Wheat powdery mildew resistance gene Pm64 derived from wild emmer(Triticum turgidum var.dicoccoides) is tightly linked in repulsion with stripe rust resistance gene Yr5
Dyun Zhng, Kyu Zhu,Lingli Dong, Yong Ling, Gnqio Li,d,Tilin Fng,d,Gungho Guo, Qiuhong Wu,Jingzhong Xi, Yongxing Chn, Ping Lu, Miomio Li,Huizhi Zhng, Zhnzhong Wng, Yn Zhng,Qixin Sun, Zhiyong Liu,*
aCollege of Agronomy and Biotechnology, China Agricultural University,Beijing 100193,China
bState Key Laboratory of Plant Cell and Chromosome Engineering,Institute of Genetics and Developmental Biology,the Innovative Academy of Seed Design, Chinese Academy of Sciences,Beijing 100101,China
cUniversity of Chinese Academy of Sciences,Beijing 100049,China
dDepartment of Plant Soil Sciences,Oklahoma State University,Stillwater,OK 74078,USA
eChina Rural Technology Development Center,Beijing 100045,China
Keywords:Blumeria graminis Genetic linkage map Yellow rust Triticum aestivum Triticum dicoccoides
ABSTRACT Stripe rust and powdery mildew are both devastating diseases for durum and common wheat.Pyramiding of genes conferring resistance to one or more diseases in a single cultivar is an important breeding approach to provide broader spectra of resistances in wheat improvement.A new powdery mildew resistance gene originating from wild emmer (Triticum turgidum var.dicoccoides)backcrossed into common wheat(T.aestivum)line WE35 was identified.It conferred an intermediate level of resistance to Blumeria graminis f.sp.tritici isolate E09 at the seedling stage and a high level of resistance at the adult plant stage. Genetic analysis showed that the powdery mildew resistance in WE35 was controlled by a dominant gene designated Pm64.Bulked segregant analysis(BSA)and molecular mapping indicated that Pm64 was located in chromosome bin 2BL4-0.50-0.89. Polymorphic markers were developed from the corresponding genomic regions of Chinese Spring wheat and wild emmer accession Zavitan to delimit Pm64 to a 0.55 cM genetic interval between markers WGGBH1364 and WGGBH612,corresponding to a 15 Mb genomic region on Chinese Spring and Zavitan 2BL,respectively.The genetic linkage map of Pm64 is critical for fine mapping and cloning. Pm64 was completely linked in repulsion with stripe rust resistance gene Yr5.Analysis of a larger segregating population might identify a recombinant line with both genes as a valuable resource in breeding for resistance to powdery mildew and stripe rust.
1. Introduction
Powdery mildew caused by Blumeria graminis f. sp. tritici (Bgt)and stripe rust caused by Puccinia striiformis f.sp.tritici(Pst)are devastating diseases that reduce wheat production worldwide. Due to rapid evolution and emergence of virulent pathogen races, it is an ongoing process to identify new resistance genes in order to maintain diversity of resistance in breeding programs. Many of the 81 powdery mildew resistance genes located at 54 loci (Pm1 to Pm60) in wheat came from cultivated or wild relatives[1,2].For example,resistance gene Pm12,transferred from Aegilops speltoides[3],and Pm21 in a series of wheat-Dasypyrum villosum translocation(T6VS·6AL)lines [4]confer resistance to all isolates of B. graminis f. sp.tritici in China[5],Europe[6]and the USA[7].
Wild emmer (Triticum turgidum var. dicoccoides) (AABB, 2n =4x = 28), the progenitor of both cultivated tetraploid and hexaploid wheat,is reported to be highly resistant to powdery mildew [8,9]. Several powdery mildew resistance genes were identified in wild emmer and transferred to durum and/or common wheat.Genes Pm16[10]and Pm30[11]on chromosome 5BS and may be identical or allelic [12]. Recessive resistance genes Pm26 and Pm42,are located on chromosome 2BS,but are separated by 36.8 cM [13,14]. Pm36 and Pm41, located on chromosomes 5BL and 3BL,were identified in tetraploid wheat backgrounds [15,16]. Temporarily designated resistance genes
MlZec1, MlIW72, Ml3D232, PmG16, PmG3M, MlIW170, and PmAs846 were located on chromosomes 2BL [17], 7AL [18], 5BL[19],6BL[20],7AL[21],2BS[22],and 5BL[23],respectively.
