miRNA-128-3p inhibits malignant behavior of glioma cells by downregulating KLHDC8A expression
YU Zhengtao,LI Jiameng,JIANG Junwen,LI You,LIN Long,XIA Ying*,WANG Lei*
1Department of Neurosurgery,Affiliated Haikou Hospital of Xiangya School of Central South University,Haikou 570208,China;2Department of Neurosurgery,Hunan Cancer Hospital and Affiliated Cancer Hospital of Xiangya School of Medicine,Central South University,Changsha 410006,China
Abstract: Objective To determine whether miRNA-128-3p regulates malignant biological behavior of glioma cells by targeting KLHDC8A.Methods Dual-luciferase reporter assays,qRT-PCR and Western blotting were used to verify the targeting of miRNA-128-3p to KLHDC8A.Edu assay,flow cytometry,Transwell assay and would healing assay were used to determine the effects of changes in miRNA-128-3p and KLHDC8A expression levels on malignant behavior of glioma cells.Rescue experiment was carried out to verify that miRNA-128-3p regulated glioma cell proliferation,apoptosis,invasion and migration by targeting KLHDC8A.Results The expression level of KLHDC8Awas significantly increased in high-grade glioma tissue and was closely related to a poor survival outcome of the patients.Overexpression of KLHDC8A promoted glioma cell proliferation,migration and invasion,and miRNA-128-3p overexpression inhibited proliferative and metastatic capacities of glioma cells.Mechanistically,KLHDC8A expression was directly modulated by miRNA-128-3p,which,by targeting KLHDC8A,inhibited malignant behavior of glioma cells.Conclusion Upregulation of miRNA-128-3p inhibits uncontrolled growth of glioma cells by negatively regulating KLHDC8A expression and its downstream effectors,suggesting that the miRNA-128-3p-KLHDC8A axis may serve as a potential prognostic indicator and a therapeutic target for developing new strategies for glioma treatment.
Keywords: glioma;miRNA-128-3p;KLHDC8A;biomarker;migration;proliferation
INTRODUCTION
Gliomas are the deadliest brain malignancies and their treatment remains clinically challenging in spite of the current advances in cancer therapies[1,2].The molecular complexity[3],high invasiveness,heterogeneity,and blood-brain barrier (BBB) penetrability[4],as well as the unique immune environment of gliomas[5]all contribute to a poor prognosis of glioma patients.The recent National Comprehensive Cancer Network guidelines (v1.2022)classified adult gliomas into 5 categories,namely adult glioma (WHO grade 1),oligodendroglioma (IDH mutant with 1p/19q co-deletion),IDH-mutant astrocytoma,glioblastoma (GBM),and intracranial/spinal ependymoma.The histological features of these glioma subtypes are analyzed in conjunction with their molecular diagnosis to provide prognostic confirmation for more personalized therapies[6].However,early diagnosis and prediction of the clinical behaviors,treatment responses,and prognosis of invasive gliomas remain challenging and require the discovery of more effective biomarkers.
KLHDC8A,a member of the Kelch domaincontaining (KLHDC) protein superfamily,is encoded by a gene located at the 1q32.1 locus[7].Kelch-repeat proteins are important regulators of cellular processes(migration,morphogenesis,and interaction) and some molecular mechanisms (gene activation and protein degradation)[8]and participate in tumorigenesis and neurological dysfunctions[9].Previous studies showed that KLHDC8A expression was increased in gliomas in association with the WHO grade[10]and in glioblastoma tissues[11],but its association with the upstream regulatory factors has not been fully understood.
Micro-RNAs(miRNAs)have been shown to regulate mRNA stability and translational turnover of their target RNAs under both physiological and pathological conditions[12,13]and participate in the regulation of a wide variety of cellular and biological processes[14-16].Aberrant miRNA expressions have been observed in oncogene activation in several life-threatening cancers,including gliomas[17-20],suggesting the possibility of using miRNAs as biomarkers for cancer diagnosis and targets for cancer therapy.
