A new species of the genus Typhlomys Milne-Edwards,1877 (Rodentia: Platacanthomyidae) from Chongqing,China
DEAR EDITOR,
A new species of the genusTyphlomysMilne-Edwards, 1877(Rodentia: Platacanthomyidae) is described based on 10 specimens collected from Chongqing in southwestern China using integrated taxonomy.Phylogenetic analysis showed that these specimens formed a distinct sister clade toT.daloushanensisWang and Li, 1996, and differed from all six knownTyphlomysspecies based on Kimura-2-parameter genetic distances of the cytochromeb(cytb) gene (ranging from 0.122 to 0.215).The new speciesTyphlomys fengjiensissp.nov.is most similar toT.daloushanensisin morphology.However, it differs from the latter by the following morphological characters: (1) larger body and skull; (2) deeper incurved zygomatic arch; and (3) mesofossette on first upper molar open on both buccal and lingual sides (vs.inner edge of endoloph closed inT.daloushanensis).The discovery of this new species supports thatTyphlomysdiversity remains underestimated in southern China.
TyphlomysMilne-Edwards, 1877 is one of the two extant genera in the family Platacanthomyidae (Musser & Carleton,2005).Typhlomysspecies (soft-furred tree or blind mice) live in subtropical mountainous forests and temperate broadleaf forests (Cheng et al., 2017; Su et al., 2020), primarily in southern China, north to the Qinling Mountains and south to the Chapa regions of northern Vietnam (Cheng et al., 2017;Su et al., 2020).These species are characterized by an external mouse-like form, with small-sized eyes and tails ending in a unique terminal brush (Cheng et al., 2017; He et al., 2021).They are the only confirmed group of rodents to have evolved echolocation ability as an adaptation to their complex and varied environments (He et al., 2021).Typhlomyswas originally considered a monotypic genus(Wang et al., 1996).However, recent studies have confirmed that the genus contains five species (i.e.,T.cinereus,T.chapensis,T.daloushanensis,T.nanus, andT.huangshanensis), as well as one candidate species from Mts.Laojun and Dawei in Yunnan Province (Typhlomyssp.2 hereafter) (Cheng et al., 2017; Hu et al., 2021; Su et al.,2020).
During recent fieldwork (2020-2021), we collected 10 blind mice from Fengjie in northeast Chongqing, China (Figure 1A,locality 14; Supplementary Table S1).We initially assigned these 10 rodents toT.daloushanensis,following Liu et al.(2007).However, preliminary molecular analysis revealed that they were genetically distinct fromT.daloushanensisand more distinct from otherTyphlomysspecies.Therefore, they were assigned into a putative new species (Typhlomyssp.1 hereafter).Voucher specimens were deposited at the College of Life Sciences, Sichuan Normal University, Chengdu, China.In addition, two blind mice collected from Mt.Guangwu in Sichuan and Kaizhou in Chongqing (Figure 1A, localities 23 and 24) were used to amplify mitochondrial gene fragments for phylogenetic analysis.Muscle and liver tissues were collected and stored at -75 °C in 95% ethanol for subsequent molecular studies.The five recognized species of the genusTyphlomys, and one unconfirmed candidate species from Mts.Laojun and Dawei in Yunnan Province, were also included in our study.