Various methods have been employed to identify and distinguish genetic loci and alleles conferring disease resistance in wheat. The current method of locating genes with molecular markers not only enables genes with small or large effects on phenotype to be localized on chromosomes, but if the linkage between the phenotype of interest and molecular marker(s) is sufficiently close, the marker can be used as a genetic proxy to expedite transfer of the trait of interest without the need for phenotyping. Currently, molecular markers linked to more than 40 wheat powdery mildew resistance loci are reported [6,24,25]and some of these markers have been used in marker-assisted selection (MAS)and pyramiding of disease resistance genes.
Pyramiding of disease resistance genes into a single genotype is an important breeding approach to obtain more durable resistance. Closely linked genes for resistance to wheat rusts and powdery mildew occur at the Pm1-Lr20-Sr15 loci on chromosome 7AL [26]and in the Pm5-Sr17-Lr14 region in 7BL[27]. Stripe rust resistance gene Yr5, originally derived from T.spelta var.album and cytologically located on chromosome 2BL[28], is effective against many isolates of Pst in China [29,30],Europe[31],and the USA[32]and likely has been widely used in wheat breeding programs. Co-segregating or closely linked disease resistance gene-like sequences developed either from resistance gene analog (RGA) or polymorphic AFLP fragments suggested that Yr5 is a member of an R-gene cluster[32-34]and this was confirmed with the recent cloning of Yr7,Yr5,and YrSP with the latter two being allelic and different from Yr7[35].
WE35 is a common wheat introgression line with powdery mildew resistance from T. turgidum var. dicoccoides accession G-573-1. It has an intermediate level of resistance to Bgt isolate E09 at the seedling stage and a high level of resistance at the adult plant stage. In this paper we report 1) the identification of a new powdery mildew resistance gene PmWE35 originating from wild emmer; and 2) close repulsion linkage of the powdery mildew resistance gene PmWE35 and stripe rust resistance gene Yr5.
2. Materials and methods
2.1. Plant materials
Common wheat line S2199,a selection from Chang 3/BY16[36],was used as the source of stripe rust resistance gene Yr5.S2199 is highly resistant to prevailing Chinese Pst races,but is highly susceptible to Bgt race E09.WE35 is a BC3F6derivative(87-1*4/G-573-1)of wild emmer accession G-573-1(originally provided by Dr. Z.K. Gerechter-Amitai, the Volcani Centre, Israel) and common wheat line 87-1,and is susceptible to many Pst races including CYR29, CYR30, and CYR32, but has an intermediate seedling response (infection type (IT) 1-2) and a high level of adult plant resistance(IT 0;-1)to isolate of Bgt race E09.The F2population and F3progenies of cross S2199 × WE35 were used for disease phenotyping and molecular marker genotyping.
2.2. Evaluation of stripe rust and powdery mildew responses
Bgt isolate E09 and Pst race CY32 were used to inoculate S2199,WE35,S2199 × WE35 F1hybrid plants,the F2population and F3families, under controlled greenhouse conditions. Two 25-seedling sets of each F3family were used for separate powdery mildew and yellow rust tests. Highly susceptible common wheat cultivars Xuezao and Mingxian 169 were used as the powdery mildew and stripe rust susceptible controls,respectively. Inoculation of Bgt isolate E09 was performed when the first leaf was fully expanded as described [37].Inoculation of Pst race CY32 was performed at the two-leaf stage [36]. Infection types recorded 14 days after inoculation for both diseases were based on 0-4 IT scales, with 0 representing immunity, 0; for necrotic flecks, and 1-4 for highly resistant, resistant, susceptible and highly susceptible reactions, respectively. Reactions were classified into two groups,resistant(R,IT 0-2) and susceptible(S,IT 3-4)[36,37].
