Artificial simulated saliva,gastric and intestinal digestion and fermentation in vitro by human gut microbiota of intrapolysaccharide from Paecilomyces cicadae TJJ1213
Juanjuan Tian,Xiaomeng Wang,Xueliang Zhang,Xiaohong Chen,Mingsheng Dong,Xin Rui,Qiuqin Zhang,Mei Jiang,Wei Li*
College of Food Science and Technology,Nanjing Agricultural University,Nanjing 210095,China
Keywords:Paecilomyces cicadae TJJ1213 Polysaccharide Fermentation Gut microbiota
ABSTRACT In this study,the in vitro digestion and fermentation of two intra-polysaccharide fractions (IPS1 and IPS2)from Paecilomyces cicadae TJJ1213 were investigated.The constituent monosaccharides of IPS1 and IPS2 were not changed after simulated saliva,gastric and small intestinal digestion.However,they can be hydrolyzed and utilized by gut microbiota,and short-chain fatty acids (SCFAs) level were increased after IPS1and IPS2 treatments.Furthermore,16S rRNA sequencing analysis of fermentation samples were performed.Alpha-diversity,beta-diversity and taxonomic composition differences analysis revealed that IPS1 and IPS2 promoted the proliferation of beneficial bacteria and modulated the overall structure of gut microbiota.Taxonomic comparison analysis found that IPS1 increased the relative abundances of benef icial bacteria including Megamonas,Bifidobacterium and Lactobacillus,while IPS2 could increase the abundance of Bacteroides,Parabacteroides and Phascolarctobacterium.In addition,they can also decrease the levels of pathogenic bacteria containing Escherichia-Shigella,Klebsiella and Fusobacterium.These results indicated that IPS from Paecilomyces cicadae TJJ1213 could be used as potential candidates for new functional foods.
1.Introduction
It has been reported that there are 1011-1014bacteria habituated in the intestine of a health adult,which were called gut microbiota [1].It is estimated that there are more than 1 000 bacterial species in the human gut [2].However,it is limited at the level of phylum.The huge variety of bacterial species mainly belong to four phyla,including Firmicutes (predominantlyClostridia;50% to 70% total bacterial population),Bacteroidetes (10% to 30%),Proteobacteria(up to 10%) and Actinobacteria (up to 5%),with 90% believed to be obligate anaerobes [1,3].The gut microbiota co-exist with the host,maintaining a dynamic balance in the gut,which means that not all bacterial members can permanently colonize in the gut [4].It has been proved that the gut microbiota can contribute to human health and provide many functions for the host,such as production of nutrients and vitamins,digestion of complex dietary macronutrients,maintenance of the immune system against pathogens [5].However,emerging data showed that the dysbiosis of gut microbiota was associated with some diseases,such as metabolic diseases [6],immunological diseases [7]and neurological diseases [8].The gut microbiota composition is also affected by many factors,such as diet,health status,different regions,age,mode of birth delivery and genetics,but diet is considered as the most important factor [9].Zhang et al.[10]reported that diet changes explained 57% of the total structural variation in gut microbiota,whereas genetic mutation accounted for no more than 12% .Thus,dietary interventions may have potentials to modulate the composition of gut microbiota.
It is noted that dietary carbohydrates including non-starch polysaccharides and oligosaccharides can be used as substrates for gut microbial growth [11].Polysaccharides are the most complex compounds,and many monosaccharide units link together by glycosidic bonds and form chains and branches.The gut microbiota can utilize the complex polysaccharides because the carbohydrateactive enzymes (CAZymes) are present in the human gut bacteria [12].Recently,theinvitrobatch-culture fermentation modelling inculcated with human fecal samples was widely accepted to simulate the human intestinal environment.It provides a simple,rapid and inexpensive method to give an insight into the fermentation of carbohydrates by complex gut microbiota [13].Through theinvitrobatchculture fermentation systems,many polysaccharides have been demonstrated to have a role in the regulation of intestinal flora.For example,Lyciumbarbarumpolysaccharide can enhance the relative abundances of genera includingBacteroides,Bifidobacterium,Phascolarctobacterium,ClostridiumXlVb,PrevotellaandCollinsellain the small intestinal [14].Mao et al.[15]reported that fungus exopolysaccharide from theCordycepssinensisincreased the relative abundance ofBifidobacteriumandClostridium.
C.cicadae,a valuable parasitic fungus,belongs to theClavicipitaceousfamily,which was formed byPaecilomyces cicadaeparasitises on larvae of cicada and has been used as a tonic food and herbal medicine for hundreds of years [16].Many studies showed thatC.cicadaecontains many active ingredients such as polysaccharide,cordycepic acid,ergosterol and effective nucleosides [17].As a traditional medicine,these active ingredients has been widely used to prevent and treat various diseases.It is frequently recommended for the treatment of lung diseases.The anti-tumor and antibacterial effects have also been reported [18].Recently,some studies indicated that polysaccharides fromP.cicadaepresented improvement of renal function and inhibition of oxygen radicals [19].Yang et al found thatP.cicadaepolysaccharide was effective in treating immune diseases [20].However,the regulatory effect of polysaccharide fromP.cicadaeon gut microbiota is unknown.
Recently,two intra-polysaccharide fractions (IPS1 and IPS2)were obtained from theP.cicadaeTJJ1213.Their structural features and immunomodulatory activity were detailed reported [21].The backbone of IPS1 was →4)-α-D-Glcp(1→ and →3,4)-α-DManp(1→ residues with a side chain consisted ofT-α-D-Galp.IPS2 was consisted of →4)-α-D-Glcp-(1→,→3,4)-α-D-Manp-(1→and →2,6)-α-D-Manp-(1→ residues,and the branches were also consisted ofT-α-D-Galp.However,the digestibility of IPS1 and IPS2 and its effects on the gut microbiota remain unclear.Therefore,in this study,we investigated theinvitrodigestibility and effects on the gut microbiota composition of IPS.In addition,the abundance and change of gut microbiota and production of short chain fatty acids (SCFAs) were examined during the fermentation by using 16S rRNA gene sequencing amplification and high performance liquid chromatography (HPLC),respectively.
2.Materials and methods
2.1 Materials
Monosaccharides standards including glucose (Glc),galactose(Gal),mannose (Man),fructose (Fru),rhamnose (Rha) and arabinose(Ara) were purchased from Aladdin Chemical Reagent Co.,Ltd.(Shanghai,China).Formic acid,acetic acid,propionic acid,butyric acid and lactic acid standards were purchased from Sigma-Aldrich Co.,Ltd.(St.Louis,Mo,USA).Inulin (content >95%) were obtained from Yuanye Bio-Technology Co.,Ltd.(Shanghai,China) with a molecular weight (mw) of approximately 5 000 Da.All other chemical reagents were analytical grade and purchased from Sinopharm Chemical Reagent Co.,Ltd.(Shanghai,China).
2.2 Isolation and purification of IPS
IPS was isolated according to our previous reported study.Briefly,P.cicadaeTJJ1213 was cultured in liquid medium (glucose 20 g/L,yeast extract 3 g/L,peptone 4 g,K2HPO41g/L,MgSO40.5 g/L) at 25 °C with shaking 160 r/min for 7 days.Then,the mycelium was obtained by centrifuging at 10 000 r/min for 10 min.Freeze-dried mycelia were crushed by high speed pulverizer.Crude polysaccharide was obtained by a series of procedures including hot water extraction,deproteinization,concentration,ethanol precipitation,dialysis and lyophilization.Furthermore,the cured polysaccharide was purified by DEAE-52 anion exchange column(2.6 cm × 30 cm).As a result,two fractions were obtained and their structures were detected by HPLC,Fourier transform infrared spectrum (FTIR) and nuclear magnetic resonance (NMR).
