橡胶树橡胶生物合成调控相关基因的表达相关性分析
杨署光 杨秀光 赵悦 史敏晶 李言 邓小敏 晁金泉 田维敏


摘 要:該文通过qPCR获得了正常割胶条件下,9个茉莉酸信号途径关键环节基因HbCOI1、HbJAZ1、HbJAZ2、HbJAZ3、HbMYC1、HbMYC2、HbMYC3、HbMYC4、HbMYC5和6个橡胶生物合成相关基因HbHRT2、HbSRPP、HbREF、HbHMGR1、HbHRT1、HbGAPDH在5个橡胶树魏克汉种质和5个1981IRRDB种质胶乳中的表达数据;通过皮尔逊相关系数分析了这15个基因彼此间的表达相关性,分别获得105对基因的双变量相关系数(r12)和偏相关系数(r12.3),所有105对基因的双变量相关系数(︱r12︱)和偏相关系数(︱r12.3︱)的平均值分别为0.486 ± 0.220和0.304 ± 0.211,达到0.05的差异显著水平。其中,r12与r12.3方向相同的63对(60%),方向相反的42对(40%);︱r12︱<︱r12.3︱的23对(21.905%),︱r12︱>︱r12.3︱的82对(78.095%);双变量相关显著性P<0.05的76对(72.38%),P<0.01的59对(56.19%);偏相关显著性P<0.05的21对(20%),P<0.01的16对(15.24%)。结果显示,这两条途径中的基因表达彼此相关,这为基因表达谱分析中普遍采用的前提假设“功能相关基因的表达相关”提供了进一步的实验证据,可用于挖掘、筛选和预测与橡胶生物合成相关的未知基因,为研究橡胶树产量形成的分子调控机理提供理论基础。
关键词:巴西橡胶树, 割胶, 茉莉酸信号途径, 橡胶生物合成, 基因表达, Pearson相关
中图分类号:Q943.2
文献标识码:A
文章编号:1000-3142(2020)12-1790-10
Abstract:Tapping-enhanced rubber biosynthesis is closely related to the activation of jasmonic acid signaling in laticifer cells of rubber tree.The expression level of genes related to jasmonic acid signaling pathway and rubber biosynthesis were both positively correlated with the dry rubber yield.However, the exact relationship between the expression of genes involved in jasmonic acid signaling and rubber biosynthesis is not to be elucidated yet.In the present study, qPCR was used to analyze the expression of nine jasmonic acid signaling genes, HbCOI1, HbJAZ1, HbJAZ2, HbJAZ3, HbMYC1, HbMYC2, HbMYC3, HbMYC4, HbMYC5 and six rubber biosynthesis genes, HbHRT2, HbSRPP, HbREF, HbHMGR1, HbHRT1, HbGAPDH, in laticifer cells of five Wichham germplasms and five 1981IRRDB germplasms following tapping them with S/2D d3 tapping system, Hb18S was used as internal reference gene.The correlation between these 15 genes was analyzed through Pearson correlation coefficient.The bivariate correlation coefficient (r12) and partial correlation coefficient (r12.3) of 105 gene pairs were obtained with the mean ± standard deviation of 0.486 ± 0.220 and 0.304 ± 0.211, respectively.Among them, 63 gene pairs (60%) with r12 in the same direction as r12.3 and 42 genes pairs (40%) in the opposite direction; In the degree of correlation, 23 gene pairs (21.905%) with | r12 | less than | r12.3 |, and 82 gene pairs (78.095%) with | r12 | greater than | r12.3 |.There 76 gene pairs (72.38%) bivariate correlation coefficient (r12) were significant correlation at P<0.05 level and 59 gene pairs (56.19%) at P<0.01 level.In contrast, there less partial correlation coefficient (r12.3) were significant correlation at P<0.05 level (21pairs, 20%) and P<0.01 level (16 pairs, 15.24%).These results suggested that the expression of genes involved in these two pathways were related to each other, which provides a further evidence for the assumption that “expression correlation of functional related genes”, which is widely adopted in gene expression profile analysis.It can be used for excavating, screening and predicting unknown genes related to rubber biosynthesis, as well as provides a theoretical basis for studying the molecular regulation mechanism of rubber yield formation in Hevea brasiliensis.