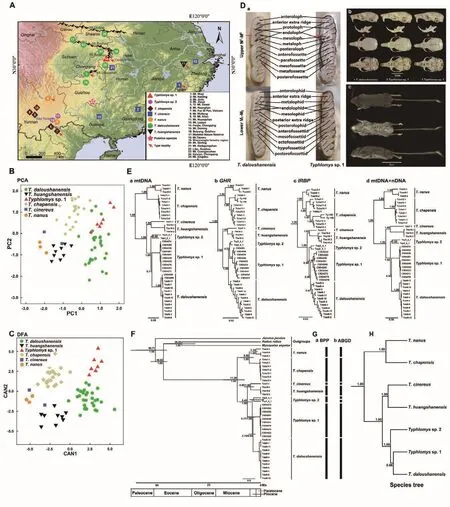
Figure 1 Sample localities, principal component (PCA) and discriminant function analysis (DFA), phylogenetic tree, divergence times,species delimitation and species tree of Typhlomys, with skull and molar comparisons of Typhlomys sp.1 and T.daloushanensis, and dorsal, ventral, and lateral views of Typhlomys sp.1
Tissue samples were extracted, amplified, and sequenced for three mitochondrial genes (i.e., cytb(1 143 bp),cytochrome c oxidase subunit I (COI, 1 396 bp), and NADH dehydrogenase sub-unit 2 (ND2, 1 010 bp)) and two nuclear genes (i.e., interphotoreceptor retinoid-binding protein (IRBP,1 214 bp) and growth hormone receptor (GHR, 860 bp)) for molecular analysis, as detailed in the Supplementary Methods.All generated DNA sequences were deposited in GenBank under accession Nos.OL691088-691 090,OL693252-693 261, OL753439-753 481, OM840094-840 097,
OM736189 and OM736190 (Supplementary Table S1).
Morphological analysis was performed using 11 cranial measurements based onTyphlomyssp.1 and five recognizedTyphlomysspecies.Principal component analysis (PCA) and canonical discriminant function analysis (DFA) were used to compare morphometric divergence among species (see Supplementary Methods).We also compared and described the cheek teeth and zygomatic arch betweenT.daloushanensisandTyphlomyssp.1, considering their morphological similarity and close distribution (Liu et al.,2007).
Bayesian phylogenetic analyses were performed separately for each nuclear gene (GHR,IRBP), concatenated mitochondrial segments (cytb+COI+ND2), and concatenated data of all five genes.We calculated the Kimura-2-parameter(K2P) distances for species/putative species based on the cytbgene.Bayesian Phylogenetics and Phylogeography (BPP)analysis and Automatic Barcode Gap Discovery (ABGD) were run to infer putative species boundaries.We also estimated a coalescent species tree and divergence times using all five genes.See Supplementary Methods for details.
The PCA results extracted two principal components (PC1 and PC2; Supplementary Table S2), the eigenvalues of which exceeded 1.0.All measurements had positive scores in PC1,which explained 64.62% of total variation, suggesting a relationship with the overall size of the specimens.In addition,PC2, which explained 13.46% of total variation, was dominated by interorbital breadth (IOB) and braincase height(BCH).The PCA results separatedTyphlomyssp.1 from all other species, exceptT.daloushanensis(Figure 1B).Discriminant cross-validation analysis showed that 95.80% of species were correctly classified based on cranial measurements.The canonical axes 1-4 (CAN 1-4) explained 50.60%, 30.10%, 14.60%, and 3.70% of total variation,respectively (Supplementary Table S2).CAN1 and CAN2 plots (Figure 1C) showed thatTyphlomyssp.1 was separated from all other species and occupied positive regions in both CAN1 and CAN2.
As shown in Figure 1D-a, the cheek teeth ofTyphlomyssp.1 andT.daloushanensiswere generally consistent.On the first upper molar (M1), the mesofossette ofT.daloushanensiswas open on the buccal side and closed on the inner edge of the endoloph, whereas forTyphlomyssp.1, the mesofossette was open on both the buccal and lingual sides and penetrated the entire surface of M1from the middle.About 70% (n=10) of theTyphlomyssp.1 specimens had this characteristic, which was distinguishable from all other species, except forT.huangshanensis( Hu et al., 2021).However,T.huangshanensiscould be distinguished fromTyphlomyssp.1 by the missing M2anterofossette (Hu et al., 2021).In addition,the zygomatic arch was more rounded and only slightly curved inward inT.daloushanensis(Figure 1D-b-1) but was deeply incurved on the posterior two-fifths inTyphlomyssp.1(Figure 1D-b-2, 3), with the curvature approaching a folded state in one individual (Figure 1D-b-3).The diagnostic characteristics ofTyphlomyssp.1 andT.daloushanensisare indicated by red arrows in Figure 1D, D.
