SSR-based hybrid identification, genetic analyses and fingerprint development of hybridization progenies from sympodial bamboo (Bambusoideae, Poaceae)
YUAN Jinling, MA Jingxia, ZHONG Yuanbiao, YUE Jinjun
(1.Research Institute of Subtropical Forestry, Chinese Academy of Forestry, Hangzhou 311400, China;2. Anji Bamboo Exposition Garden Co., Ltd., Huzhou 313300, China)
Abstract: 【Objective】 The study aims to accurately verify sympodial bamboo hybrids, clarify genetic relationships between hybrids and their parents, and develop applicable fingerprints. 【Method】We screened, tested and analyzed 30 pairs of randomly selected published bamboo simple sequence repeat (SSR) primers using hybrids derived from Bambusa multiplex × Dendrocalamus latiflorus and B. multiplex × B. chungii crosses. 【Result】According to the results, six SSR primer pairs (P8, P9, P18, P19, P20 and P27) were suitable for the hybrid identification and genetic analysis of the B. multiplex × D. latiflorus cross, which revealed that all 34 progenies were true hybrids (100%). Six SSR primer pairs (P2, P9, P10, P11, P20 and P28) were suitable for the hybrid identification and genetic analysis of the B. multiplex × B. chungii cross, which revealed that 54 of the 59 progenies were true hybrids (91.5%). Hybrids of the B. multiplex × D. latiflorus cross inherited more maternal alleles, while hybrids of the B. multiplex × B. chungii cross inherited more paternal alleles. The alleles detected using SSR primers were used to construct hybrid fingerprints of the two crosses for variety protection. 【Conclusion】 The SSR primers could be used to effectively verify sympodial bamboo hybrids as the progeny inherited alleles from different parents at varying probabilities in different crosses. In addition, the development of hybrid fingerprints could facilitate variety protection.
Keywords:sympodial bamboo; hybridization progenies; simple sequence repeat (SSR); hybrid identification; genetic analyses; fingerprint
BambusamultiplexandB.chungiibelong to the subgeneraLelebaandLingnaniaof the Gramineae family, respectively. They are characterized by long internodes, cold-resistance, and straight stalks.Dendrocalamuslatiflorusbelongs to the Gramineae family, and it is characterized by a large stalk, desirable shoot and timber traits for use by humans[6]. To lay a foundation for future breeding activities for novel bamboo varieties and research on bamboo genetic diversity, we established distant hybridization and inbred populations usingB.multiplex,B.chungiiandD.latiflorusas parental material[6]. Hybrid identification is vital during plant cross-breeding. Phenotypic traits of hybrids distinct from those of parental lines are always used for hybrid identification. Furthermore, various molecular makers based on DNA sequences are often used because they are more accurate. For example, random amplification of polymorphic DNA (RAPD), simple sequence repeat (SSR), inter-simple sequence repeats(ISSR), amplified fragment length polymorphism (AFLP), and single nucleotide polymorphism (SNP) techniques have been used for hybrid identification in bamboo and other plants[9-14]. Compared to other molecular techniques, SSR markers are based on tandem repeat sequences of dozens of nucleotides composed of several nucleotides as repeat units, which are informative, stable and co-dominant. In addition, the costs of SSR markers in experimental applications are considerably lower than the costs associated with DNA sequencing-based SNP markers. Therefore, SSR markers have been used extensively in hybrid identification, fingerprint development, and the analysis of the genetic structures of different populations[15-23].
In the present study, SSR molecular markers were used to analyze the two populations and verify hybrids, clarify the genetic relationships between parents and their progeny, then develop fingerprints for the selection and conservation of excellent novel varieties.