We previously demonstrated,by comparing mRNA and miRNA expression profiles between glioblastoma and paired adjacent tissue,that miRNA-128-3p expression was significantly reduced in glioblastoma and KLHDC8A mRNA was one of its targets in the gene regulatory network[21].We therefore hypothesized that miRNA-128-3p is capable of regulating KLHDC8A expression in glioblastoma.In this study,we investigated the cross-talk between miRNA-128-3p and KLHDC8A in glioma cells,and the results suggest that the oncogenic expression of KLHDC8A is instrumental for the malignant phenotype of glioblastoma and associated with poor treatment responses of the patients.
METHODS
Bioinformatic analysis
Bioinformatic analysis was performed as described previously[21].Briefly,miRNA expression profile dataset(GSE90603) and transcriptome profile dataset(GSE90598) were downloaded from combined miRNA-mRNA microarray chips in the Gene Expression Omnibus (GEO) database.The expressions of miRNA and mRNA were detected using the platform GPL21582 and GPL17692.The differentially expressed miRNAs(DEMs) and differentially expressed genes (DEGs) were identified using the R limma software package.FunRich was used to perform functional enrichment analysis;the miRNAs and their target gene regulatory network was constructed using Cytoscape 3.6.1 software.
Survival analysis and glioma grading analysis
The Chinese Glioma Genome Atlas (CGGA) database(http://www.cgga.org.cn/) was used for glioblastoma grading and survival analyses.This database of over 2000 glioma tissue samples contains miRNA microarrays(198 samples) and matched clinical data that can be used to assess the impact of certain miRNAs on glioma patient survival.The miRNA expression levels between grade II and grade IV gliomas were analyzed using CGGA database.The median expression value of each DEM in glioma specimens was calculated to define the high expression (above the median) group and low expression(below the median) group.Combined with the prognostic results obtained from the CGGA database (including overall survival data),Kaplan-Meier survival curves of the two groups of patients were drawn,and log-rank test was used to evaluate the relationship between the gene expression level and the patients'survival.A P value less than 0.05 was considered statistically significant.
Cell culture
Human glioma lines U87-MG (#BNCC 100646;BNCC,China) and U251 (#BNCC 337874;BNCC,China) and normal human astrocytes (NHAs)(#QCB849;QCHENG,China) were cultured routinely in DMEM medium(#R8758,Sigma,USA) supplemented with 10% fetal bovine serum (FBS;Gibco,USA) and 1% Pen-Strep(#SV30010,Beyotime,China) at 37 ℃in 5% CO2in a humidified chamber.
Cell transfection
OE-KLHDC8A,OE-NC,KLHDC8A siRNA,and siRNA NC were purchased from RiboBio.miRNA-128-3p mimic,mimic NC,miRNA-128-3p inhibitor,and inhibitor NC were purchased from HonorGene.For cell transfection,the cells were seeded in a 6-well plate,and when the cell confluence reached 60%,the culture medium was removed.95 μL of serum-free culture medium was added to each sterile centrifuge tube,followed by addition of 5 μL of each of the 8 plasmids(at a concentration of 20 μmol/L and 5 μL of Lipofectamine 2000 reagent (Invitrogen,USA).After gentle mixing,the mixtures were incubated at room temperature for 20 min.In the 6-well plate,800 μL of serum-free culture medium was added in each well,and the aforementioned mixtures were added to each well and gently mixed.The plate was incubated at 37 ℃in 5% CO2for 6 h.Fresh complete culture medium was then changed,and the cells were further incubated for 48 h for subsequent analysis.The sequences of KLHDC8A siRNAs and miRNA-128-3p mimic and inhibitor are listed in Tab.1.