Key words:Hevea brasiliensis, tapping, jasmonate signaling, rubber biosynthesis, gene expression, Pearson correlation
茉莉酸是植物逆境响应的关键激素信号(Qi et al., 2011),COI1、JAZ和MYC是茉莉酸信号途径的关键环节(Chini et al., 2009);JA-Ile、COI1、JAZ和MYC通过精细的互作(Donnell et al., 1996; Chini et al., 2007; Fonseca et al., 2009; Qi et al., 2011)参与植物次生代谢的调节(Qi et al., 2011; Schweizer et al., 2013)。巴西橡胶树中的天然橡胶生物合成是一种典型的植物类异戊二烯代谢,受茉莉酸信号途径的调节(Deng et al., 2018);橡胶生物合成途径包括一系列橡胶生物合成关键酶(Koyama & Tanaka, 1996),如REF(Dennis & Light, 1989)、HMGR(Chye et al., 1992)、HRT(Asawatreratanakul et al., 2003)、SRPP(Collins, 2009)等;茉莉酸信号途径对这些酶的精细调节机制尚不完全清楚。橡胶树种质PR107、RRIM600、熱垦628、热垦525和热垦523的干胶产量普遍高于RO/CM/10 44/160、MT/IT/13 29/8、RO/C/8 24/104、RO/I/103 107和RO/CM/10 44/454,并且茉莉酸信号途径关键环节基因HbCOI1、HbJAZ1、HbJAZ2、HbJAZ3、HbMYC1、HbMYC2、HbMYC3、HbMYC4、HbMYC5的表达和橡胶生物合成酶基因HbHRT2、HbSRPP、HbREF、HbHMGR1、HbHRT1、HbGAPDH的表达与他们的橡胶产量正相关(杨署光等,2019a,b)。但是,这些基因表达的株间差异以及这些基因间的表达相关性尚不清楚。本研究通过分析这些基因在这10份种质中的表达相关性,可为研究橡胶树产量形成的分子调控机理提供理论基础。
1 材料与方法
1.1 材料
10份巴西橡胶树(Hevea brasiliensis)种质、试剂、耗材与先前的报道相同(杨署光等,2019a,b)。其中,RO/C/8 24/104为2株、RO/I/103 107为4株,其他种质各3株,共30株树。
1.2 方法
1.2.1 材料处理 生产中按S/2D d3正常割胶,在8月份的某次割胶时,分别收集前10 min流出的胶乳,用于提取胶乳总RNA。
1.2.2 总RNA的提取与cDNA的合成
用曾日中等(2003)的方法提取胶乳总RNA。cDNA第一链的合成与先前的报道相同(杨署光等,2019a,b)。
1.2.3 基因表达分析
1.2.3.1 qPCR引物合成 引物为序列来自文献(Tian et al., 2013;杨署光等,2019a,b),委托Invitrogen公司合成。
1.2.3.2 qPCR反应 qPCR反应按先前报道的方法完成(杨署光等,2019a,b)。
1.3 数据处理
以Hb18S作内参,按先前报道的方法(杨署光等,2019a,b)计算目的基因的相对表达值。用SPSS软件分析基因表达值的Pearson 相关性,P<0.01表示极显著相关,P<0.05表示显著相关;相关性的密切程度参照文献报道(朱婉丽,2019)的标准,根据相关系数(r)的数值大小,分为四个等级:0<|r︱≤0.3为微弱相关;0.3<︱r︱≤0.5为低度相关;0.5<︱r︱≤0.8为中度相关;0.8<︱r︱≤1为高度相关。