1 Materials and Methods
1.1 Materials
All experimental materials were obtained from the Hybrid Bamboo Experimental Base located in Guilin, Guangxi Province, China. Nine clones (A1-A9) ofD.latiflorus, 10 clones (C1-C10) ofB.chungii, and 10 clones (E1-E10) ofB.multiplexwere derived from self-pollinated plants. Thirty-four clones (B1-B34) of suspectedB.multiplex×D.latiflorushybrids and 59 clones (D1-D59) of suspectedB.multiplex×B.chungiihybrids were selected based on their phenotypes. Fresh leaves of each clone with no signs of pest damage or disease were sampled for genomic DNA extraction using the CTAB method[24]. Each clone refers to a plant from a seed.
1.2 Methods
1.2.1 Primer screening and SSR detection
Thirty SSR primer pairs were selected randomly from published literature[25-26](Table 1) and synthesized by the Beijing Liuhe Huada Gene Technology Co., Ltd.
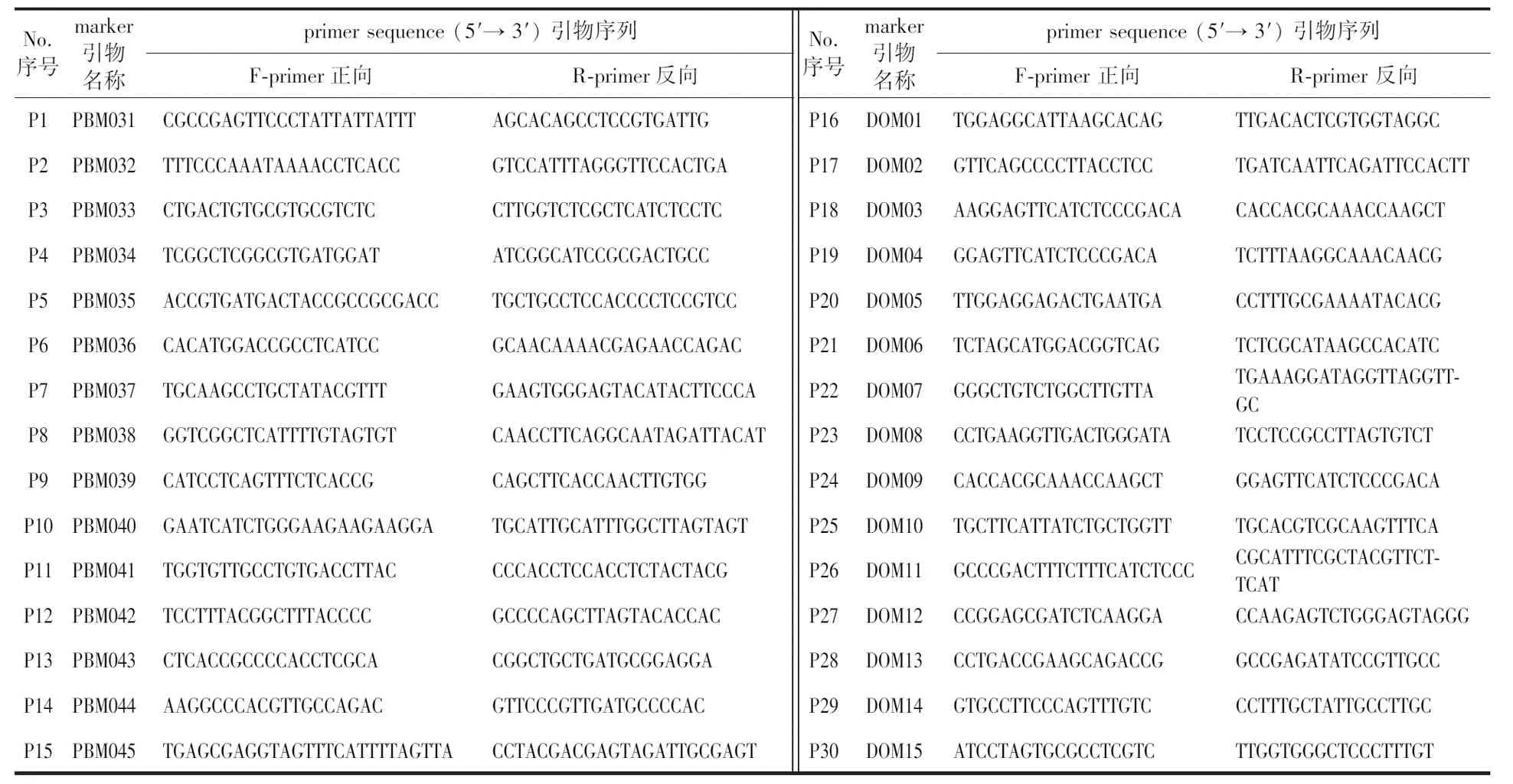
Table 1 Thirty SSR primers randomly selected and their sequence表1 随机选取的30对SSR引物及其序列等信息
Thirty SSR primer pairs were used for the PCR amplification of the genomic DNA of nine clones (A1-A9) ofD.latiflorusand 10 clones (E1-E10) ofB.chungii. PCR products were detected using gel electrophoresis. Primer pairs that generated clear polymorphic products between the two groups of clones were selected. Subsequently, equal DNA amounts of the 9 clones (A1-A9) ofD.latifloruswas pooled to form bulk A and the DNA of the 10 clones (E1-E10) ofB.chungiiwas pooled to form bulk E. The bulks and DNA of the 34 clones (B1-B34) of the suspectedB.multiplex×D.latiflorushybrids were used as PCR amplification templates along with the selected primers. The PCR products were detected in gels.
Additionally, equal DNA amounts of the 10 clones (C1-C10) ofB.chungiiwere pooled to form bulk C. Thirty SSR primer pairs were used in the PCR amplification of the DNA from bulks E and C. SSR primer pairs that produced clear polymorphic products between the two bulks were selected. The selected SSR primer pairs were used in the PCR amplification of the DNA from bulks E and C, as well as the 59 clones (D1-D59) of the suspectedB.multiplex×B.chungiihybrids. The PCR products were detected in gels.