Tab.1 Sequences of miRNA-128-3p mimic and inhibitor and 3 KLHDC8A siRNAs used in this study
Quantitative real-time-PCR(qRT-PCR)
The total RNA was extracted from the cells using TRIzol®method (Thermo,USA).The miRNAs were reverse transcribed into complementary DNA (cDNA) using Super RT reagent,and compatible SYBR Master mix(#CW2569,and#CW2601,CWBIO,China)was used for qRT-PCR following the manufacturer's instructions.SnU6 and β-actin served as the references for quantitation of miRNA and KLHDC8A mRNA expressions,respectively.The primer pairs were synthesized by Shanghai Genomics Institute(Tab.2).The amplification system consisted of 2 μL cDNA,1 μL Primer R,1 μL Primer F,11 μL ddH2O,and 15 μL 2×SYBGREEN PCR Master Mix.Each of the 30 μL mixture for each sample was added in 3 wells(10 μL per well)for qRT-PCR detection.The amplification was initiated at 95 ℃for 10 min,followed by 40 cycles of 95 ℃for 15 s and 60 ℃for 30 s.The 2-ΔΔCtmethod was used to determine the relative expressions of the mRNAs and miRNAs.
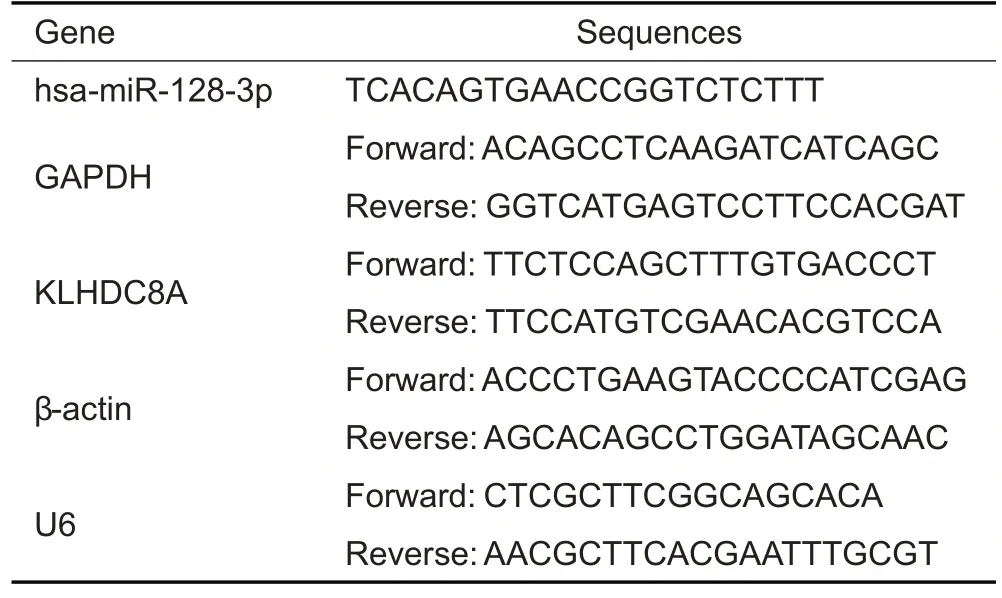
Tab.2 Primer sequences for reverse transcription quantitative polymerase chain reaction
Western blotting for detecting KLHDC8A protein expression
Cell lysates were prepared in RIPA(#P0013B,Beyotime,China) buffer,and the protein concentrations were measured using a BCA kit (Thermo,USA).After 10%SDS-PAGE,the total proteins were transferred from the gel to polyvinylidene difluoride membranes,which were blocked with 5%skim milk for 1.5 h at room temperature.The membranes were then incubated with the primary antibodies anti-KLHDC8A (ab235419,Abcam,UK) and anti-β-actin (#66009-1-Ig,Ptg,USA) overnight at 4 ℃,and then with the secondary antibodies HRP-conjugated goat anti-mouse IgG (1:5000;SA00001-1,Proteintech,US) and HRP-conjugated goat anti-rabbit lgG (1:6000;SA00001-2,Proteintech,US) for 1 h at room temperature.The protein bands were visualized by exposure for 60 s to enhanced chemiluminescence(ECL)reagent.