2.3 Simulated saliva,gastric and intestinal digestion
Saliva digestion was carried out by previous report [22]with some modifications.The saliva was collected from three volunteers,who had not taken antibiotics for at least three months.The collected saliva was immediately centrifuged at 5 000 r/min for 10 min.Then,4.0 mL IPS solution and 4.0 mL saliva solution were mixed completely.4.0 mL distilled water and 4.0 mL saliva solution were used as controls.After mixing completely,the test tubes were digested at 37 °C for 15 min and then incubated for 5 min in boiling water to inactivate human salivary amylase.
The basic medium of gastric digestion was prepared according to the published method [23].For simulated gastric digestion,10.0 mL IPS solution (8.0 mg/mL) with 10.0 mL simulated gastric liquid and 10 mL distilled water with 10 mL simulated gastric fluid were prepared and incubated at 37 °C.After digestion for 2 h,2.0 mL digestion solution was taken out and immediately inactivated enzymes in boiling water bath for 5 min.
The basic medium of intestinal digestion was prepared according to the published method [25].For simulated intestinal digestion,the mixture of 3.0 mL of simulated intestinal fluid with 10.0 mL digested simulated gastric solution and 3.0 mL of simulated intestinal fluid with 10.0 mL simulated gastric fluid were incubated at 37 °C for 6 h.Then,2.0 mL digestion solution was taken out and immediately inactivated enzymes in boiling water bath for 5 min.
2.4 In vitro fermentation
Theinvitrofermentation was carried out according to the previous method with some modifications [26].The basal nutrient medium contained (per liter) 2 g of peptone,2 g of yeast extract,0.1 g of NaCl,0.04 g of K2HPO4,0.04 g of KH2PO4,0.01 g of MgSO4·7H2O,2 g of NaHCO3,0.01 g of CaCl2·6H2O,0.02 g of hemin,0.5 g ofL-cysteine hydrochloride,0.5 g of bile salts,1 mg resazurin,2 mL of Tween 80,10 μL of vitamin K1and distilled water.After mixing thoroughly,the medium was sterilized at 121 °C for 15 min.Six healthy volunteers (three female and three males) aged 20-30 who had not taken antibiotics for at least 3 months provided fecal samples.20 g of fresh fecal sample of each donor was mixed with 20 mL modified sterile saline (cysteine-HCl 0.5 g/L and NaCl 9.0 g/L).The suspension was centrifuged at 500 r/min for 10 min at 4 °C,and the supernatant was mixed for further experiments.Then,1.0 mL of the fecal supernatant was added into 9.0 mL of basal nutriment medium containing 100 mg IPS,and the mixture was incubated at 37 °C for 24 h in an Anaero Pack System (Mitsubishi Gas Chemical Co.,Inc.,Tokyo,Japan).Under the same condition,the culture without IPS and with inulin was acted as the blank control (BLK) and the positive control (INL),respectively.Each fermentation experiment was carried out in triplicate and incubated at the same conditions.
2.5 Determination of pH for in vitro fermentation
To better understand the fermentation situation,the pH of culture before and after fermentation was measured using a micro-pH meter.
2.6 Determination of mw change
As previously reported,themwdistribution of IPS was determined by a Waters HPLC equipped with an evaporative light-scattering detector (ELSD) [26].20 μL of the saliva digestion products,gastric digestion products,intestinal digestion productions,fermented sample solutions and unfermented polysaccharide samples were injected after filtered through a 0.22 μm membrane and eluted with ultrapure water at a flow rate of 0.8 mL/min in a TSK GEL G4000 PW column(300 mm × 7.8 mm,Tosoh Corp.,Tokyo,Japan).The column temperature was 30 °C;the drift tube temperature was 90 °C;the carrier gas was N2.
2.7 Analysis of constituent monosaccharides
The constituent monosaccharides of IPS before and after digestion and fermentation were analyzed according to previous method with sight modifications [27].The 1 mL of sample was hydrolyzed with 1 mL 4 mol/L trifluoroacetic acid (TFA) at 120 °C for 2 h.Then,excess TFA was removed by adding methanol and evaporated at deduced pressure.The hydrolysate was dissolved with 0.4 mL distilled water and mixed with 0.4 mL of 0.5 mol/L PMP solution and 0.2 mL of 0.3 mol/L NaOH solution reacting for 30 min at 70 °C.After neutralization with 0.3 mol/L HCl,the excess PMP was extracted with chloroform for three times.The upper aqueous phase was the PMP derivative product and analyzed by using a Waters HPLC equipped with a PDA detector and a C18column (250 mm × 0.4 mm i.d.,0.5 μm) after filtering through a 0.45 μm membrane.The flow rate was 0.8 mL/min.Mobile phases were consisted of 0.1 mol/L ammonium acetate (pH 5.0),acetonitrile and tetrahydrofuran in a ratio of 81:17:2 (V/V).
2.8 Analysis of gut microbiota
After fermentation for 24 h,the total bacterial DNA was extracted immediately by using a QiAamp Fast DNA Stool Mini Kit (Sangon Biotech Co.,Ltd.Shanghai,China) according to the manufacturer’s instructions and stored at -20 °C for 16S rRNA gene pyrosequencing.The sequencing of the bacterial 16S rDNA V3-V4 was performed by Majorbio Inc.(Shanghai,China) on Illumina MiSeq platform.All the results were based on sequenced reads and operational taxonomic units (OTUs).
2.9 Analysis of SCFAs
SCFAs concentrations in the fermentation samples were calculated as previously described in the literature [13].Briefly,every sample was centrifuged at 12 000 r/min for 5 min and filtered through a 0.22 μm membrane.Then,the supernatant was determined by HPLC system (Waters) with a C18column (4.6 mm × 250 mm,5 μm,Agilent Technologies Inc.USA).The column temperature was 30 °C and the flow rate was 0.8 mL/mim.The mobile phase was consisted of A and B phase.A was a 20 mmol/L KH2PO4aqueous solution(pH 2.5 was regulated by H3PO4) and B was chromatographic methanol.The elution procedures were as follows: 0-16 min,95% A,5% B;16-30 min,95% -70% A,5% -30% B;30-40 min,70% A,30% B;30-40 min,70% A,30% B;40-50 min 95% A,5% B.
2.10 Statistical analysis
All the experiments were carried out in triplicate.All the data were expressed as means ± standard deviation after passing a Duncan’s test and processed with SPSS Statistics (SPSS 20.0).Significant differences among groups were considered ifP<0.05.
3.Results and discussion
3.1 Changes of mw after saliva,gastric and intestinal digestion
Previous study reported that the structure and chemical properties of polysaccharide might be affected by the enzymes,pH and salts during human digestion [28].Therefore,the digestion properties of IPS1 and IPS2 were evaluated.As shown in Fig.1a,the retention time of IPS1 did not change before and after saliva,gastric and intestinal digestion,indicating that saliva,pH and salts had no effect on IPS1.Therefore,this result implied that IPS1 could reach the colon and could be utilized by human gut microbiotal.Our results were consistent with previous report,which found that some polysaccharides were not degraded by saliva,and gastric [29].
From the Fig.1b,after saliva digestion,the retention time of IPS2 did not change,indicating that the polysaccharide was not hydrolyzed by saliva amylase.However,two peaks appeared in the chromatogram after gastric digestion,which means that IPS2 may be hydrolyzed and themwwas reduced.It is known that polysaccharide tend to from aggregates in aqueous system.However,pH might cause polymers dissociation or breakdown [30].To found out the influence of pH on the digestion of polysaccharide,Hu et al.[30]reported that themwof polysaccharide was obvious reduced after digestion at pH 1.5,while themwof polysaccharide was not changed under the digestion at pH 6.8.Therefore,it could be considered that the pH of the digestion medium would be a main reason that causes themwloss of IPS2 during digestion.Our results were similar with previous reported that themwof polysaccharides fromPlantago asiaticaL.seeds and longan pulp exhibited some extent of change duringinvitrodigestion [24,30],which breakdown covalent bonds of polysaccharides.