The four Bayesian phylogenetic trees showed similar topologies.Typhlomyssp.1 formed a monophyletic clade with high nodal support (PP≥0.98) in theIRBPand concatenated(mtDNA+nDNA) phylogenetic trees (Figure 1E-c, d).The mtDNA tree also showed thatTyphlomyssp.1 formed a monophyletic clade, but without high support (PP=0.71;Figure 1E-a).In theGHRgene tree,Typhlomyssp.1 was clustered withTyphlomyssp.2, but with low support(PP=0.22; Figure 1E-b).These differences in topology may be due to incomplete lineage sorting.In addition, low-supported internodes in mtDNA trees are not necessarily unstable,depending on the relative stability or instability of their terminal taxa (Thorley & Wilkinson, 1999).
The K2P genetic distances betweenTyphlomyssp.1 and other species ranged from 0.122 to 0.215 (Supplementary Table S3), indicating species-level divergence ofTyphlomyssp.1 from other species.The BPP results supported valid species status for all seven species with high support (PP≥0.99) (Figure 1G-a; Supplementary Table S4).ABGD analysis based on differentXvalues (0.5, 1.0, 1.5, and 2.0) obtained identical results and identified eight species, withT.cinereus,T.chapensis,T.daloushanensis,T.nanus,T.huangshanensis,andTyphlomyssp.1 supported as separate species andTyphlomyssp.2 identified as two species(Figure 1G-b; Supplementary Figure S1).Both the BPP and ABGD results supportedTyphlomyssp.1 as a separate species to all otherTyphlomysspecies.
All seven clades within the genusTyphlomyswere well supported (PP≥0.96) in the species tree (Figure 1H).The estimated age of the most recent common ancestor ofTyphlomyswas ~11.39 million years ago (Ma) (95%CI=7.85-18.04; Figure 1F).The seven putative species diverged ~3.79-6.49 Ma.Furthermore,Typhlomyssp.1 andT.daloushanensisdiverged ~3.79 Ma (95% CI=2.59-5.8).Both morphological and molecular analyses indicated a species status forTyphlomyssp.1.Thus, we considerTyphlomyssp.1 to be a new species, which we formally describe below.
Taxonomy
Typhlomys fengjiensis sp.nov.Pu, Chen, and Liu
Holotype:SCNU02616 (collection number: csd4274).Adult male (Figure 1D-c) collected in March 2021 by Rong-Hui Fan,Bu-Qing Peng, and Jing Li.The specimens were deposited at the College of Life Sciences, Sichuan Normal University,Chengdu, China.
Type locality:Shiruguan in Xinglong Town, Fengjie County,Chongqing, China (N30.663°, E109.521°, 1 883 m above sea level (a.s.l.)).
Measurements of holotype (mm):BM=34.60 g; HB=105.00;TL=124.00; HL=24.00; EL=18.00; GLS=26.71; CBL=23.2;BL=21.92; BCH=8.89; IOB=5.26; ZMW=13.52; LUIM=12.50;UML =4.00; M1-M1=5.74; HCV=4.77; and LNM-FLM=7.18.
Paratypes:Two adult males (SCNU02583 and SCNU02623),two adult females (SCNU02591 and SCNU02618), and two adult specimens of unknown sex (SCNU02580 and SCNU02615) were collected with the holotype in Fengjie County, Chongqing, China (N30.663°, E109.521°, 1 883 m a.s.l.).Three adult specimens (SCNU02544, SCNU02564,and SCNU02565) of unknown sex were collected inNovember 2020 from the same mountain and type locality(N30.649°, E109.490°, 1 579 m a.s.l.).Two adult male paratypes from Fengjie (SAF97118 and SAF97119, 1 880 m a.s.l., no record of latitude and longitude) were collected in May 1997 by Shao-Ying Liu and stored at the Sichuan Academy of Forestry.These two specimens were not included in the above analysis due to specimen aging and lack of preserved tissue samples.
Diagnosis:The new species is most similar toT.daloushanensisbut can be distinguished based on its large body size and skull within the genus.It differs fromT.chapensisandT.nanusby more flattened braincase; from all knownTyphlomysspecies, except forT.cinereus, by zygomatic arch with distinct deeper incurve; fromT.cinereusby mesofossette on M1open on both buccal and lingual sides rather than open on buccal side only; and fromT.nanusby posterofossettid on M1present.The new species further differs from other species, exceptT.daloushanensis, by anterofossette on M2present.