分别用SPSS软件的Duncan检验和Excel TTEST(Array 1,Array 2,Tails 2,Type 2)完成多重比较分析和双样本比较分析:P<0.01表示组间差异极显著,P<0.05表示组间差异显著。用GraphPad Prism 5作图。
2 结果与分析
2.1 橡胶树种质胶乳中橡胶生物合成调控相关基因的表达分析
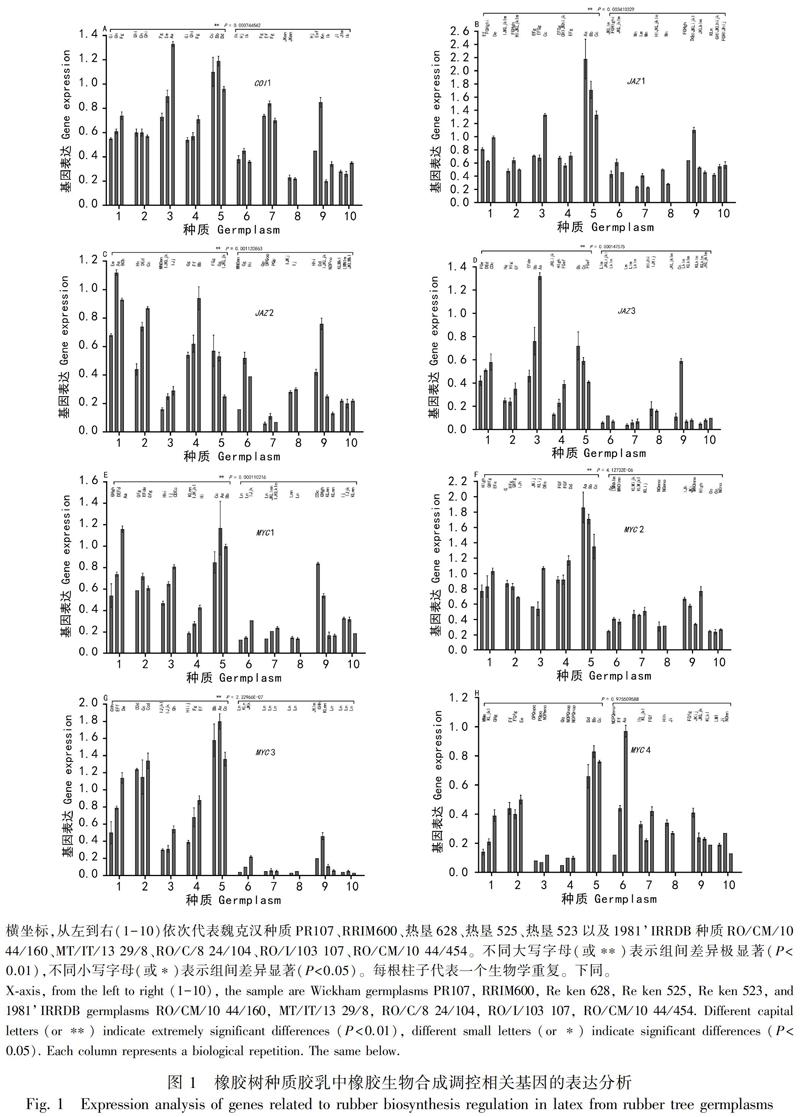
qPCR分析表明,不同种质中的基因表达差异明显,相同种质的生物学重复间也能达到极显著差异,并且高产种质中的基因表达普遍高于低产种质(图1、图2)。在高低产种质组间,COI1(图1:A)、JAZ1(图1:B)、JAZ2(图1:C)、JAZ3(图1:D)、MYC1(图1:E)、MYC2(图1:F)、MYC3(图1:G)、MYC5(图2:I)、GAPDH(图2:J)、HMGR1(图2:K)、REF(图2:M)、HRT2(图2:O)差异极显著,HRT1(图2:N)差异显著,SRPP(图2:L)差异不大,MYC4(图1:H)差异不显著;高低产之比分别为1.74、1.84、2.17、3.73、2.58、2.38、8.72、0.34、2.58、1.36、2.28、3.02、1.84、1.36、1.01。15个基因在高低产种质组间的平均表达差异极显著,两者之比为2.08(图2:P)。
2.2 橡胶树种质胶乳中橡胶生物合成调控相关基因表达的相关性分析
共分析了15个基因间的表达相关性,分别获得105对基因的双变量相关系数(r12)和偏相关系数(r12.3)(图3、图4)。其中,r12与r12.3方向相同的占60%,相反的占40%;微弱相关、低度相关、中度相关、高度相关的r12分别为20.952%、 33.333%、37.143%、8.571%,在r12.3中分别为54.286%、22.667%、18.095%、0.952%;︱r12︱<︱r12.3︱的为21.905%,︱r12︱>︱r12.3︱的为78.095%;显著相关和极显著相关的r12分别为72.38%和56.19%,在r12.3中分别为20%和15.24%;在30个植株中,9个茉莉酸信号途径相关基因表达的平均值与6个橡胶生物合成相关基因表达的平均值间的r12为0.892,极显著正相关。
分析了单个基因分别与其余14个基因间Pearson 相关系数的平均值(图4:P)。所有105对基因的︱r12︱和︱r12.3︱的平均值分别为0.486和0.304,两者差异显著;HRT2、REF、COI1、JAZ1、JAZ3、MYC1、GAPDH、MYC2、MYC3的︱r12︱与︱r12.3︱差异显著,而HMGR1、SRPP、HRT1、JAZ2、MYC4、MYC5的︱r12︱与︱r12.3︱差异不显著;各组中︱r12︱普遍大于︱r12.3︱,但彼此间的差异显著性具有广泛的交集;5个MYC家族
成员中,MYC1,2,3与其他14个基因的相关性相当,且极显著高于MYC4,5;3个JAZ家族成员中,JAZ1,2,3与其他14个基因的相关性差异不大,仅JAZ1,2间的︱r12︱达到0.05的显著水平;相对而言,REF、GAPDH、MYC1、MYC2和MYC3与其他14个基因的相关性高于MYC4和MYC5。
3 讨论与结论
橡胶树的橡胶生物合成是一种典型的植物类异戊二烯代谢,茉莉酸信号途径与类异戊二烯代因的表达与橡胶产量的相关性;同时,该研究结果显示,这两条途径中的基因表达彼此间具有一定的相关性,说明功能相关的蛋白,基因表达倾向相关;这为基因表达谱分析中普遍采用的前提假设“功能相关基因的表达相关”提供了理论依据,可用于挖掘、筛選和预测与橡胶生物合成相关的未知基因,为研究橡胶树产量形成的分子调控机理提供理论基础。