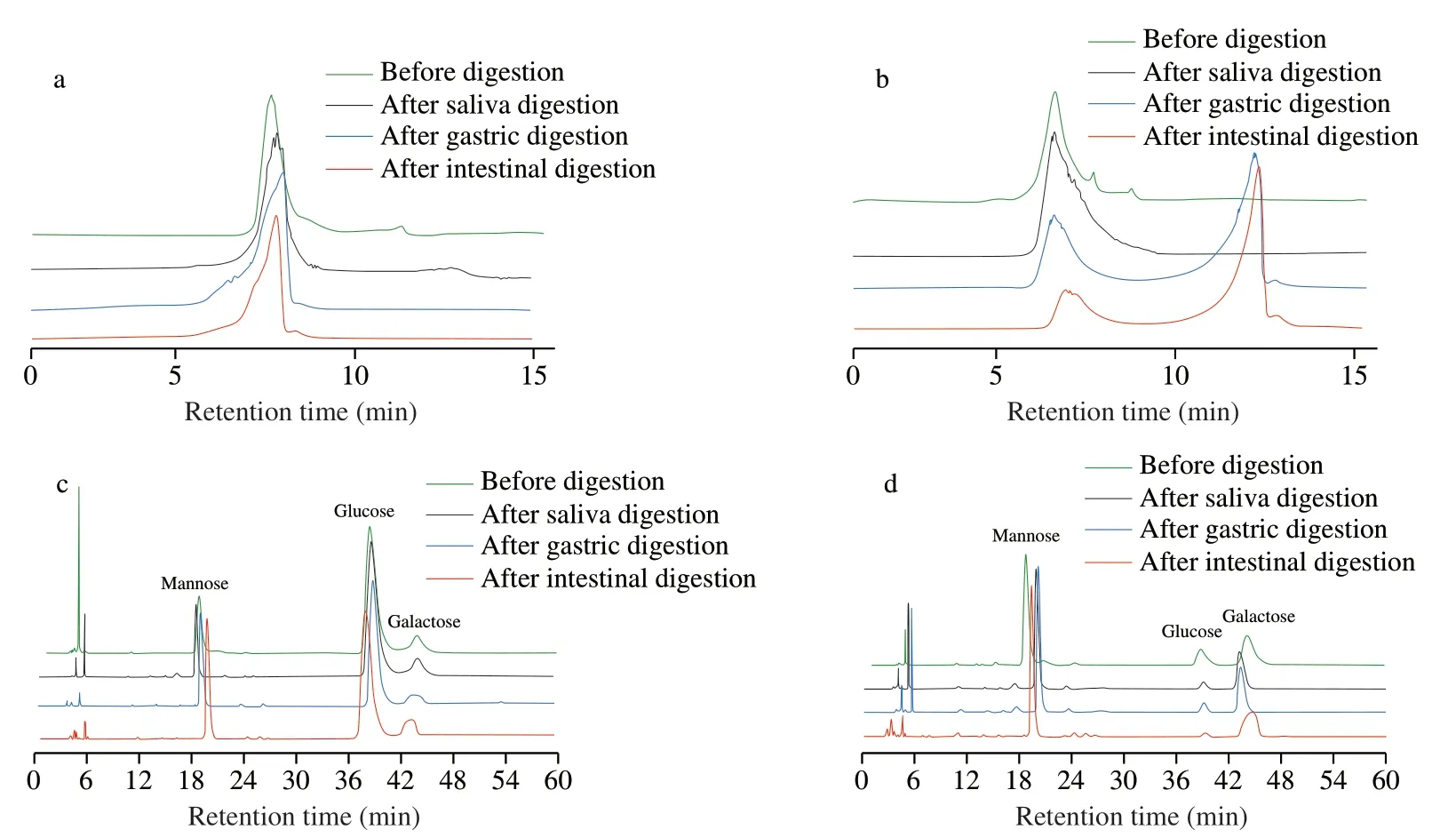
Fig.1 Changes of mw and monosaccharide composition before and after digestion by HPLC.Change in mw of IPS1 (a),change in mw of IPS2 (b),change in monosaccharide composition of IPS1 (c),change in monosaccharide composition of IPS2 (d).
3.2 Analysis constituent monosaccharides after saliva,gastric and intestinal digestion
Monosaccharide is the basic unit of polysaccharide,which impacts the distinct structures and properties.Thus,the changes of monosaccharide constituent of IPS duringinvitrodigestion were investigated.As shown in the Figs.1c and 1d,the constituent monosaccharides of IPS duringinvitrodigestion were similar,and Man,Glc and Gal were detected in all digestion samples.The changes of molar ratios of monosaccharides duringinvitrodigestion were shown in the Table.S1.Results indicated that the constituent monosaccharides and molar ratios of IPS were stable during digestion,which was similar with the reported study [31].
3.3 Changes of mw after fermentation
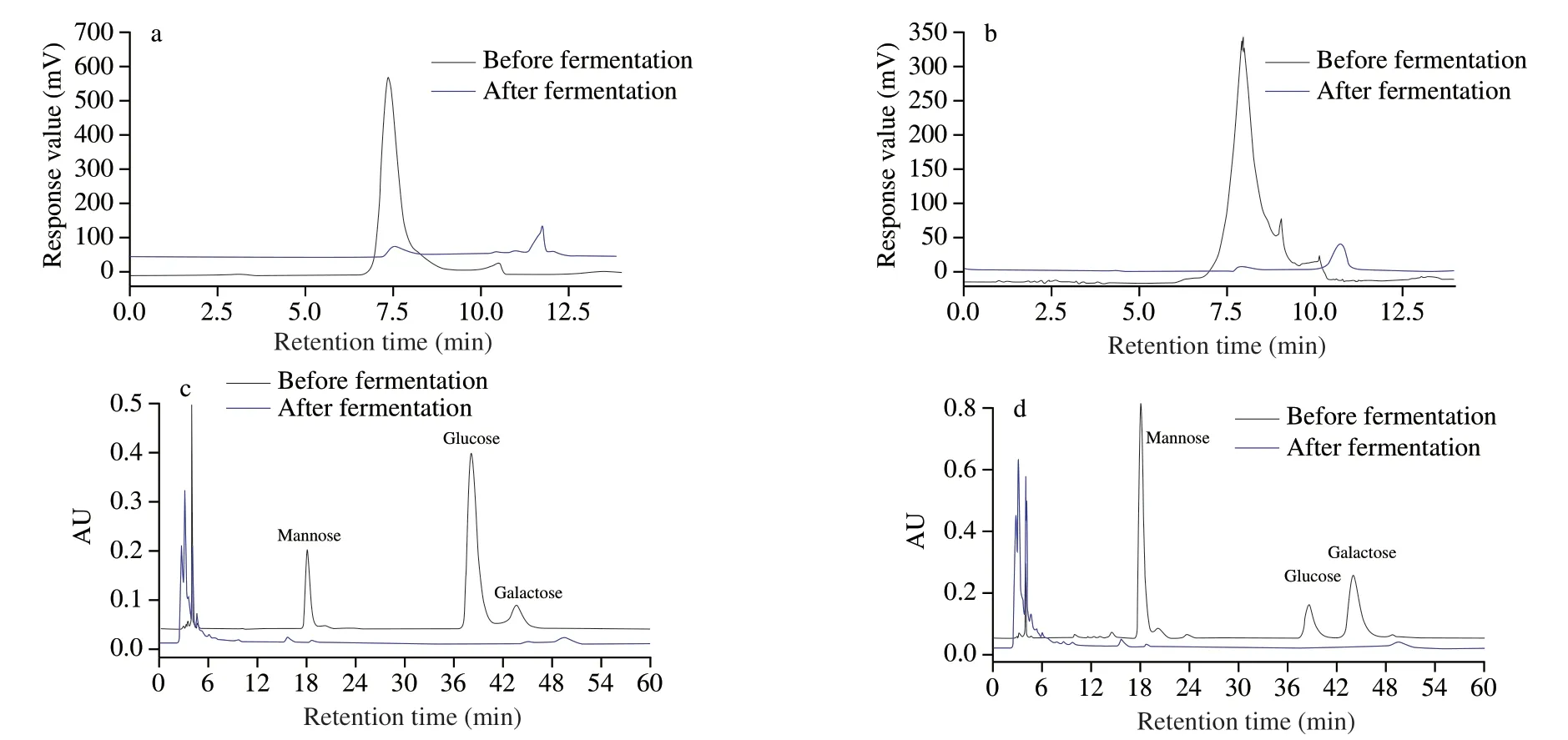
Fig.2 Changes of mw and monosaccharide composition before and after fermentation by HPLC.Change in mw of IPS1 (a),Change in mw of IPS2 (b),change in monosaccharide composition of IPS1 (c),change in monosaccharide composition of IPS2 (d).