The PCR reaction system (20 μL) included 2 μL (20 ng/μL) DNA template, 0.3 μL (20 μmol/L) F-primer, 0.3 μL (20 μmol/L) R-primer, 2 μL 10× Buffer, 1.2 μL (25 mmol/L) MgCl2, 0.4 μL dNTPs (2.5 μmol/L each), 0.2 μL (5 U/μL)TaqDNA polymerase, and 14.8 μL ddH2O.
Before long, however, the door opened and a tiny man stepped into the room, not more than three feet high, clothed in a dressing-gown, and with a small wrinkled face, and a grey beard which reached down to the silver buckles11 of his shoes
The PCR thermal program consisted of an initial denaturation at 94 ℃ for 5 min, followed by 35 cycles of denaturation at 94 ℃ for 30 s, annealing at 55-62 ℃ (depending on the specific primer pairs) for 35 s, extension at 72 ℃ for 40 s, and a final extension at 72 ℃ for 10 min.
Electrophoresis and staining of the PCR products were mixed with loading buffer, denatured at 94 ℃ for 10 min, and loaded onto a 6% denaturing polyacrylamide gel. Vertical electrophoresis included pre-electrophoresis at 90 W for 30 min prior to sample loading and electrophoresis at 90 W for 60 min after sample loading. Afterwards, the PCR products were visualized using the silver staining method[27]; the procedure included fixation in 10% glacial acetic acid for 3 min, rinsing once in ddH2O for no more than 10 s, staining in AgNO3solution (the 2 L solution included 6 g AgNO3, 200 mL absolute ethanol, and 10 mL glacial acetic acid) for 5 min, developing in developer solution (the 2 L solution included 40 g NaOH and 6 mL formaldehyde), and final fixing in 10% glacial acetic acid.
1.2.2 Data processing
Based on the relative positions of the amplified products displayed in the gel, we used 1 and 0 to denote the presence and absence of a fragment, respectively, in the order of the fragment size from large to small. Data were recorded in Excel 2010. Similarity coefficient calculations, cluster analysis, and dendrogram construction were processed using NTsyspc 2.10[28].