从多个角度研究了“功能相关基因的表达相关”这一现象。在线虫(Roy et al., 2002; Lercher et al., 2003)、酵母(Cohen et al., 2000; 王晨光等,2006)、果蝇(Spellman & Rubin,2002)和人类(Lercher et al., 2002;郭政等,2003)等不同物种上陆续发现了染色体上邻近基因表达相关这一现象。酵母中互作蛋白质的编码基因(王磊等,2004;王晨光等,2006;欧阳玉梅,2008)、蛋白质复合物的编码基因(Jansen et al., 2002;王磊等,2004)表达显著相关,并且功能联系越强基因表达相关性越高(王磊等,2004),相互作用蛋白质更倾向于对具有相同的亚细胞定位(王晨光等,2006;欧阳玉梅,2008);对基因数据库的分析表明,同一代谢通路上的基因表达倾向于高度相关(Harris et al., 2004; Kanehisa et al., 2004;Curtis et al., 2005),在酵母(李传星等,2004;王晨光等,2006)和狗(华琳等,2008)的研究中进一步证实了这种相关性。以上结果表明真核生物中的基因存在模块化的共表达趋势。
“功能相关基因的表达相关”具有实验条件依赖性:不同的实验条件对这种相关性有一定的影响(Bader et al., 2004;王晨光等,2006)。橡胶生物合成途径中也存在类似的实验条件依赖性,如在该研究的S/2D d3的割胶制度下,MYC1和MYC2显著正偏相关,而在S/2D d1的割胶制度下相反(Zhao et al., 2011)。研究基因组内和跨基因组的基因表达谱与蛋白质相互作用之间的相关性,发现与人类、小鼠等物种相比,酵母的蛋白质相互作用与基因表达谱的相关性最弱(Bhardwaj & Lu, 2005),推测“功能相关基因的表达相关”具有进化程度依赖性。
“HbCOI1-HbJAZs-HbMYCs”彼此间、家族成员间以及他们与FPS、SRPP、REF、HRT2等橡胶生物合成关键酶的相互作用(马弗明, 2010;赵悦,2011;刘伟,2011;何鑫, 2013;包杰,2014;肖华,2015;王靖等,2016;姚笛,2016;邓小敏等, 2018;Deng et al, 2018)表明他们在调控橡胶生物合成过程中具有协同作用,为这些基因的表达相关性提供了理化基础。
橡胶树的产量性状是一种典型的次生产量性状,由反复合理的收获胁迫(割胶)形成,是一个“形成-收获-再形成-再收获……”的过程(林位夫,2012);2次割胶之间的橡胶再生以及相关基因的表达是一个动态过程;由于基因表达的动态性、瞬时性、多样性和复合性,不同mRNA 的稳定性和降解速率不同,可能会影响特定实验条件下的基因表达水平(Kruglyak & Tang, 2000)。因此,系统追踪这些相关基因在排胶过程中的表达相关性是今后的一个研究方向。
参考文献:
ASAWATRERATANAKUL K, ZHANG YW, WITITSUWANNAKUL D, et al., 2003.Molecular cloning, expression and characterization of cDNA encoding cis-prenyltransferases from Hevea brasiliensis —A key factor participating in natural rubber biosynthesis[J].Eur J Biochem, 270(23):4671-4680.