As shown in the Figs.2a and 2b,the response peak of IPS1 and IPS2 delayed over the fermentation and two little peaks emerged at 11.1 and 10.8 min,respectively,demonstrating that IPS1 and IPS2 were degraded to a lowmwpolysaccharides or oligosaccharides by microbiota during the fermentation.In general,previous studies also reported that themwof polysaccharide would sharply reduce duringin vitrofecal fermentation [32,33].Themwsignificantly reduced,which was attributed to microbial metabolism,including hydrogen carbon dioxide and SCFA generation [22].
3.4 Analysis constituent monosaccharides after fermentation
It has been confirmed that the constituent monosaccharide of natural polysaccharide duringinvitrofermentation can be alter by gut microbiota [34].Hence,the constituent monosaccharide changes of IPS1 and IPS2 were analyzed by HPLC.From the Figs.2c and 2d,we could see that IPS1 and IPS2 were mainly composed of Man,Glc and Gal before fermentation,while there was almost no detectable monosaccharide after fermentation.The inability to detect monosaccharide after fermentation might be that IPS were degraded and utilized by intestinal microbiota [14].It has been reported that polysaccharide could be digested by the gut microbiota and produced some beneficial metabolites like SCFAs [22].
3.5 Change in pH during fermentation
The change of pH during fermentation was detected by a micropH meter.According to Fig.3,the pH of IPS and INL groups after 24 h of fermentation by human fecal microbiota were significantly decreased.It has been reported that pH value would decrease during polysaccharide fermentation process,which was consistent with the previous studies [22,29].Furthermore,the lower pH could inhibit the growth of Gram-negative bacteria such asEscherichia coli[35].It has been reported that people with colon cancer usually have a higher pH in the intestine than healthy people [36].Therefore,the decrease in pH value of the intestine can prevent the occurrence of colon cancer to some extent.It was reported that polysaccharide was utilized by microbiota in the fermentation and degraded into SCFAs to lower the pH [37].Besides,apart from the SCFAs production,other factors also can affect intestinal pH value,such as the culture medium variations,the types and amounts of microbiota in human fecal medium,and the types of generated SCFAs [22].
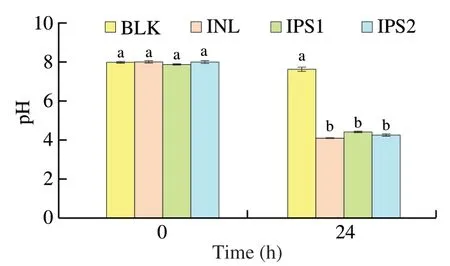
Fig.3 Changes in pH during in vitro fermentation by human fecal microbiota in different groups.Different lowercase letters (a and b) at the same time point indicate significant difference (P <0.05).
3.6 Gut microbiota analysis
3.6.1 Effects of IPS1 and IPS2 on the overall structure of intestinal bacteria
Tens of trillions of microorganisms reside in the human gut and are termed as gut microbiota or gut microbiome.The gut microbiota is important for maintaining normal physiology and energy production throughout life [38].It is important to note that the inter-individual variability of gut microbiota is easily affected by age,gender,geographic location and diet,etc.[39].To minimize the impact of individual differences,six healthy volunteers (three female and three males) aged 20-30 were selected.High throughput sequence analysis of bacterial 16S rRNA were performed to investigate the effects of IPS on gut microbiota.The results showed that the average sequencing depth of total 12 samples was 42 077 ± 8 450 reads.From the Fig.S1,we can see that the Shannon index of the 12 samples were stable,demonstrating that the sequencing data had covered the phylotypes and diversity.
As shown in the Table 1,the index value of Chao1 and ACE reflected the community richness,while the index value of Shannon and Simpson reflects the community diversity.Compared with the BLK group,the community richness and community diversity of IPS1,IPS2 and INL groups were lower.The similar results on gut community diversity of polysaccharide from Chinese Wolfberry andAscophyllumnodosumwere observed,and these can likely be explained by the competitive role of dominant microbiota [40,41].
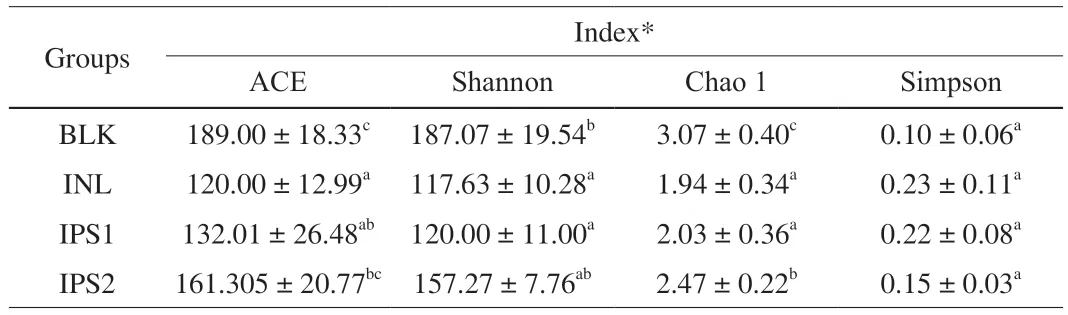
Table 1 Alpha diversity of samples among different groups.
3.6.2 Effects of IPS1 and IPS2 on microbial composition at the phylum level
In order to further clarify the effect of polysaccharide on intestinal microorganisms,the composition of microflora at the phylum level was analyzed.Many microorganisms exist in the human gut,but the level of phylum on intestinal microbiota are limited.The most common bacterial phyla are Bacteroidetes and Firmicutes,Proteobacteria,and Actinobacteria in healthy adults [1,12].At the phylum level,we also mainly identified these four phyla (Supporting information Fig.S2).The relative proportions of these phyla were changed under different treatments.From the Table 2,the ratio of Firmicutes/Bacteroidetes was changed after INL or IPS1 treament.Notably,the Firmicutes was increased and Bacteroidetes was decreased with INL or IPS1 fermentation,however,a completely opposite result was found for fermentation with IPS2,meaning that IPS2 could significantly reduce the ratio of Firmicutes/Bacteroidetes.It has been reported a decreased ratio of Bacteroidetes to Firmicutes might lead to a reduction in bacterial energy use,and thereby to a reduction in the risk of obesity in humans [40].The Proteobacteria has been regarded as a sign of gut microbial dysbiosis and linked to some diseases,such as diabetes,inflammation and cancer [42].Both the IPS1 and IPS2 treatment groups can decrease the level of Proteobacteria compared with BLK groups.