2 Results and Analyses
2.1 SSR primer screening
Thirty SSR primer pairs were used to amplify the DNA of nine clones (A1-A9) derived from self-pollinatedD.latiflorusplants and 10 clones (E1-E10) derived from self-pollinatedB.multiplexplants to screen polymorphic markers. According to the results, a total of six primer pairs (P8, P9, P18, P19, P20 and P27) yielded clearly distinct polymorphic PCR products between the clones of the two parents, which could be used for hybrid identification and parentage analysis. In the parental clones, the loci were usually heterozygous. Fig. 1 shows that primer pairs P8, P9 and P19 detected polymorphic sites between theD.latiflorusclones, P9 and P19 detected polymorphic sites betweenB.multiplexclones. To comprehensively represent the genetic characteristics of the parents, bulks A and E were used as the parental DNA in the parentage test. Similarly, 30 SSR primer pairs were screened by amplifying bulks E and C. According to the results, six primer pairs (P2, P9, P10, P11, P20 and P28) yielded clearly distinct polymorphic products between the parental bulks, which were used in subsequent analyses.

Fig.1 Electropherogram profiles of E1-E10 and A1-A9 under PCR amplification with primer pairs P8, P9 and P19图1 利用SSR引物P8、P9和P19的扩增样品E1-E10和A1-A9的电泳图
2.2 SSR marker-based identification of intergeneric and interspecific hybrids, genetic analyses of parent-progeny relationships, and development of hybrid fingerprints
2.2.1 Identification of E × A hybrids, genetic analyses of parent-progeny relationship, and development of hybrid fingerprints
The six SSR primer pairs (P8, P9, P18, P19, P20 and P27) were used in the analysis of 36 DNA samples (A, E and B1-B34) using PCR amplification and polyacrylamide gel electrophoresis. Thirty-four suspected hybrids contained alleles from female parent E and male parent A, indicating that they were true hybrids. The amplification results of 36 samples, including A, E and B1-B34, using P8 and P27 were presented in Fig. 2.
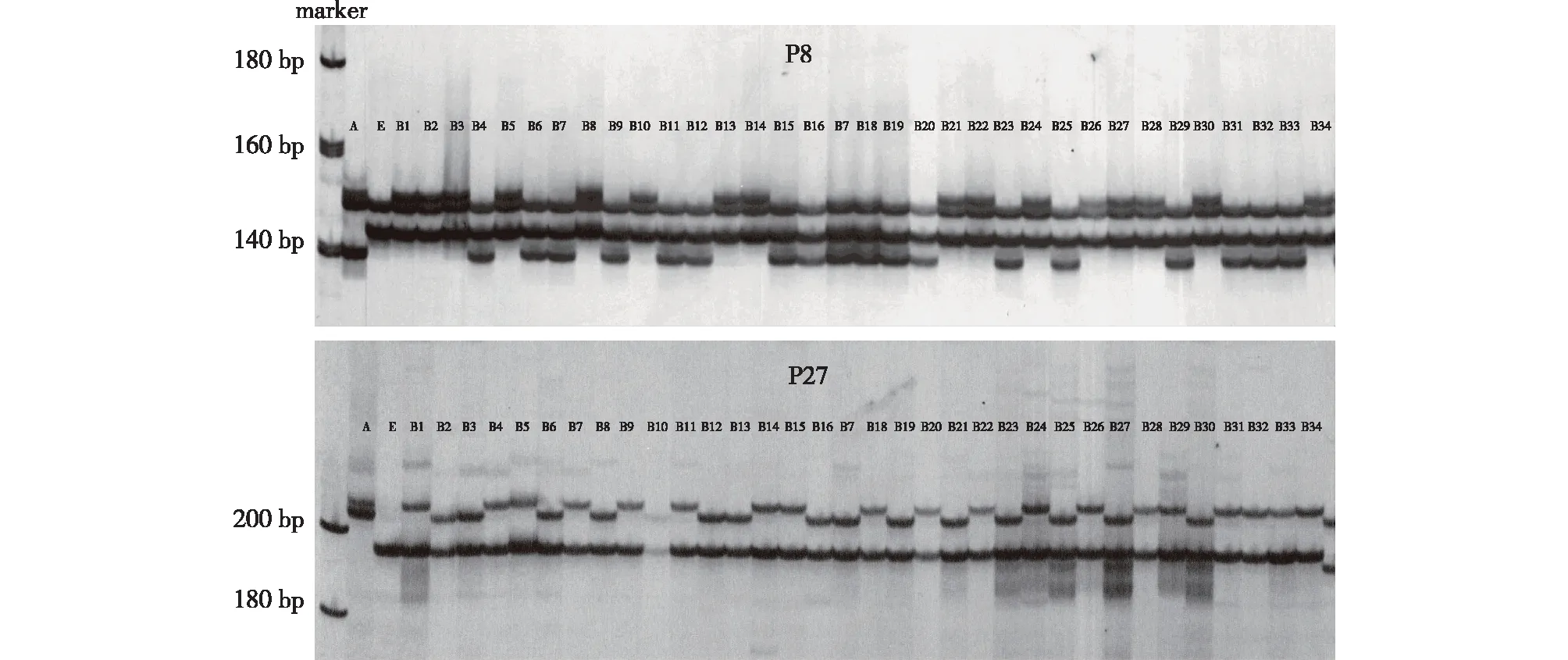
Fig.2 Electropherogram profiles of E, A and B1-B34 under the PCR amplification of primer P8 and P27图2 利用SSR引物P8和P27扩增样品E、A和B1-B34的电泳图