2.3. Genomic DNA isolation and marker analysis
Genomic DNA was extracted from parental lines and F2:3families following the CTAB method[38].Two DNA bulks were constructed for bulked segregant analysis (BSA) [39]by pooling equal amounts of DNA from 10 homozygous stripe rust resistant but powdery mildew susceptible F2:3families(SRPSbulk) and 10 homozygous stripe rust susceptible but powdery mildew resistant F2:3families (SSPRbulk), respectively. Genomic SSR, EST-SSR and EST primers (Graingenes,http://wheat.pw.usda.gov/) located on chromosome 2BL were screened for polymorphisms between the parental lines and contrasting SRPSand SSPRDNA bulks. The resulting polymorphic markers were evaluated across the entire F2population for linkage determination.
PCR were performed in 10 μL volumes containing 10 mmol L-1Tris-HCl, 50 mmol L-1KCl, 1.5 mmol L-1MgCl2,200 μmol L-1dNTPs, 20 ng of each primer, 50 ng genomic DNA, and 0.75 U Taq DNA polymerase. PCR conditions were initial denaturation at 94 °C for 5 min followed by 35 cycles of 94 °C for 45 s, 50-60 °C (depending on specific primers) for 45 s, and 72 °C for 60 s, with a final extension at 72 °C for 10 min. PCR products were mixed with 2 μL loading buffer(98%formamide,10 mmol L-1EDTA,0.25%bromophenol blue,and 0.25% xylene cyanol) and separation in 8% nondenaturing polyacrylamide gels (39 acrylamide:1 bisacrylamide) as described by Li et al. [16]. Gels were silverstained and photographed.
2.4. Genetic linkage map construction
Chi-squared(χ2)tests were used to evaluate the goodness-offit of observed data with expected segregation ratios.Linkages between molecular markers and resistance genes were determined using MapMaker 3.0b [40]and a LOD score of 3.0 as threshold. The genetic map was drawn with the software Mapdraw V2.1[41].
2.5. Chromosome arm and bin assignments of polymorphic markers
Chromosomal and bin locations of polymorphic markers flanking the disease resistance genes were determined using Chinese Spring homoeologous group 2 nullisomic-tetrasomic,ditelosomic, and chromosome 2BL deletion lines. Deletion lines enable markers to be localized to a chromosome bin flanked by the breakpoints of the largest deletion possessing the fragment and the smallest deletion lacking it after comparing amplification patterns.
2.6. Development of polymorphic SSR and STS markers
Genomic regions near the locations of the resistance genes for marker development were identified in the T. aestivum cv.Chinese Spring[42]and T.turgidum ssp.dicoccoides cv.Zavitan[43]whole genome assembly sequences. Firstly, these sequences were used to screen simple sequence repeat (SSR)motifs using BatchPrimer3 to develop SSR markers[44].Then,STS (sequence-tagged site) primer pairs were developed to identify length polymorphisms using DNAMAN software with the following parameters: amplification product size 200-500 bp with an optimal 300 bp, primer length 18-22 bp,temperature 55-65 °C, and GC content 40%-60%.
2.7. Gene annotation
The TriAnnot pipeline from URGI (http://wheat-urgi.versailles.inra.fr/Tools/Triannot-Pipeline) [45]was used for sequence annotation, where sequences were analyzed with various integrated coding region predictions, homology search analysis, and repetitive DNA analysis programs.The functions of predicted genes were described according to the best hit of BLASTP in the National Center for Biotechnology Information database (https://www.ncbi.nlm.nih.gov/).
3. Results
3.1. Genetic analysis of the powdery mildew and stripe rust resistances
When inoculated with Pst race CY32 and Bgt isolate E09,S2199 was highly susceptible(IT 4)and resistant(IT 0;),respectively.WE35 was intermediate (IT 1-2) in response to powdery mildew and highly susceptible to stripe rust (IT 4) (Fig. 1). F1plants of S2199 × WE35 gave an intermediate seedling response (IT 1-2) to powdery mildew and highly resistant response to stripe rust (IT 0;), indicating dominance of resistance to both diseases. Genetic analysis showed that both the powdery mildew and yellow rust resistances were inherited as single genes and the powdery mildew resistance gene, temperately designated PmWE35, co-segregated in repulsion with stripe rust resistance gene Yr5(Table 1,Fig.2).