Luciferase reporter assay
The coding sequence of miRNA-128-3p was obtained from TargetScan(http://www.targetscan.org/)and MiRDB(http://www.mirdb.org/miRDB/).The pmirGLO vector(Promega,USA)was used for cloning the wild-type(WT)and mutant (MUT) KLHDC8A sequences to assess the binding of miRNA-128-3p to its target mRNA.The vectors were subsequently transfected into HEK-293A cells,in parallel to miRNA mimic,inhibitor and negative control,and luciferase activity was measured 48 h later using the luciferase reporter activity assay kit (Promega,USA).
EdU assay
The EdU Apollo-567 kit (RiboBio,China) was used for cell staining with 50 μmol/L EdU overnight,followed by fixing and imaging following the manufacturer's instructions.For quantitative analysis,3 visual fields were randomly captured,and the cell nuclei were counterstained with DAPI.
Apoptosis analysis
The cells transfected with the plasmids or miRNAs were stained with 5 μL Annexin V-APC and 5 μL propidium iodide (#KGA108,KeyGen BioTECH,China) in the binding buffer for 10 min in darkness.Flow cytometry was performed for each sample in triplicates to detect both late and early apoptotic cells.
Wound-healing assay
Six hours after transfection,U87-MG and U251 cells were suspended in FBS-free medium and seeded at the density of 5×105cells/well into 6-well plates,and a vertical scratch was made using a sterile pipette tip.The cells were cultured for another 24-48 h,and the width of the wound was photographed at 0,24,and 48 h.The healing abilities of the glioma cells were assessed by measuring the area of migrating cells.
Transwell assay
Transwell assay was used to evaluate the invasion and migration abilities of the cells.The upper layers of the Transwell plates were coated with Matrigel (BD Biosciences,Franklin Lakes,NJ,USA) at the final concentration of 200 μg/well,diluted with serum-free DMEM medium,and the plates were incubated at 37 ℃for 30 min.Following coating,the cells were digested with trypsin and resuspended in 100 μL serum-free medium (2×106cells/mL).The cell suspensions were added into the upper layer,and 500 μL medium containing 10% FBS was added into the lower compartment.The cells were cultured in a cell incubator at 37 ℃for 48 h in 5%CO2for 48 h,and the invaded or migrated cells in the lower compartment were fixed in 4%paraformaldehyde and stained with 0.1% crystal violet.The cells inside the upper layer were removed using cotton balls,while the cells that penetrated the upper layer were counted under a microscope.Finally,500 μL of 10%acetic acid was added to the lower compartment,and the absorbance value at 550 nm was measured with a microplate reader.
Statistical analysis
The data were analyzed using GraphPad Prism 8.0.All data were presented asMean±SD.Shapiro-Wilk test and Levene's test were performed to assess the normality and homogeneity of variances,respectively.One-way analysis of variance (ANOVA) was used to determine statistical differences among different groups.Two-way ANOVA was employed to determine the significance of differences between different time points and different groups.APvalue less than 0.05 was considered to indicate a statistically significant difference.
RESULTS
KLHDC8A is upregulated with WHO Grade of gliomas
To explore the correlation between WHO grades and KLHDC8A expression in gliomas,we compared KLHDC8A expressions among patients with different WHO grades (II-IV) based on data from the CGGA database.The results showed that KLHDC8A expression was significantly higher in high-grade (WHO grade III and IV) than in low-grade (WHO grade II) gliomas(Fig.1A).A higher KLHDC8A expression level was associated with a poorer overall survival (OS) rate of the glioma patients,irrespective of the tumor grade(Fig.1B).The results of RT-PCR and Western blotting showed that the expression of KLHDC8A was significantly elevated in U251 and U87 cells as compared with that of NHAs(Fig.1C).These results indicate that altered expressions of KLHDC8A might be involved in glioma pathogenesis.