BADER JS, CHAUDHURI A, ROTHBERG JM, et al., 2004.Gaining confidence in high-throughput protein interaction networks[J].Nat Biotechnol, 22 (1):78-85.
BAO J, 2014.Studies on the transcriptional regulation of key enzymes of natural rubber biosynthesis by MYC and Myb transcription factors[D].Haikou:Hainan University:1-70.[包杰, 2014.MYC和Myb转录因子对橡胶生物合成关键酶转录调节的研究[D].海口:海南大学:1-70.]
BHARDWAJ N, LU H, 2005.Correlation between gene expression profiles and protein-protein interactions within and across genomes[J].Bioinformatics, 21(11):2730-2738.
CHINI A, BOTER M, SOLANO R, 2009.Plant oxylipins:COI1/JAZs/MYC2 as the core jasmonic acid-signalling module[J].Febs J, 276(17):4682-4692.
CHINI A, FONSECA S, FERN NDEZ G, et al., 2007.The JAZ family of repressors is the missing link in jasmonate signalling[J].Nature, 448(7154):666-671.
CHYE ML, TAN CT, CHUA NH, 1992.Three genes encode 3-hydroxy-3-methylglutaryl-coenzyme A reductase in Hevea brasiliensis:hmgr1 and hmgr3 are differentially expressed[J].Plant Mol Biol, 19(3):473-484.
COHEN BA, MITRA RD, HUGHES JD, et al., 2000.A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression[J].Nat Genet, 26(2): 183-186.
COLLINS JE, 2009.The role of small rubber particle proteins in rubber biosynthesis[D].Reno, USA:University of Nevada:1-112.
CURTIS RK, ORESIC M, VIDAL-PUIG A, 2005.Pathways to the analysis of microarray data[J].Trends Biotechnol, 23(8): 429-435.
DENG XM, GUO D, YANG SG, et al., 2018.Jasmonate signalling in regulation of rubber biosynthesis in laticifer cells of rubber tree (Hevea brasiliensis Muell.Arg.)[J].J Exp Bot, 69(15):3559-3571.
DENG XM, WANG J, CHEN DK, et al., 2018.Preliminary screening of candidate interactors of HblJAZ3 from Hevea brasiliensis laticifer by using yeast two-hybridization methods[J].Chin J Trop Crop, 39(2):287-292.[鄧小敏, 王靖, 陈多坤, 等, 2018.橡胶树乳管细胞HblJAZ3互作蛋白的分离鉴定[J].热带作物学报, 39(2):287-292.]
DENNIS MS, LIGHT DR, 1989.Rubber elongation factor from Hevea brasiliensis identification, characterization, and role in rubber biosynthesis[J].J Biol Chem, 264(31):18608-18617.
DONNELL PJ, CALVERT C, ATZORN R, et al., 1996.Ethylene as a signal mediating the wound response of tomato plants[J].Science, 274(5294):1914-1917.
FONSECA S, CHINI A, HAMBERG M, et al., 2009.(+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate[J].Nat Chem Biol, 5(5):344-350.
GUO Z, ZHANG TW, LI X, et al., 2003.Analysis of expression correlation within adjacent genes along the human chromosomes by GeneHub[J].Chin J Bioinform, 1(1):11-14.[郭政, 张田文, 李霞, 等, 2003.利用GeneHub软件分析人类染色体相邻基因的表达相关性[J].生物信息学, 1(1):11-14.]