3.2. Genetic linkage map for PmWE35 and Yr5
Because Yr5 was previously located on chromosome 2BL, 48 genomic SSR markers located on 2BL were screened to identify polymorphisms between the parental lines and DNA bulks.Eight markers were polymorphic and linked to Yr5 and PmWE35;Xgwm388,Xbarc128,Xgwm120,Xwmc175,Xgwm526,and Xwmc317 were co-dominant,and the other two were dominant.Xwmc332 was in coupling with the Yr5 allele and Xgwm47 was in coupling with the PmWE35 allele. PmWE35 and Yr5 were mapped to a 3.3 cM interval between markers Xwmc175 and Xwmc332, and Xgwm47 co-segregated with PmWE35 and Yr5 (Fig. 2-b). To find more polymorphic markers linked to PmWE35 and Yr5,five ESTSSR and 47 EST-STS primer pairs previously mapped on the 2BL were examined.Five markers were polymorphic and linked to Yr5 and PmWE35, among which CA648434, Pwc5-1/6, and CD490485 were co-dominant; BE444894 and BE637228 were dominant.PmWE35 and Yr5 were thus delimited to a 2.45 cM genetic interval between Xwmc175 and Xpwc5-1/6.EST-SSR marker CA648434 cosegregated with both PmWE35 and Yr5(Fig.2-b).
3.3. Bin mapping of PmWE35 and Yr5
Chinese Spring homoeologous group 2 nullisomictetrasomics, ditelosomics and deletion lines were used to assign the chromosomal and physical bin locations of Yr5 and PmWE35-linked SSR markers.SSR marker Xgwm47 was absent in N2B-T2A,and deletion line 2BL-4(Fig.3).This indicated that PmWE35, Yr5 and linked SSR marker Xgwm47 were located in bin 2BL-4(0.50-0.89)(Fig.2-a).
3.4. Comparative genomics analysis

Fig.1-Seedling responses of WE35 and S2199 at 15 days post inoculation with Bgt isolate E09 and Pst race CY32.
The sequences of the STS, EST-SSR, and EST-STS markers flanking the target resistance genes were used as queries to search the rice(Oryza sativa)and Brachypodium distachyon genomic sequences to identify orthologous loci. Orthologous gene pairs between rice and Brachypodium were compared within homologous genomic regions. The sequences of ESTs CA648434,CD490485, BE444894, and BE637228-2 were used as queries to search for orthologous genes in the rice and Brachypodium genomic sequences. Comparative analyses indicated that the genomic region flanking PmWE35 and Yr5 showed a good syntenic relationship with rice chromosome 4 and Brachypodium chromosome 5. CA648434, CD490485, and BE444894 detected orthologs on the long arm of rice chromosome 4(LOC_Os04g53740, LOC_Os04g57300, and LOC_Os04g57440) and Brachypodium chromosome 5 (Bd5g22770, Bd5g25500, and Bd5g25637),respectively(Fig.2-c,d).Therefore,PmWE35 and Yr5 were delimited to a genomic region orthologous to collinear genomic regions of 2091 kb(LOC_Os04g53740 to LOC_Os04g57440)on rice chromosome 4L, and 1791 kb (Bradi5g22770 to Bradi5g25500) on Brachypodium chromosome 5L. These orthologous genomic regions could provide further assistance for fine mapping and gene cloning of PmWE35.