To clarify the genetic relationship between parents and their progeny from the E × A cross, the amplification data generated using six primer pairs (P8, P9, P18, P19, P20 and P27) were used for the similarity coefficient calculations, cluster analysis and dendrogram construction (Fig. 3). Female parent E had a closer genetic relationship with the hybrids than male parent A, indicating that the hybrids inherited more maternal alleles. Moreover, 25 amplified products generated by using the six primer pairs were unable to distinguish the clones B15 and B31, indicating that the genetic backgrounds of the two clones were relatively close. Furthermore, to identify the different hybrid clones and provide a molecular basis for variety conservation, fingerprints of 34 hybrids from E × A cross were developed using the PCR amplification loci from primer P8, P20 and P27 (Table 2).
Fig.3 Dendrogram of A, E and their 34 E×A hybrids under cluster analysis using alleles detected from six SSR primers (P8, P9, P18, P19, P20 and P27)图3 利用6对引物(P8、P9、P18、P19、P20 和 P27)扩增的位点对A、E及其34个E×A 杂种聚类分析的树状图

Table 2 Fingerprints of 34 hybrids from E × A cross using alleles detected from SSR primers of P8, P20 and P27表2 利用SSR引物P8,P20和P27扩增的位点构建34个E×A 杂种的指纹图谱
2.2.2 Identification of E × C hybrids, genetic analyses of the parent-progeny relationship, and development of hybrid fingerprints
The six primer pairs (P2, P9, P10, P11, P20 and P28) were used to amplify 61 DNA samples (E, C and D1-D59) and their products were observed following polyacrylamide gel electrophoresis, and the amplification results of 61 samples based on primer pair P9 were presented in Fig. 4. When using D48 DNA, all six primer pairs generated some PCR products that were different from parents C and E, indicating that D48 was neither the progeny of C nor E, but potentially a clone of other bamboo species crosses contaminated during the experiment. D21, D45, D57 and D58 contained alleles only from female parent E and no alleles from male parent C. Additionally, some of the primer pairs generated non-E products using the four clones as templates (D21/P9, D45/P20, D57/P9 and D58/P9), suggesting that the four clones were derived from a cross between female parent E and a non-C male parent. Therefore, D48, D21, D45, D57 and D58 were determined to be untrue hybrids. The remaining 54 clones all contained alleles from the female and male parents and were true hybrids.