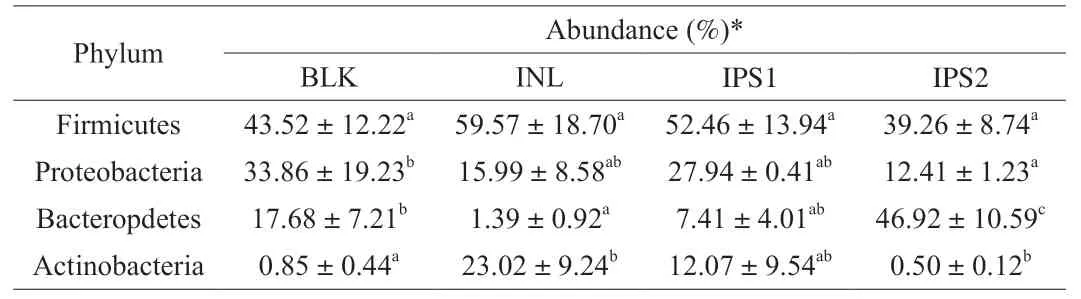
Table 2 Relative abundance of phylum levels in different treatments.
3.6.3 Effects of IPS1 and IPS2 on microbial composition at the genus level
To better show the microbial changes in different treatment groups,we performed heat map at the genus level.In the heat map,top 50 genera with high relative abundance were identified (Supporting information Fig.S3).All the genera belonged to the seven phyla including Actinobacteria,Bacteroidetes,Firmicutes,Fusobacteria,Proteobacteria,Verrucomicrobiota and unclassified.Significant differences in gut microbiota at the genus in each group were shown in the Table 3.
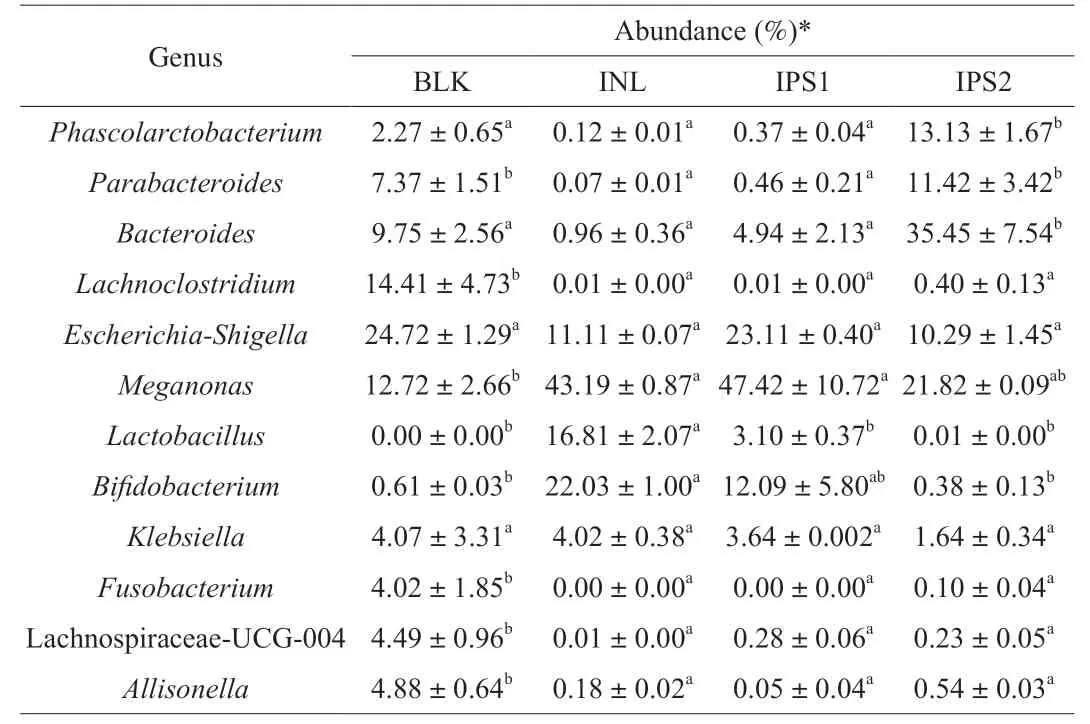
Table 3 Differences in gut microbiota at the genus in each group.
After IPS1and INL treatment,the abundance ofMegamonaswas significantly increased (P<0.05) to 47.12% and 43.19%,respectively.It becomes the dominant microbiota in IPS1 and INL groups,demonstrating thatMegamonasmight be the major gut microorganism to utilize IPS1 and INL.By contrast,Bacteroideswas significantly increased from (9.75 ± 2.56)% to (35.45 ±7.54)% after IPS2 treatment (P<0.05),but the same trend was not observed in the IPS1 group.Bacteroidesspecie is one of anaerobic,bile-resistant,non-spore-forming,gram-negative rods [43].It has been reported that the level ofBacteroideswas strongly associated with long-term diets rich in protein and animal fat [44].Now,Bacteroideshas been considered as a kind of beneficial gut microbiota,which plays an important role in maintaining a complex and beneficial relationship with the host [45].Meanwhile,Bacteroidescan decompose and use polysaccharides to produce SCFAs that are good for the human body [30].In addition,IPS2 can also promote the genus proliferation ofParabacteroidesandPhascolarctobacterium.Wu et al.[46]reported thatHirsutellasinensispolysaccharide notably enriched the gut bacteriumParabacteroidesgoldsteiniiand it represented a novel probiotic that may be used to treat obesity and type 2 diabetes.Phascolarctobacterium,belong to the Firmicutes,can produce SCFAs,including acetate and propionate and it was found to be positively correlated to the positive mood of the human [4,47].Meanwhile,Phascolarctobacteriumwas also demonstrated to provide nutrition to the colonic cell,protect the colonic mucosa,reduce the colonic inflammation and reduce the risk of colon cancer [48].BifidobacteriumandLactobacilluswere increased following treatment with IPS1 and INL groups,compared with the BLK group.BifidobacteriumandLactobacillusare two well-known probiotics and widely used for improving human health,such as modulation enteric nervous system [49],resistance to harmful bacteria [50],and stimulation the immune system [51].Finally,IPS1,IPS2 and INL reduced the levels of pathogens such asEscherichia-ShigellaandKlebsiella,which have a great relationship with colon cancer and pneumonia,respectively [52].Some pathogenic strains belong to the genusFusobacteriumexist in oral cavity can cause oral cancer,Fusobacteriumhas been closely associated with other diseases such as adverse pregnancy,cardiovascular disease,rheumatoid arthritis and lung abscesses [53].Its efficient pathogenicity was due to its ability to adhere with Gram-negative and Gram-positive microorganisms in
biofilms,leading to a highly invasive pathogen.As expected,after the treatment of IPS1 and IPS2,the level ofFusobacteriumwas decreased to 0 and (0.10 ± 0.04)%,respectively.Taking the above results into account,it could be concluded that IPS1 and IPS2 had an obvious prebiotic effect on promoting the proliferation of probiotics.