3.5. Development of closely linked genetic markers
STS and SSR markers were developed from the released T.aestivum cv. Chinese Spring [42]and T. turgidum ssp. dicoccoides Zavitan [43]whole genome assembly sequences, and screened for polymorphism between parental lines WE35 and S2199, as well as the resistant and susceptible DNA bulks. Ten co-dominant markers, WGGBH134, WGGBH686, WGGBH1212,WGGBH1260,WGGBH1364,WGGBH218,WGGBH1099,WGGBH612,WGGBH913,and WGGBH252,were developed and mapped to the PmWE35 genomic region after genotyping the mapping population (Figs. 2-b, 4, Table 2). The recently cloned Yr5 gene specific marker [35]was also tested in this population. The Yr5-derived marker co-segregated with stripe rust resistance and markers WGGBH218, WGGBH1099 and CA648434 that also co-segregated with the Yr5 and PmWE35. Therefore, PmWE35 and Yr5 were delimited to a 0.55 cM genetic interval between markers WGGBH1364 and WGGBH612, and WGGBH218 and WGGBH1099 co-segregated with PmWE35 and Yr5(Fig.2-b).
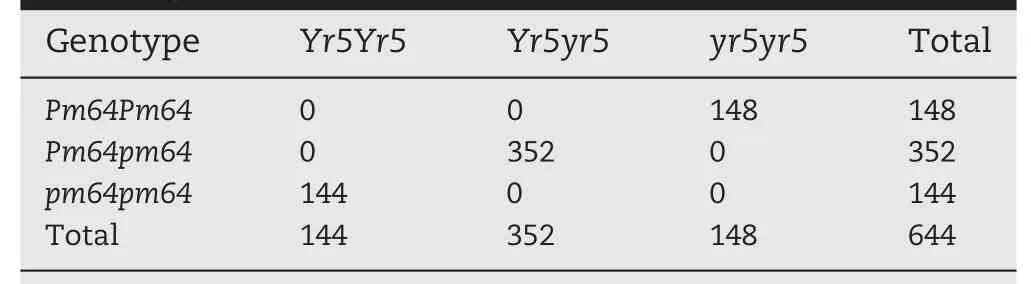
Table 1-Distribution of F3 family genotypes in cross S2199 × WE35 tested for response to stripe rust and powdery mildew.
3.6. Gene annotation of the PmWE35 genomic region
Flanking markers WGGBH1212 and WGGBH612,closely linked to PmWE35 and Yr5, were used to identify the corresponding genomic region in T.aestivum cv.Chinese Spring and T.turgidum ssp. dicoccoides Zavitan, respectively. Finally, PmWE35 and Yr5 were delimited to a 15 Mb physical region in Chinese Spring and Zavitan.After gene annotation,there were 206 predicted protein coding genes in Chinese Spring and 202 predicted protein coding genes in Zavitan (data not shown). The gene density was 13.8 genes Mb-1in the genomic region of Chinese Spring, and similarly,13.5 genes Mb-1in the genomic region of Zavitan.
4. Discussion
4.1. Relationship between PmWE35 and known powdery mildew resistance genes

Fig.2- Genetic and comparative genomics linkage maps of wheat powdery mildew resistance gene Pm64 and stripe rust resistance gene Yr5.(a)Wheat 2BL physical bin map;(b)genetic linkage map of Pm64 and Yr5 on wheat chromosome arm 2BL,genetic distances are shown to the left in cM;(c)orthologous genomic region on Brachypodium chromosome 5 with orthologous genes shown on the right;(d)orthologous genomic region on rice chromosome 4 with orthologous genes shown on the right.
Seven powdery mildew resistance genes, Pm6 [46], Pm33 [47],MlZec1[17],PmJM22[48],MlAB10[49],Pm51[50],and Pm52[51],are reported on chromosome arm 2BL.In common wheat,the T. timopheevii-derived Pm6 is located in a large linkage block that spans the centromere and rarely undergoes recombination. In this respect, it behaves as an alien segment and mapping studies are more likely to detect the break-point between wheat and alien chromatin, or rare recombinants,than give a true indication of actual location in the wheat chromosome[52].It can be predicted that in a recombination test PmWE35 and Pm6 would be allelic,but such a result need not be indicative of a truly allelic or even orthologous relationship. A RFLP-derived Pm6-specific STS marker NAU/STSBCD135-2 was located 0.8 cM from Pm6 [53]. This marker amplified in WE35,S2199,and Coker 747,which carries Pm6.A 230 bp Pm6-specific DNA band was present in Coker 747, but not in WE35 and S2199 (data not shown). This suggests absence of T. timopheevii chromosome 2G chromatin or Pm6 in wheat lines used in this study assuming that the marker was specific to T. timopheevii chromatin.