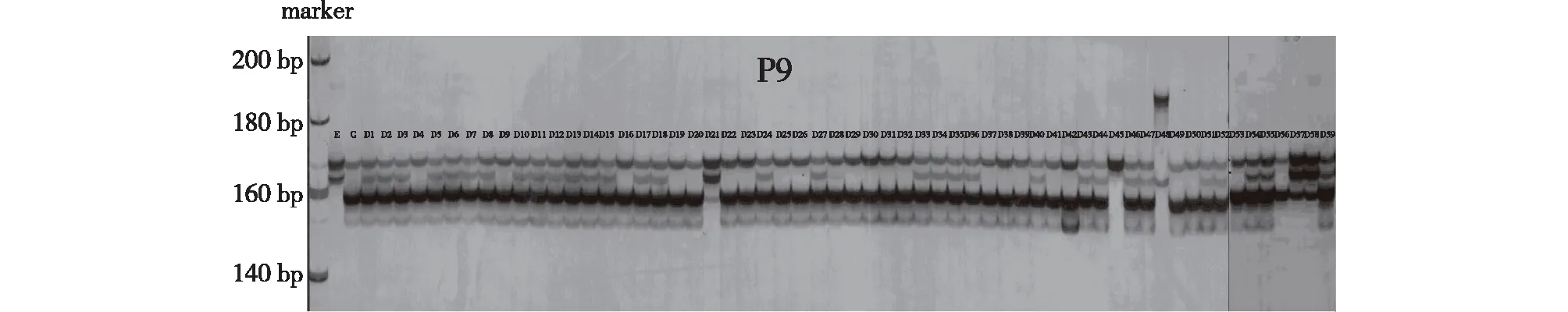
Fig.4 Electropherogram profiles of E, C and D1-D59 under the PCR amplification of primer P9图4 利用SSR引物P9扩增样品E,C和D1-D59的电泳图
To elucidate the genetic relationships between parents and their progeny from the E × C cross, similarity coefficients of E, C and 59 hybrids were calculated, a cluster analysis was conducted, and a dendrogram was constructed (Fig. 5).
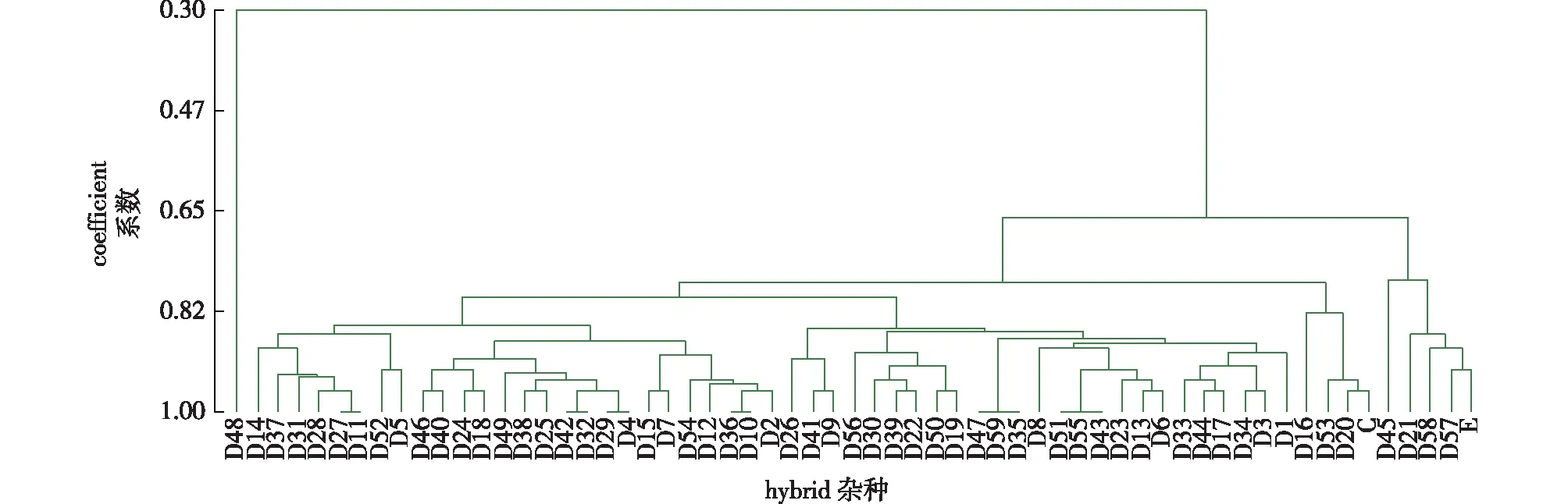
Fig.5 Dendrogram of E, C and their 59 suspected hybrids under cluster analysis using alleles detected with six SSR primers(P2, P9, P10, P11, P20 and P28)图5 利用6对引物(P2、P9、P10、P11、P20和P28)扩增的位点对E、C和59个疑似杂种进行聚类分析的树状图
D48 was independent from the parents and hybrids. Female parent E and four false hybrids, D21, D45, D57 and D58, were in the same group, while the true hybrids were in another groups. Male parent C was closer to the true hybrids, indicating that the hybrids inherited more paternal alleles. The genetic relationship between the parents, true hybrids, and false hybrids indicated by the dendrogram was consistent with the parentage analysis results. The six primer pairs were unable to distinguish D43/D55 and D51, D35/D59 and D47, D10 and D36, D4 and D29, D32 and D42, and D11 and D27, indicating that their genetic characteristics were relatively close. Moreover, to facilitate the clone identification and variety conservation, fingerprints of 54 true hybrids from the E × C cross were developed using the PCR amplification loci from primer P2, P9 and P11 (Table 3).

Table 3 Fingerprints of 54 true hybrids from E × C cross using alleles detected with SSR primers of P2, P9 and P11表3 利用SSR引物P2、P9和P11扩增的位点构建54个真实杂种的指纹图谱
3 Discussions and Conclusions
In the present study, two hybrid populations established fromB.multiplex×D.latiflorusandB.multiplex×B.chungiicrosses were analyzed using SSR molecular markers. Subsequently, hybrid identification, genetic analysis and fingerprinting were performed. Results revealed that among the 30 randomly selected bamboo SSR primer pairs, six primers pairs (P8, P9, P18, P19, P20 