3.6.4 Analysis of intestinal microbial Beta diversity
Hierarchical clustering tree,principal component analysis (PCA)with the unweighted unifrac distance and principal co-ordinates analysis (PCoA) were analyzed to show the beta diversity of different treatments.Cluster tree displayed that the different groups aggregated into two branches,INL and IPS1 groups were grouped into one branch,while BLK and IPS2 groups were grouped into another branch (Fig.4a).As shown in the Fig.4a,PCA revealed significant differences in the microbial composition of the four groups,PC1 and PC2 explained 73.89% of the total variance.The results of PCoA exhibited that the gut microbiota of the IPS1 group was close to the INL group and keep away from BLK group,while IPS2 was not only far from INL Group but also BLK Group,which consistent with the results of PCA.Adonis revealed significant differences in the structure of gut microbiota among different groups.The corresponding Adonis value (R2=0.80,P=0.001) was calculated based on the Bray-Curtis.These results indicated that there were significant differences between the different groups.
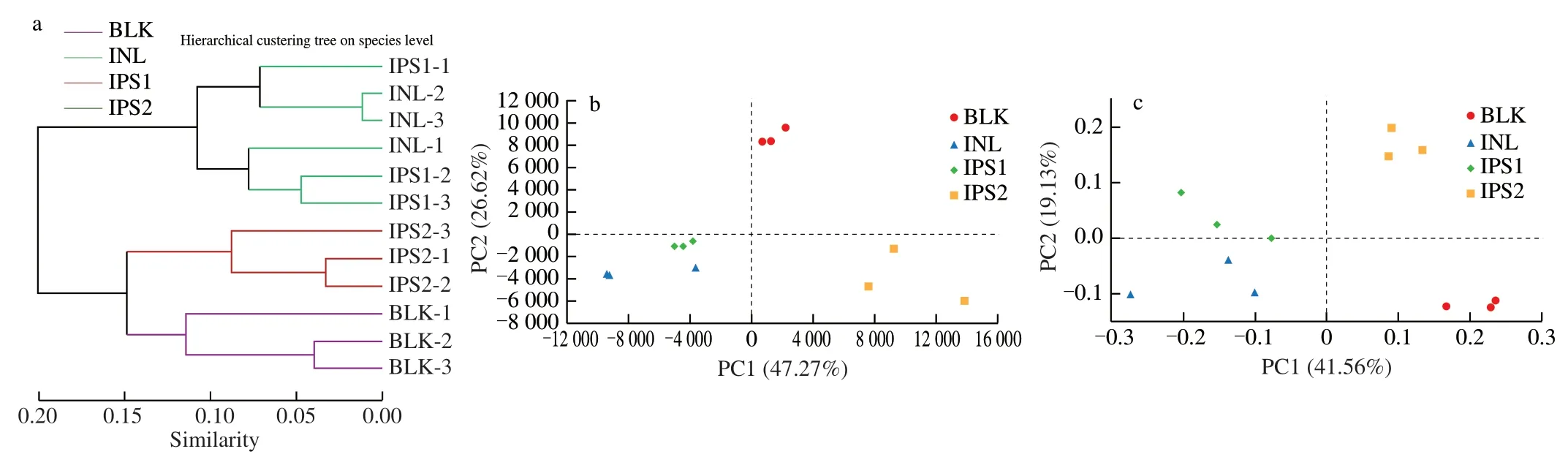
Fig.4 Analysis of intestinal microbial Beta diversity.Multivariate analysis of variance from matrix scores based on Bray-Curtis method (a);principal component analysis of gut microbiota at the OTU level (b) and PCoA (c) were analyzed based on the unweighted-unifrac method.
3.6.5 Analysis of species differences between groups
The top 15 high abundance species were compared among different groups.As shown in the Fig.5a,there were 8 statistically significant differences between BLK and INL groups at the species level.The relative abundance ofBifidobacteriumsp.(P<0.05) andLactobacillusruminis(P<0.05) was significantly increased in the INL group.In the Fig.5b,Megamonas(P<0.05) andLactobacillus ruminis(P<0.01) were clearly higher in the IPS2 group,while they decreased in the INL group exceptBilophilawadsworthia.In the Fig.5c,compared with the BLK group,IPS2 could markedly increase the levels ofBacterodiescoprocolaDSM 17136 (P<0.01) andPhacolarctobacterium(P<0.001).The Fig.5d showed that IPS1 and IPS2 could promote the growth of different intestinal microorganisms,which may be due to the different structures of the two fractions.
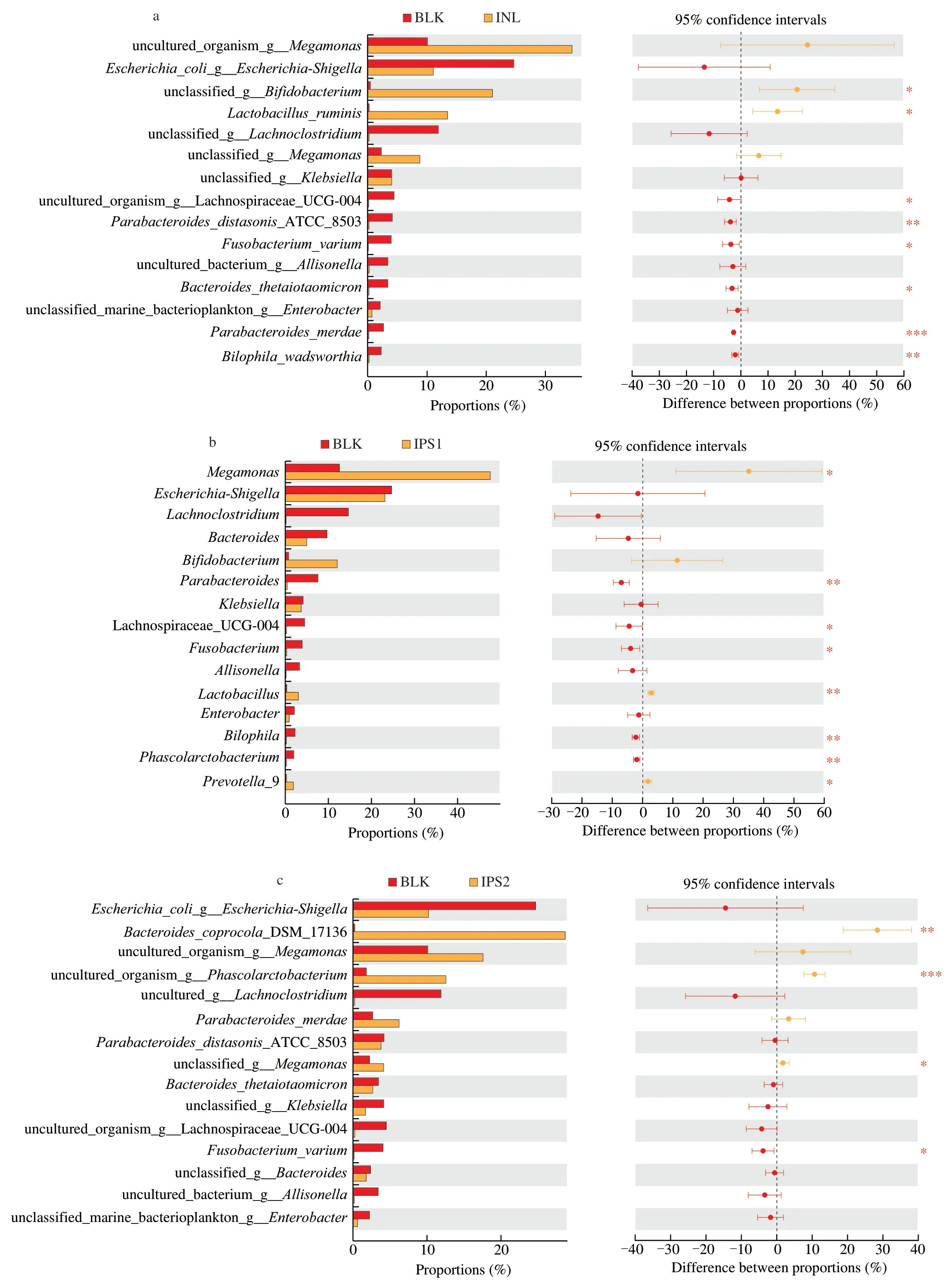
Fig.5 Relative abundance of species showed significant differences among different samples.Comparison of high abundance species in the BLK and INL groups(a),comparison of high abundance species in the BLK and IPS1 groups (b),comparison of high abundance species in the BLK and IPS2 groups (c),comparison of high abundance species in the IPS1 and IPS2 (d).The method of student’s T test was used to evaluate the significance differences between two groups.*P <0.05,**P <0.01 and ***P <0.001.