Fig.1 Analysis of the expression of KLHDC8A in glioma tissues and glioma cell lines.A: KLHDC8A expression is positively correlated with glioma grade.B: Prognostic value of KLHDC8A in gliomas of all grades.C: Protein expressions of KLHDC8A in glioma cell lines (U251 and U87 cells) detected with Western blotting (*P<0.05 vs NHAs group).D: qRT-PCR and Western blotting of KLHDC8A mRNA and protein levels in U251 and U87 cells transfected with KLHDC8A siRNA or KLHDC8A-overexpressing plasmid(*P<0.05 vs control group;#P<0.05 vs OE-KLHDC8A NC).
KLHDC8A overexpression promotes proliferation,invasion and migration of glioma cells
Of the 3 siRNA sequences for KLHDC8A knockdown,siRNA-1774 was found to produce the strongest effect for lowering KLHDC8A expression in U251 and U87 cells and was therefore used for subsequent experiments(Fig.1D).KLHDC8A overexpression in U251 and U87 cells transfected with the KLHDC8A-overexpressing plasmid were verified by qRT-PCR and Western blotting(Fig.1D).EdU assay showed that KLHDC8A knockdown significantly inhibited proliferation of the glioma cells,and KLHDC8A overexpression significantly enhanced the proliferation of U251 cells (Fig.2A).Flow cytometry showed that KLHDC8A knockdown caused increased cell apoptosis,while KLHDC8A overexpression suppressed apoptosis in both of the glioma cell lines(Fig.2B).
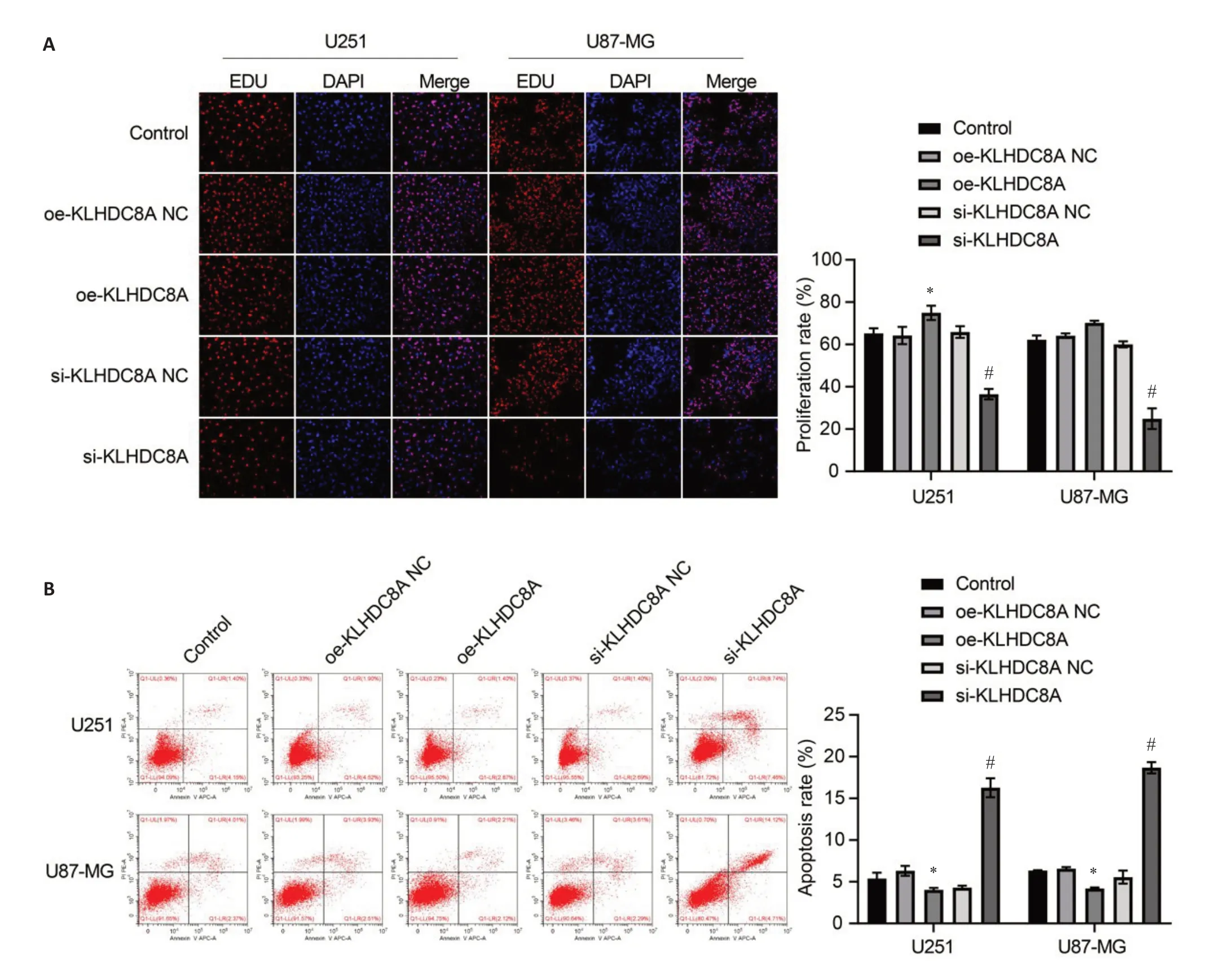
Fig.2 Effect of KLHDC8A knockdown and overexpression on proliferation and apoptosis of glioma cells.A:Cell proliferation assay(Edu assay)of U251 and U87 cells transfected with OE-KLHDC8A NC,OE-KLHDC8A plasmid,KLHDC8A siRNA-NC,or KLHDC8A siRNA.B: Flow cytometry for detecting apoptosis of U251 and U87 cells with KLHDC8A knockdown and overexpression.Each assay was repeated for at least 3 times.*P<0.05 vs control group;#P<0.05 vs KLHDC8A siRNA-NC group.