and P27) were suitable for the identification ofB.multiplex×D.latiflorushybrid populations, and 6 primer pairs (P2, P9, P10, P11, P20 and P28) were suitable for the identification of theB.multiplex×B.chungiihybrid population. Therefore, 20% of the primer pairs were selected after screening. Primer pairs P9 and P20 were suitable for the identification of both populations, accounting for 6.66% of the primer pairs. The results indicated that the published SSR primer pairs had good applicability.
The hybrid identification results indicated that all 34 progeny from theB.multiplex×D.latifloruscross were true hybrids (100%) and 54 of the 59 progeny from theB.multiplex×B.chungiicross were true hybrids (91.5%), indicating that hybrid identification based on phenotype had a high success rate due to large parental phenotypic differences; however, in the case of intrageneric crossing combinations, phenotype-based hybrid identification may be inaccurate and require supplementation with molecular marker technologies. Results of the parent-progeny genetic relationship analyses revealed that, in theB.multiplex×D.latiflorushybrid population, female parent E was the closest and male parent A was the farthest, indicating that the progeny inherited more maternal alleles. In contrast, in theB.multiplex×B.chungiihybrid population, the genetic relationship between male parentB.chungii, and the hybrids was close, indicating that the progeny inherited more paternal alleles. Therefore, in different crossing combinations with the same female parent and different male parents, the progeny inherited maternal and paternal alleles at different proportions, and should be treated differently in genetic relationship analyses between parents and their progeny.
In conclusion, in the present study, we used SSR markers to test the authenticity of hybrids derived from two crossing combinations of sympodial bamboo, clarified the inheritance of parental alleles, and developed fingerprints for hybrid populations, which could facilitate the breeding of sympodial bamboo.