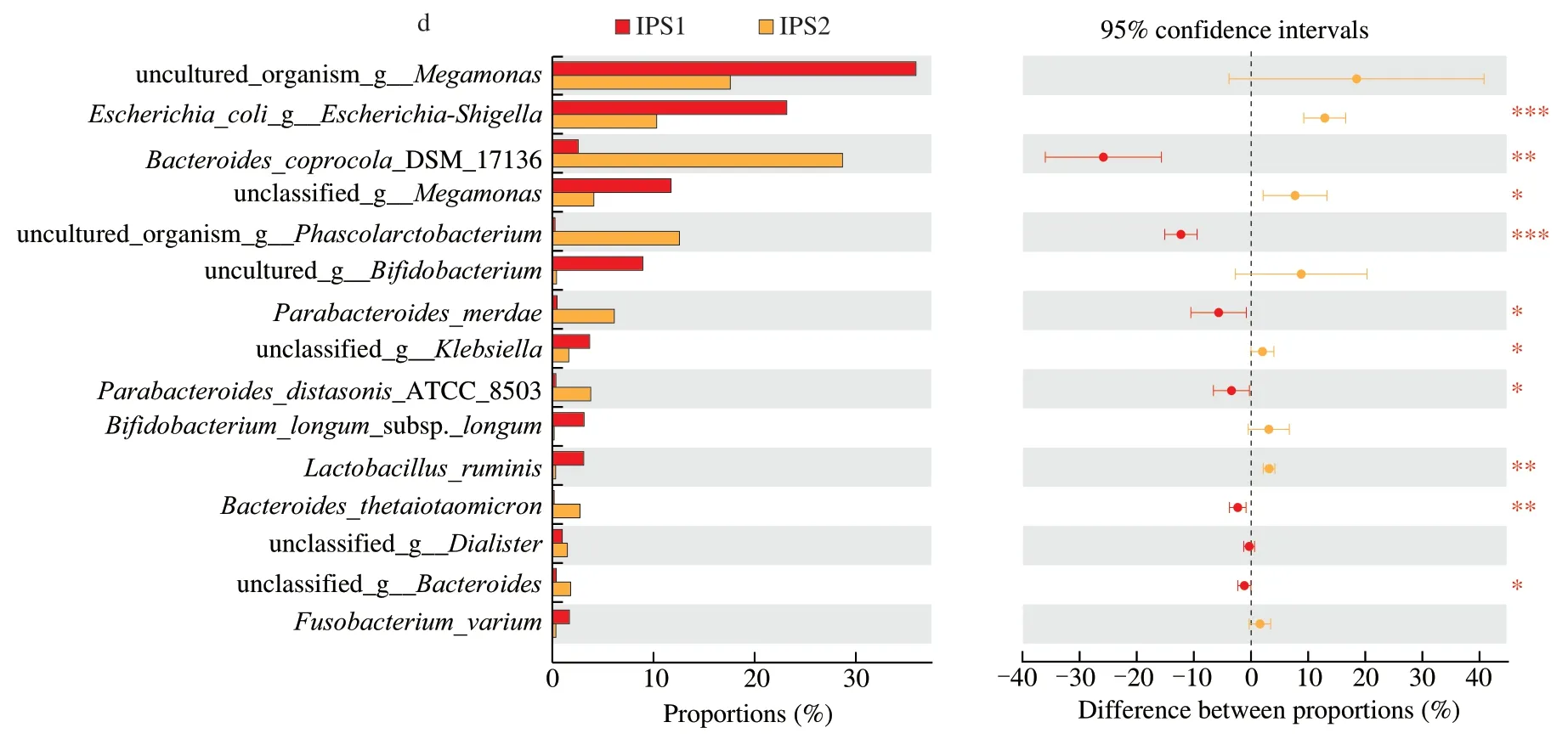
Fig.5 (Continued)
In order to better display the microbiota with significant differences in abundance in each group,the method of liner discriminant analysis effect size (LEfSe) was used (Fig.6).Compared with BLK group,Firmicutes (Megamonas) and Bacteroidetes(Prevotella9) were absolutely significant in the IPS1 group.Firmicutes (including the species ofMegasphaera,Dialister,Phascolarctobacterium,Selenomonadales,Holdemanella) and Bacteroidetes (such asBacteroides,Parabacteroides) were higher in the IPS2 group.In the INL group,the species ofCollinsella,Bifidobacterium,LactobacillusandLactococcuswere dominant.Moreover,OTUs (the relative abundance over 0.1%) were selected for linear discriminant analysis (LDA) according to the LDA scores(lg).In the BLK group,Escherichia-Shigella(OTU 56,OTU 19),Lachnospiraceae UCG-004 (OTU 53),Parabacteroides(OTU 231),Fusobacterium(OTU 64) andBacteroides(OTU 88,OTU 205,OTU 211,OTU 185,OTU 119,OTU 127 and OTU 184) were more abundant.However,in the INL group,Bifidobacterium(OTU 225 and OTU 223),Lactobacillus(OTU 5 and OTU 13),Collinsella(OTU 17),Streptococcus(OTU 26) andBlautia(OTU 44 and OTU 158) were the major bacteria.In the IPS1 group,Megamonas(OTU 28 and OTU 192),Bifidobacterium(OTU 163),Prevotella9 (OTU 42),Bacteroides(OTU 226 and OTU 155) were more emblematic.The relative abundance ofMegamonasaccounted for 47.48% of total microbial population in the IPS1 group and was higher than those in other groups.This might be that IPS1 contains higher a monosaccharide of glucose than IPS2 andMegamonascan utilize glucose [54].In the IPS2 group,Bacteroides(OTU 183,OTU 170,OTU 239 and OTU 232),Phascolarctobacterium(OTU 189 and OTU 196),Parabacteroides(OTU 179,OTU 105,OTU 169,OTU 128,OTU 203 and OTU 165),Dialister(OTU 201) andHoldemanella(OTU 200) were the represent bacteria.The result was similar to a recent report which found thatC.sinensispolysaccharides(CSP) increased the abundance of probiotics (Lactobacillus,Bifidobacterium,Bacteroides) and decreased pathogenic bacteria(ClostridiumandFlexispira) [55].