Description:(measurements in mm): Largest species withinTyphlomys(HB=89.86± 8.76; GLS=25.38±0.77).Vibrissae long; ears prominent, almost bare; eyes vestigial(Supplementary Figure S2).Dorsal body dark gray, entire ventral body from chin to anus, including inner side of limbs to wrists and knees, creamy white due to dark gray hair base and white tip.Fingers four, toes five; hind feet slender and elongated (HL=22.86±0.69); palms of all limbs light brown;fingers pale yellow; skin on dorsal surfaces of hind feet brown,covered with dark hair.Tail long, exceeding length of head and body (TL=123.00±4.40), with scale rings; first third of tail covered with extremely short and sparse hairs, back part covered with long hairs; tail tip tufted with long white hair.
Braincase flattened, but relatively high due to its large size(BCH=8.35±0.37).Rostrum straight beyond upper incisors.Tympanic bulla small.Zygomatic plate narrow; zygomatic arch incurvate at posterior two-fifths, anterior part narrower than posterior part.Two pairs of long, oval palatal foramina relatively developed, with one or two pairs of symmetrical or asymmetrical small round foramina.Dental formula 1.0.0.3/1.0.0.3=16.M1, with wider anteroloph and narrow posteroloph.Endoloph of three upper molars relatively wider than endolophid of lower molars.First and second molars of upper and lower jaws with five fossette-shaped structures;third molars with three to four.Mesofossette on M1open on both buccal and lingual sides.M2with anterofossette.M1with posterofossettid and closed mesofossettid.Anterofossettids present in M2, but relatively short.
Variations:Tail tip was without tuft of long white hair in one individual (SCNU02615).Zygomatic arch showed varying degrees of deep incurvature.In specimens with at least one intact zygomatic arch (n=7), three (SCNU02583, SCNU02623,SCNU02615) had similar degrees of incurvature as the type specimen (Figure 1D-b-2), one (SCNU02580) approximated a folded state (Figure 1D-b-3), two (SCNU02564, SCNU02618)were more incurved than type specimen but not as curved as SCNU02580.The mesofossette on M1was open on both the buccal and lingual sides in all specimens, except SCNU02544,SCNU02583, and SCNU02618.Six specimens had two pairs of long, oval palatal foramina of similar size, and one(SCNU02564, SCNU02565, SCNU02591, SCNU02615) or two (SCNU02618, SCNU02616) pairs of symmetrical or asymmetrical small round palatal foramina.Palatal foramina of the other four specimens (SCNU02544, SCNU02583,SCNU02623, and SCNU02580) were too damaged to be quantified.
Comparisons:Species in the genusTyphlomysare highly similar in external appearance.Herein, we used characteristics presented in previous research (Cheng et al.,2017; Hu et al., 2021) and the morphological diagnosis results of specimens obtained in this study to distinguish the six recognized species, as shown in Supplementary Table S5.
Etymology:The specific Latin name of the new species refers to its type locality in Fengjie County, Chongqing, China.The sampling location is adjacent to the famous and historic Baidi(White Emperor) City, therefore, we suggest “Baidi blind mouse” as the English common name and “白帝猪尾鼠” as the Chinese common name.
Distribution and ecology:The new species is currently known only from the type locality, but may also occur in the adjacent mountainous areas in southwestern Hubei of the southern Yangtze River.Moreover, it may also be distributed in northern Chongqing and northwestern Hubei around the Mt.Bashan region if it can disperse across the Yangtze River,such asT.daloushanensis.The study specimens were captured in mountainous forests with bamboo underbrush adjacent to roads at mid-altitudes (1 579-1 883 m a.s.l.).Sympatric species includeBlarinellacf.quadraticauda,Chodsigoa smithii,Anourosorex squamipes,Crocidura vorax,Uropsilus gracilis,Apodemus draco,Niviventer lotipes, andEothenomys eleusis.