Transwell and wound-healing assays showed that KLHDC8A overexpression significantly enhanced the migration and invasion capacities of glioma cells,and KLHDC8A knockdown produced the opposite effects(Fig.3).Collectively,these findings demonstrate that KLHDC8A overexpression promotes proliferation,migration and invasion and inhibits apoptosis of glioma cells,suggesting the possibility of KLHDC8A as an oncogene in glioma cells.

Fig.3 Transwell (A) and wound healing (B) assays for assessing invasion and migration of glioma cells with KLHDC8A overexpression or knockdown(×100).Each assay was repeated for at least 3 times.*P<0.05 vs control group;#P<0.05 vs KLHDC8A siRNA-NC group.
miRNA-128-3p modulates malignant behaviors of glioma cells
In both U251 and U87 cells,transfection with miRNA-128-3p mimic resulted in at least a 2-fold increase in miRNA-128-3p expression as shown by qRT-PCR (Fig.4A),miRNA-128-3p expression was significantly diminished upon treatment with the inhibitor (Fig.4A).EdU assay showed a decreased proliferation rate in the mimic-treated cells,while the inhibitor produced the opposite effect (Fig.4B).Flow cytometry revealed an increased apoptosis rate in the glioma cells transfected with miRNA-128-3p mimic and a lowered apoptosis rate in the cells transfected with the inhibitor(Fig.4C).

Fig.4 Expression and function of miRNA-128-3p in glioma cells.A:RT-qPCR analysis of miRNA-128-3p expression in U251 and U87 cells treated with miRNA-128-3p mimic,miRNA-338-3p inhibitor,or scrambled negative control RNA(*P<0.05 vs control group;#P<0.05 vs inhibitor-NC).B,C: Cell proliferation assay and flow cytometry for analyzing apoptosis of the transfected U251 and U87 cells.Each assay was repeated for at least 3 times(×100).*P<0.05 vs control group;#P<0.05 vs miRNA-128-3p inhibitor-NC group.
As shown by Transwell and wound-healing assays,transfection with miRNA-128-3p mimic significantly lowered the migration and invasion capacities of the glioma cells,which were strongly enhanced by transfection with miRNA-128-3p inhibitor(Fig.5).These findings demonstrate that miRNA-128-3p was capable of regulating proliferation,metastasis and apoptosis of glioma cells.