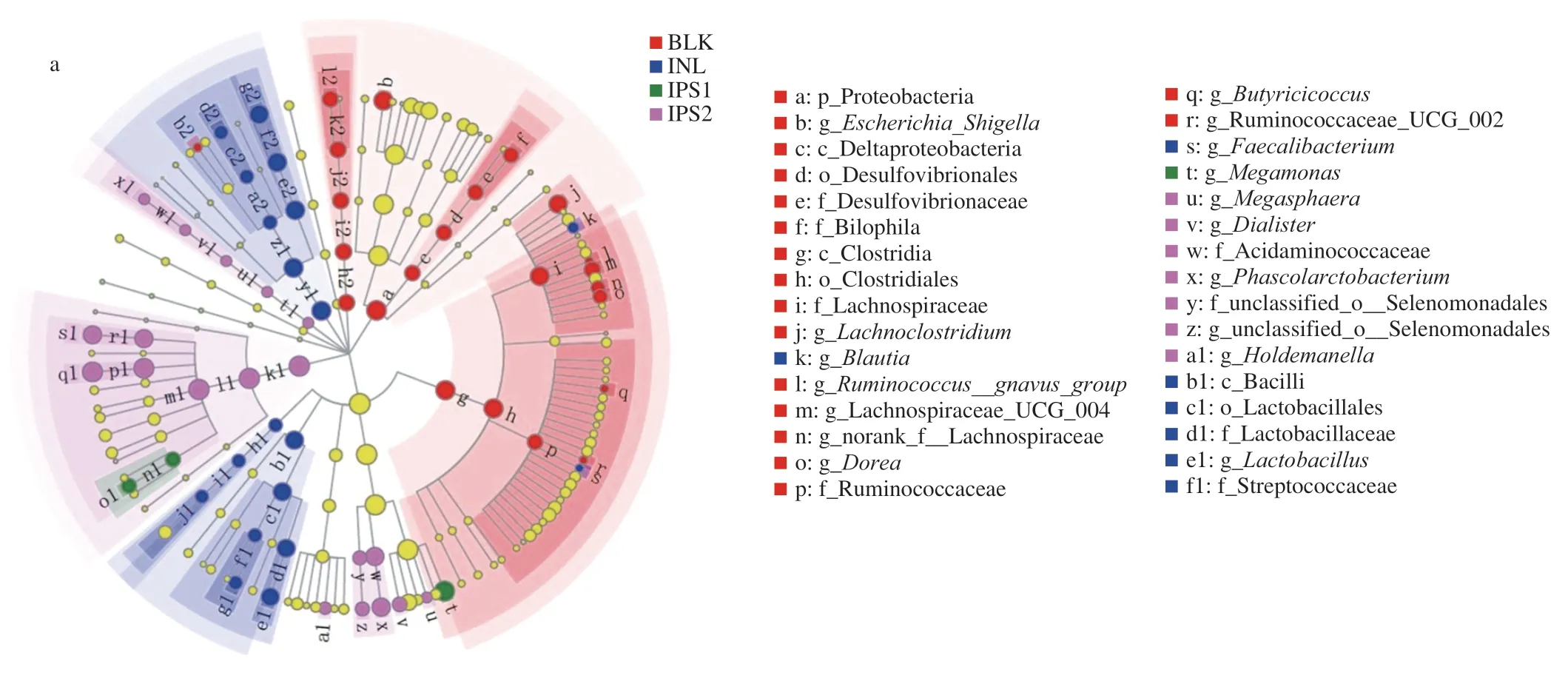
Fig.6 Comparison of microbiota among BLK,INL,IPS1 and IPS2 groups based on linear discriminant analysis effect size (LEfSe) (a).LDA scores of 3.0 or greater in bacterial communities,different colored regions represent different groups (b).Circles indicate phylogenetic levels from phylum to genus.The diameter of each small circle was proportional to the abundance of the group and yellow circles represent microbiota was no difference BLK,INL,IPS1 and IPS2 groups.
Fig.6 (Continued)
3.7 SCFAs analysis
SCFAs are the final products of the fermented polysaccharides by specific anaerobic gut microbiota.Increasing evidence demonstrated that SCFAs play an important and beneficial role in the host physiology and energy homeostasis [56].As shown in Fig.7,the total SCFAs in all samples increased with the proceeding of fermentation.It was noted that the content of propionic acid in IPS1 and IPS2 treatment groups reached to (66.86 ± 7.53) mmol/L and (87.24 ± 6.56) mmol/L,respectively.The level of propionic acid was significantly higher than that BLK group.The production of propionic acid was mainly due to fermentation of glucose,mannose and arabinose [57].It reported that propionic acid can be found in many Bacteriodetes and Firemicures [34],which was also consistent with our previous description of the changes in the phylum levels (Table 3).The increase of propionic acid might be attributed to the increasePhascolarctobacteriumin the IPS2 group according to the previous studies [14].Furthermore,Bacteroides-Prevotellais a dominant group of the gut microbial communities and is the known producer of propionic acid [58].The level of acetic acid accumulated was higher in the IPS1 and IPS2 group,which may be related to the fermentation of galactose in the IPS1 and IPS2 [59].Acetic and lactic acid a major end-product ofBifidobacteriumandLactobacillusfermentation and the most abundant SCFAs in the peripheral circulation,and they are able to pass through the blood-brain barrier to inhibit appetite via a central homeostatic mechanism [56].For sample groups,large amount of lactic and acetic acid was produced during fermentation,which resulted in a high number ofBifidobacteriaandLactobacillus.In general,the different SCFAs compositions and content were not only related to the structural properties,monosaccharide composition,molecular weight,glycosidic bond and branched chain,but also related to the type and amounts of selective gut microbiota and the fermentation parameters such as time,temperature,and anaerobic environment [60].Moreover,the microbial activity also influences the composition and content of SCFAs [61].Therefore,the content and distribution of SCFAs are associated with various factors.
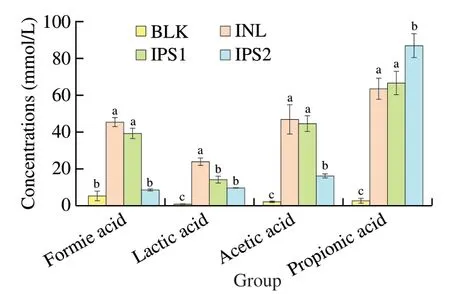
Fig.7 Concentrations of SCFAs after fermentation.Data were shown as mean ± standard deviation (n =3).The letters of a-c represented significant differences among different groups by one-way ANOVA procedure followed by Duncan test (P <0.05).
4.Conclusion
In this study,we evaluated the effects of IPS fromP.cicadaeTJJ1213 on intestinal microorganisms byinvitrofecal fermentation.The results showed that IPS1 and IPS2 were hydrolyzed and promoted the production of SCFAs especially formic acid,lactic acid,acetic acid and propionic acid after 24 h fermentation by human fecal microbiota.Furthermore,IPS1 could enhance the relative abundances of beneficial bacteria includingMegamonas,BifidobacteriumandLactobacillus,while IPS2 could increase the proliferation of beneficial bacteria containingBacteroides,ParabacteroidesandPhascolarctobacterium.Considering the above results,we can conclude that IPS fromP.cicadaeTJJ1213 could be regarded as a candidate prebiotic.In order to further explain the prebiotic effects of the polysaccharide,invivoanimal experiments will be studied in the future.
Declaration of conflicting interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was co-financed by National Natural Science Foundation of China (U1903108,31871771 and 31571818),Natural Science Foundation of Jiangsu Province (BK20201320),Jiangsu Agriculture Science and Technology Innovation Fund (CX (20)3043),Postgraduate Research&Practice innovation Program of Jiangsu Province (KYCX19_0589),Qing Lan Project of Jiangsu Province and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Appendix A.Supplementary data
Supplementary data associated with this article can be found,in the online version,at http://doi.org/10.1016/j.fshw.2022.07.065.
- 食品科学与人类健康(英文)的其它文章
- Colloidal nanoparticles prepared from zein and casein:interactions,characterizations and emerging food applications
- Biological factors controlling starch digestibility in human digestive system
- Preparation methods,biological activities,and potential applications of marine algae oligosaccharides: a review
- Development of hyaluronic acid-based edible film for alleviating dry mouth
- Mushroom β-glucan and polyphenol formulations as natural immunity boosters and balancers: nature of the application
- Preparation of multicore millimeter-sized spherical alginate capsules to specifically and sustainedly release fish oil