The main distribution areas of allTyphlomysspecies are shown in Figure 1A.Populations with molecular evidence were distributed in localities 1-14, 20, and 22-24.The population from Libo, Guizhou Province (locality 22), is considered a probable separate clade (Su et al., 2020), but this needs to be verified by further research.Populations without molecular evidence were distributed in localities 15-19, 21, and 25.Morphological analysis identified the Shennongjia forest population (locality 19) asT.daloushanensis(Wang et al., 1996).This population is not geographically isolated from the Kaizhou and Mt.Guangwu populations (localities 23 and 24), which were assigned toT.daloushanensisbased on our molecular results (Figure 1E-a,CSD5200 and CSD4999).Hence, we considered the Shennongjia population asT.daloushanensis.In addition, we re-examined the morphological characteristics of the only specimen collected from Maozhai Nature Reserve (locality 17), which corresponded toT.daloushanensisin body size and exhibited the same round zygomatic arch and semienclosed mesofossette of M1.Thus, we preliminarily classified the specimen asT.daloushanensis.Regarding the Suiyang,Guizhou, and Mts.Daming, Tianmu, and Xingdou populations(localities 16, 15, 18, and 25, respectively), we followed the classification of Wang et al.(1996), i.e.,T.daloushanensisorT.cinereus, based on morphological characteristics and distribution.The population from Badagongshan National Nature Reserve (locality 21) is only mentioned asT.cinereuswithout any morphological or molecular information (Xie et al.,2014).Therefore, we temporarily classified it asT.cinereusfollowed Xie et al.(2014).However, the valid species of the genusTyphlomysin southern China still require further indepth taxonomic and phylogenetic study.
NOMENCLATURAL ACTS REGISTRATION
The electronic version of this article in portable document format represents a published work according to the International Commission on Zoological Nomenclature (ICZN),and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone (see Articles 8.5-8.6 of the Code).This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN.The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information can be viewed through any standard web browser by appending the LSID to the prefixhttp://zoobank.org/.
Publication LSID:
urn:lsid:zoobank.org:pub:F913C1F5-5A48-4E7C-A6B1-8103EB2B7921
Typhlomys fengjiensisLSID:
urn:lsid:zoobank.org:act:FFB5C439-ACF8-4F27-BBB6-0260F2EFF197
SCIENTIFIC FIELD SURVEY PERMISSION INFORMATION
All samples were obtained following Chinese regulations for the Implementation of the Protection of Terrestrial Wild Animals (State Council Decree (1992) No.13), and the Guidelines for Care and Use of Laboratory Animals by the Ethics Committee at Sichuan Normal University (Chengdu,China).
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
S.D.C.and S.Y.L.conceived the study and prepared the manuscript.Y.T.P.and T.W.performed the experiments,analyzed the data, and prepared the manuscript.C.K.F.and K.Y.T.revised the manuscript and helped perform experiments.X.L.J, B.W.Z., and T.L.H.provided morphological data.R.H.F.collected the specimens and helped perform experiments.All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
We would like to acknowledge Mr.Rui Liao, Bu-Qing Peng,and Jing Li for collecting field samples.We thank Dr.Zhong-Zheng Chen for providing morphological measurement data and tissue samples and Prof.Liang Dou for providing tissue samples from Mt.Guangwu of Sichuan.We thank Dr.Sheng-Chao Shi for revising an earlier version of this manuscript.We are also thankful to Mr.Ke Li for his assistance in modifying the figure of sample localities.
- Zoological Research的其它文章
- Mechanism of hyperproteinemia-induced blood cell homeostasis imbalance in an animal model
- Identification of global alternative splicing and sexspecific splicing via comparative transcriptome analysis of gonads of Chinese tongue sole (Cynoglossus semilaevis)
- Phylogenetic relationships of the zokor genus Eospalax(Mammalia, Rodentia, Spalacidae) inferred from wholegenome analyses, with description of a new species endemic to Hengduan Mountains
- MonkeyTrail: A scalable video-based method for tracking macaque movement trajectory in daily living cages
- Unknown species from China: The case of phrurolithid spiders (Araneae, Phrurolithidae)
- A new species of the gudgeon genus Microphysogobio Mori, 1934 (Cypriniformes: Cyprinidae) from Zhejiang Province, China