Fig.5 Transwell assay (A) and wound healing assay (B) for assessing invasion and migration of glioma cells transfected with miRNA-128-3p mimic and inhibitor(×100).*P<0.05 vs control group;#P<0.05 vs miRNA-128-3p inhibitor-NC group.
KLHDC8A expression is directly modulated by miRNA-128-3p
The in-silico analysis suggested the presence of a putative binding site for miRNA-128-3p at the 3'-untranslated region (3'-UTR) of KLHDC8A mRNA(Fig.6A).We verified this miRNA target site with dualluciferase reporter assay using both WT and MUT KLHDC8A 3'-UTR sequences,and the results showed obviously reduced luciferase activities in glioma cells co-transfected with the plasmid carrying WT 3'-UTR and miRNA-128-3p mimic as compared with that in mock-treated cells.As expected,the cells transfected with MUT 3'-UTR and miRNA-128-3p mimic did not show any significant changes in luciferase activities(Fig.6B),demonstrating that KLHDC8A mRNA 3'-UTR is a direct target of miRNA-128-3p.We further tested these results in the glioma cells transfected with miRNA-128-3p inhibitor or mimic and detected the changes in the expression of KLHDC8A using qRT-PCR and Western blotting,which yielded consistent results with the luciferase reporter assays(Fig.6C).
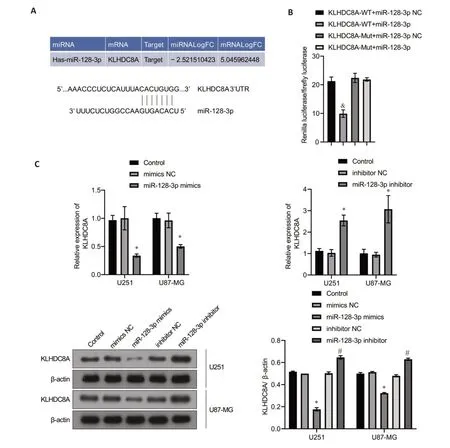
Fig.6 KLHDC8A is a direct target gene of miRNA-128-3p.A: Targeting relationship between miRNA-128-3p and KLHDC8A.B: Predicted binding sites for miRNA-128-3p in the 3'-UTR of KLHDC8A and mutations in the binding sites.Relative luciferase activity was measured in HEK293A cells co-transfected with miRNA-128-3p mimic (or miRNA-NC) and a luciferase reporter plasmid (&P<0.05 vs KLHDC8Awt.miRNA-128-3p mimic group).C: qRT-PCR and Western blotting of KLHDC8A mRNA and protein levels in U251 and U87 cells transfected with miRNA-128-3p mimic,miRNA-128-3p inhibitor,or scrambled negative control RNA(*P<0.05 vs control group;#P<0.05 vs miRNA-128-3p inhibitor-NC group).
miRNA-128-3p regulates malignant behavior of glioma cells by targeting KLHDC8A
In the rescue experiment,EdU assay showed that in the glioma cells,KLHDC8A overexpression-induced increase of cell proliferation was significantly suppressed by co-transfection of the cells with miRNA-128-3p mimic;Similarly,miRNA-128-3p inhibitor partially reversed the inhibitory effect of si-KLHDC8A on the proliferation of U87 and U251 cells (Fig.7A).Flow cytometry also confirmed that co-transfection with miRNA-128-3p mimic inhibited KLHDC8A overexpression-induced increase of apoptosis,while miRNA-128-3p inhibitor rescued KLHDC8A knockdown-mediated inhibition of apoptosis in both the glioma cell lines (Fig.7B).Consistently,Transwell assay and wound healing assay showed that modulation of miRNA-128-3p expression by transfection with its mimic or inhibitor obviously attenuated the effects of KLHDC8A overexpression and knockdown on invasion and migration of the glioma cells (Fig.8).These results indicate that miRNA-128-3p regulates biological behaviors of the glioma cells by directly targeting KLHDC8A.