Fig.3- Amplification pattern of Xgwm47 in parental lines WE35 and S2199,Chinese Spring(CS)and homoeologous group 2 nulli-tetrasomics,ditelosomics,and deletion lines.

Fig.4- PCR amplification patterns of polymorphic markers WGGBH134(a), WGGBH218(b), and WGGBH252(c).Lanes 1 and 2,WE35 and S2199;lanes 3-6,lines homozygous resistant to powdery mildew;lanes 7-10, lines homozygous susceptible to powdery mildew;lanes 11-14,heterozygous resistant lines.
Pm33, identified in T. carthlicum accession PS5 and transferred to common wheat[47],is closely linked to SSR markers Xwmc317, Xgwm111, and Xgwm382 on chromosome 2BL. The temporarily designated dominant powdery mildew resistance gene MlZec1, identified in wild emmer-derived wheat line Zecoi-1, mapped distally to SSR marker Xwmc356 in terminal bin 2BL 0.89-1.00 [17]. Xwmc356 was null-allelic in both S2199 and WE35 and therefore could not be mapped in the present study. Another temporarily designated powdery mildew resistance gene MlAB10 identified in T. dicoccoides was physically mapped in the distal bin 2BL6-0.89-1.00 [49]. It is unclear if MlZec1 and MlAB10 are allelic since both originated from wild emmer and were located in the same chromosomal bin. Pm33, MlAB10, and MlZec1 were distal to SSR marker Xgwm526 at genetic distances of 18.1 cM, 36 cM, and 14.2 cM,respectively. Since PmWE35 was located proximally to SSR marker Xgwm526 with a genetic distance of 24.1 cM and physically mapped in bin 2BL-4(0.50-0.89),it must be different from Pm6,Pm33,MlZec1,and MlAB10(Fig. 5).
Another powdery mildew resistance gene located on chromosome 2BL is Pm51. This gene is allegedly derived from Thinopyrum ponticum and 3.2 cM distal to SSR marker Xwmc332 [50]. In this study, PmWE35 was located 2.18 cM proximal to SSR marker Xgwm332. However, different genetic maps can have different genetic distances between the two genes and the same SSR marker (Fig. 5). These two genes are likely different due to uncommon origin but need to be proved by a future test of allelism.
Pm52 was identified in Chinese wheat cultivar Liangxing 99 and physically mapped to 2BL bin 0.35-0.50 [51], and PmJM22 was identified in Chinese wheat cultivar Jimai 22 and physically mapped to bin 2BL-0.89-1.00 [48]. Therefore, Pm52 and PmJM22 are supposed to be different from PmWE35 which was physically mapped in bin 2BL-0.50-0.89 (Fig. 5). Other powdery mildew resistance genes mapped on chromosome 2BS include the wild emmer-derived Pm26 [13], Pm42 [14],MlIW170 [22]and T. dicoccum derived Pm49 [54]. Therefore,PmWE35 is presented as a new powdery mildew resistance gene designated Pm64.

Table 2-Polymorphic markers linked with Pm64 and Yr5.

4.2. Gene analysis of the Pm64 and Yr5 genomic regions
Two hundred and 202 genes in the Pm64 and Yr5 genomic region were annotated in T.aestivum cv.Chinese Spring and T.dicoccoides cv. Zavitan, respectively (data not shown). Only seven genes (Ta_gene10, Ta_gene31, Ta_gene32, Ta_gene33,Ta_gene34, Ta_gene35, and Ta_gene90) in Chinese Spring were missing in Zavitan, and three genes (Td_gene52, Td_gene184,and Td_gene185) in Zavitan lacked corresponding genes in Chinese Spring. The protein sequences of the 199 shared genes relatively high similarity. These results indicated that the genomic region of Pm64 and Yr5 was evolutionarily well conserved, and provided confidence that genes could be cloned based on the reference sequences.
