Liaohe River systems.Journal of Shanghai Fisheries University, 9(3): 189-193
Liu Q, Liu H, Wu X G,et al. 2015. Genetic variation of wild and cultured populations of Chinese mitten crabEriocheir sinensisfrom the Yangtze, Huanghe, and Liaohe river basins using microsatellite marker.Oceanologia Et Limnologia Sinica, 46(4): 958-968.
Ma H T, Chang Y M, Yu D M,et al. 2007. Microsatellite variations among four populations ofEriocheir sinensis.Zoological Research, 28(2): 126-133.
Mao R X, Lu C Y, Liu F J,et al. 2009. Fractional genomic libraries constructing and microsatellites DNA screening of Chinese mitten crab (Eriocheir sinensis).Journal of Shanghai Fisheries University,18(2): 129-135.
Nei M.1978. Estimation of average heterozygosity and genetic distance from a small number of individuals.Genetics, 89(3): 583-590.
Qu L J, Li X Y, Du Z Q,et al. 2004. Thecomparision of denatured and non-denatured page-silver-staining detection of PCR product of microsatellites.Hereditas(Beijing), 26(4): 522-524.
Sui L Y, Zhang F M, Wang X M,et al. 2009. Genetic diversity and population structure of the Chinese mitten crabEriocheir sinensisin its native range.Marine Biology, 156(8): 1573-1583.
Tamura K, Peterson D, Peterson N,et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.Molecular Biology & Evolution, 28(10): 2731-2739.
Tang B P, Zhou K Y, Song D X. 2000. Biodiversity of mitten crabEriocheir.Journal of Hebei University, 20(3): 304-308.
Wang C H, Li S F. 2002. Advances in studies on germplasm in Chinese mitten crab,Erocheir sinensis.Journal of Fishery Sciences of China,9(1): 82-86.
Wang D, Yu W J. 1995. A comparative study on the isozymes inEriocheir sinensisfrom Yangtze and Liaohe Rivers.Journal of Liaoning University(Natural Sciences Edition), 22(4): 79-81.
Wang W, Li Y S, Cheng Y X. 2007. River crab breeding and crab culture.Fisheries Science & Technology Information, 34(5):25-28.
Xiong L W, Ma Z, Ma K Y,et al. 2012. Analysis of genetic diversity in cultured populations of the Chinese Mitten Crab (Eriocheir sinensis) by microsatellite markers.Journal of Agricultural Biotechnology,20(12): 1441-1448.
Xu D K, Lin Y. 1997. Theanalysis of mitten crab larvae at different rivers.Journal of Aquaculture, 3(1): 26-28.
Yin Q Q, Li D Y, Wang H Z,et al. 2008. Microsatellite maker analysis on genetic diversity in two German mirror carp (CyprinuscarpioL.)families.Journal of Anhui Agricultural University, 35(2): 211-218.
Zhao N G.1980. Experiments on the artificial propagation of the woolly-handed crab (Eriocheir sinensish. milne-edwards) in artificial sea water.Journal of Fisheries of China, 4(1): 95-104.
Zhou K Y, Gao Z Q. 1999. Identification of the mitten crabEriocheir sinensispopulations using RAPD markers.Chinese Journal of Applied and Environmental Biology, 5(2): 176-180.
Zhou M D. 2009.The comparison of culture results and growth charateristics of Erocheir sinensis under different specifications and densities. Huazhong Agricultural University, Wuhan.
Zhu Q S, Xia A J, Pan J L,et al. 2008. Analysis of biological characteristics of "Suxie No.1" (F3) selected breeding population ofEriocheir sinensis.Journal of Fisheries of China, 29(6): 8-12.
 Journal of Northeast Agricultural University(English Edition)2018年2期
Journal of Northeast Agricultural University(English Edition)2018年2期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Development of High Efficient in vitro Regeneration and Transformation System in Strawberry
- Effects of Low Temperature on Physiological Responses of Two White Clover Cultivars
- Risk Assessment of Maize Cold Damage in Heilongjiang Province in Recent 30 Years
- Evaluation of Post-operative Anti-stress Response of Dexmedetomidine in Dogs
- Effect of Lactobacillus acidophilus as a Dietary Supplement on Nonspecific Immune Response and Disease Resistance in Juvenile Common carp, Cyprinos carpio
- Design of Greenhouse Environment Control System Based on Internet of Things