HARRIS MA, CLARK J, IRELAND A, et al., 2004.The gene ontology (GO) database and informatics resource[J].Nucl Acids Res, 32:D258-D261.
HE X, 2013.Research on the expression of several members of JAZ and MYC genes family correlated with natural rubber yield in Hevea brasilensis Muell.Arg.[D].Haikou:Hainan University:1-77.[何鑫, 2013.巴西橡胶树JAZ和MYC家族几个成员基因表达和产量相关性的研究[D].海口:海南大学:1-77.]
HUA L, ZHENG WY, LIU H, et al., 2008.Relativity of gene expression and Co-regulated gene patterns in feature KEGG pathways[J].Chin J Biotechnol, 24(9):1643-1648.[华琳, 郑卫英, 刘红, 等, 2008.特征代谢通路上的基因表达相关性及共调控表达模式[J].生物工程学报, 24(9):1643-1648.]
JANSEN R, GREENBAUM D, GERSTEIN M, 2002.Relating whole-genome expression data with protein-protein interactions[J].Genome Res, 12(1):37-46.
KANEHISA M, GOTO S, KAWASHIMA S, et al., 2004.The KEGG resource for deciphering the genome[J].Nucl Acids Res, 32:D277-D280.
KOYAMA T, TANAKA Y, 1996.Isopentenyl diphosphate isomerase in rubber latex[J].Phytochemistry, 43(4):769-772.
KRUGLYAK S, TANG H, 2000.Regulation of adjacent yeast genes[J].Trends Genet, 16 (3):109-111.
LERCHER MJ, BLUMENTHAL T, HURST LD, 2003.Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes[J].Genome Res, 13(2):238-243.
LERCHER MJ, URRUTIA AO, HURST LD, 2002.Clustering of housekeeping genes provides a unified model of gene order in the human genome[J].Nat Genet, 31(2):180-183.
LI CX, LI X, GUO Z, et al., 2004.Analysis of the mRNA expression similarity of genes in the same gene expression regulatory pathway[J].Heredias, 26 (6):929-933.[李傳星, 李霞, 郭政, 等, 2004.调控通路内基因表达的相关性分析[J].遗传, 26 (6):929-933.]
LIN WF, 2012.Secondary yield characters of crops and their exploitation[J].Chin J Trop Crop, 33(4):602-604.[林位夫, 2012.作物次生产量特性及其利用对策探讨[J].热带作物学报, 33(4):602-604.]
LIU W, 2011.Screen the proteins interacting with HbJAZ of Hevea brasiliensis[D].Haikou:Hainan University:1-55.[刘伟, 2011.橡胶树HbJAZ1基因的互作蛋白筛选研究[D].海口:海南大学:1-55.]
LU SX, 2010.Structural and HblMYC1 gene correlated with natural rubber yield in Hevea brasilensis Muell.Arg.[D].Haikou:Hainan University:1-49.[卢世香, 2010.树皮结构和HblMYC1基因与橡胶树产量相关性研究[D].海口:海南大学:1-49.]
MA FM, 2010.Cloning and expression of transcription factor for cis-isopentenyl transferase gene from Hevea brasiliensis[D].Haikou:Hainan University:1-80.[马弗明, 2010.橡胶树顺式-异戊烯基转移酶基因转录因子的克隆及表达[D].海口:海南大学:1-80.]
OUYANG YM, 2008.Correlation between protein-protein interactions in yeast and gene expression profiles and subcellular localization[J].Chin J Biochem Mol Biol, 24(10):950-956.[欧阳玉梅, 2008.酵母蛋白质相互作用与基因表达谱和亚细胞定位的相关性[J].中国生物化学与分子生物学报, 24(10):950-956.]
QI T, SONG S, REN Q, et al., 2011.The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana[J].Plant Cell, 23(5):1795-1814.
ROY PJ, STUART JM, LUND J, et al., 2002.Chromosomal clustering of muscle-expressed genes in caenorhabditis elegans[J].Nature, 418(6901):975-979.
SCHWEIZER F, FERN NDEZ-CALVO P, ZANDER M, et al., 2013.Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior[J].Plant Cell, 25(8):3117-3132.
SPELLMAN PT, RUBIN GM, 2002.Evidence for large domains of similarly expressed genes in the drosophila genome[J].J Biol, 1(1):5.