All four of the currently cloned powdery mildew resistance genes in wheat produce CC-NBS-LRR (CNL) proteins.The first cloned resistance gene Pm3b located on chromosome 1AS encoded a CC-NBS-LRR protein[55].Using powdery mildew susceptible mutants and NLR capture-sequencing,Sanchez-Martin et al. [56]cloned Pm2 on 5DS and that gene also encoded a CC-NBS-LRR protein. Pm60 also encoding a CC-NBS-LRR gene derived from T.urartu on chromosome 7AL was map-based cloned [2]. The broad-spectrum resistance gene Pm21 encoding a CC-NBS-LRR protein on chromosome 6VS of Haynaldia villosa was isolated by comparative genomic analysis [57]and NLR capture-sequencing of H. villosa chromatins [58]. Also, a serine/threonine kinase gene Stpk-V on 6VS was found to be involved in the Pm21 regulation pathway[59].In the genomic region of Pm64 and Yr5 there are seven predicted NLR proteins and 21 putative Ser/Thr protein kinases (Tables S1 and S2) that provide target genes for cloning of Pm64.
4.3. Pm64 is closely linked in repulsion with stripe rust resistance gene Yr5
The powdery mildew resistance gene Pm64 in hexaploid wheat derivative WE35 originated from wild emmer accession G-573-1 collected in Israel. WE35 gave an intermediate seedling response to Bgt isolate E09, and a high level of resistance at the adult plant stage. Whether the adult plant resistance (APR) of WE35 was contributed solely by Pm64 needs further investigation. The resistance allele Pm64 segregated in repulsion with stripe rust resistance allele Yr5.Pst races virulent for Yr5 are extremely rare [30,32,60],suggesting that the gene could be very useful in breeding for resistance. Genomic regions with high gene density and frequent recombination in wheat are interspersed with relatively large regions of low gene density and infrequent recombination[61,62].No recombinant line with Pm64 and Yr5 was detected among 644 F2plants. It is likely that the relatively low gene density (13.5-13.8 genes Mb-1) led to low recombination in this genomic region. Alternatively, the genetic distance between Yr5 and Pm64 was too close to obtain a recombinant. Another reason could be a relatively large genomic difference between the chromosome segment of Yr5 originating from T.spelta and the Pm64 segment derived from T. dicoccoides. An increased mapping population might produce a recombinant line with stripe rust and powdery mildew resistance,which could be useful in breeding.
5. Conclusions
We identified a new powdery mildew resistance gene Pm64 originating from wild emmer that was completely linked in repulsion with stripe rust resistance gene Yr5. This gene will add diversity to the available genes for powdery mildew resistance. Pm64 was located to chromosome bin 2BL-0.50-0.89 and delimited to a 0.55 cM genetic interval between markers WGGBH1364 and WGGBH612, corresponding to a 15 Mb genomic region in common wheat.
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2019.03.003.
Conflict of interest
Authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2017YFD0101004), and the Science and Technology Service Network Initiative of Chinese Academy of Sciences (KFJ-STS-ZDTP-024). Gifts of aneuploid and deletion stocks from Prof. B.S. Gill and Mr. W.J. Raupp,Wheat Genetics Resource Centre,Kansas State University,USA,and STS primer PWC5-1/6 provided by Dr.X.Chen,Washington State University,USA,are gratefully acknowledged.
- The Crop Journal的其它文章
- Meta-analysis of QTL for Fusarium head blight resistance in Chinese wheat landraces
- The Crop Journal 作物学报(英文版) (Started in 2013, Bimonthly)
- Positional cloning of PmCH1357 reveals the origin and allelic variation of the Pm2 gene for powdery mildew resistance in wheat
- Wheat breeding in northern China: Achievements and technical advances
- Breed ing w heat for resistance to Fusarium head blight in the Global North: China,USA,and Canad a
- Pyramiding disease resistance genes in elite winter wheat germplasm for Western Canada