TIAN WM, ZHANG H, YANG SG, et al., 2013.Molecular and biochemical characterization of a cyanogenic β-glucosidase in the inner bark tissues of rubber tree (Hevea brasiliensis Muell.Arg.)[J].J Plant Physiol, 170(8):723-730.
WANG CG, GUO Z, LI X, et al., 2006.Multiple analysis of the relationship between correlation of gene function and expression by GeneHub software[J].J Biomed Eng, 23(6):1157-1161.[王晨光, 郭政, 李霞, 等, 2006.利用 GeneHub软件多角度分析基因功能相关与表达相关的联系[J].生物医学工程学杂志, 23(6):1157-1161.]
WANG J, DENG XM, TIAN WM, 2016.Screening and in vitro functional analysis of the HblMYC3 interacted proteins in laticifer cell of Hevea brasiliensis[J].Chin J Trop Crop, 37(7):1312-1317.[王靖, 鄧小敏, 田维敏, 2016.巴西橡胶树乳管细胞HblMYC3互作蛋白筛选与功能分析[J].热带作物学报, 37(7):1312-1317.]
WANG L, GUO Z, LI X, et al., 2004.Comparative analysis of gene expression correlation between interactive proteins and among subunits of protein complexes[J].Chin J Bioinform, (4):1-4.[王磊, 郭政, 李霞, 等, 2004.互作蛋白质与复合物亚基的基因表达相关性比较分析[J].生物信息学, 2(4):1-4.]
XIAO H, 2015.Validation and characterization of HbJAZ1 interacted proteins of Hevea brasiliensis[D].Haikou:Hainan University:1-70.[肖华, 2015.巴西橡胶树HbJAZ1互作蛋白的验证及功能鉴定[D].海口:海南大学:1-70.]
YANG SG, CHEN YY, LI Y, et al., 2019a.Correlation between the expression level of rubber biosynthesis genes and rubber yield[J].Chin J Trop Crop, 40(3):475-482.[杨署光, 陈月异, 李言, 等, 2019a.橡胶树橡胶生物合成相关基因表达与橡胶产量的相关性[J].热带作物学报, 40(3): 475-482.]
YANG SG, ZHAO Y, CHEN YY, et al., 2019b.Correlation between the expression level of genes related to jasmonate signaling and rubber yield[J].Guihaia, 39(5):641-649.[杨署光, 赵悦, 陈月异, 等, 2019b.橡胶树茉莉酸信号途径相关基因表达与橡胶产量的相关性[J].广西植物, 39(5):641-649.]
YAO D, 2016.Cloning and interactions of HbJAZs of Hevea brasiliensis[D].Haikou:Hainan University:1-55.[姚笛, 2016.巴西橡胶树HbJAZs基因的克隆与互作蛋白鉴定[D].海口:海南大学:1-55.]
ZENG RZ, DUAN CF, LI Y, et al., 2003.Construction of cDNA library and analyses of cDNA sequences in latex from JA-stimulated rubber trees[J].Chin J Trop Crop, 24(3):1-6.[曾日中, 段翠芳, 黎瑜, 等, 2003.茉莉酸刺激的橡膠树胶乳cDNA消减文库的构建及其序列分析[J].热带作物学报, 24(3):1-6.]
ZHAO Y, 2011.Involvement of jasmonate signaling pathway in regulating rubber biosynthesis in laticifer cells of Hevea brasilensis[D].Haikou:Hainan University:1-159.[赵悦, 2011.巴西橡胶树乳管细胞茉莉酸信号途径对橡胶生物合成调节的研究[D].海口:海南大学:1-159.]
ZHAO Y, ZHOU LM, CHEN YY, et al., 2011.MYC genes with differential responses to tapping, mechanical wounding, ethrel and methyl jasmonate in laticifers of rubber tree (Hevea brasiliensis Muell.Arg.)[J].J Plant Physiol, 168(14): 1649-1658.
ZHU WL, 2019.Study on application of correlation analysis principle in library and information analysis[J].Jiangsu Sci Technol Inform, (1):9-12.[朱婉丽, 2019.相关性分析原理在图书情报分析中的运用[J].江苏科技信息, (1):9-12.]
(责任编辑 李 莉)