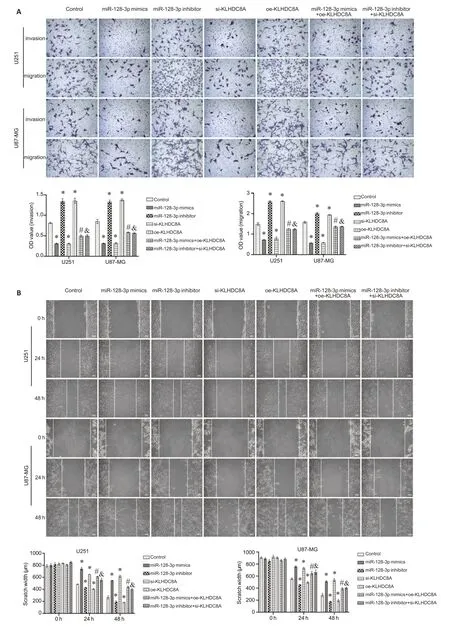
Fig.8 miRNA-128-3p modulates migration and invasion of glioma cells via targeting KLHDC8A.A:Transwell assay of U251 and U87 cells transfected with miRNA-128-3p mimic plus KLHDC8A siRNA or with miRNA-338-3p inhibitor plus KLHDC8A siRNA(×100).B: Wound healing assay of the transfected cells(×100).(*P<0.05 vs control group,#P<0.05 vs miR-128-3p mimics,&P<0.05 vs miR-128-3p inhibitor).
DISCUSSION
The difficulties in treatment of gliomas can be at least partially attributed to their high genetic heterogeneity and molecular complexity[3,22,23].Gliomas are now classified into more specific subtypes according to their biological and molecular characteristics[6,24,25].Considerable advances have been achieved in recent years in individualized and precision treatment of gliomas,including molecular pathology-guided glioma surgery and selection of chemotherapeutics[22,26,27].Molecular markers are not only intended for prognostic assessment of the patients but rather for more effective treatment and monitoring glioma progression,especially for recurrent gliomas,and screening for sufficient and effective molecular markers is essential for exploring better treatment options for high-grade gliomas.
In this study,based on data from CGGA database,we found that KLHDC8A was highly expressed in high-grade gliomas in close correlation with a poorer overall survival of the patients.We show that KLHDC8A overexpression strongly promots migration,invasion,and proliferation and inhibits apoptosis of cultured glioma cells,suggesting the important role of KLHDC8A in modulating malignant phenotype of gliomas,which was consistent with previous reports[28-30].KLHDC8A may thus serve as a potential biomarker for gliomas.We further screened the upstream regulatory genes of KLHDC8A through the gene regulatory network,and identified the miRNA-target relationship between miRNA-128-3p and KLHDC8A.
Many miRNAs have been identified to participate in tumor occurrence and progression[31,32].In this study,we demonstrate that upregulation of miRNA-128-3p suppresses the migration,proliferation and invasion and promotes apoptosis of glioma cells,indicating the possible role of miRNA-128-3p as a tumor suppressor.According to Qu et al[33],miRNA-128-3p may induce mitochondrial injury and apoptosis by targeting PDK1 in glioma cells,which show low expressions of miRNA-128-3p[34].Fu et al reported that miRNA-128-3p upregulation inhibited the metastatic potential of glioma cells by targeting GREM1[35].The expression of miRNA-128-3p was also downregulated in malignant melanoma cells,and restoration of its expression inhibited the malignant behaviors of the cells by targeting neurotrophin receptor 3 (NTF3)[15].In addition,miRNA-128-3p was also reported to enhance the sensitivity of antitumor drugs[36,37].
We further demonstrate that miRNA-128-3p modulates KLHDC8A expression by directly targeting its mRNA 3′-UTR.In the two glioma cell lines,miRNA-128-3p was found to negatively regulate KLHDC8A expression.The results of rescue experiments showed that transfection with miRNA-128-3p mimic reversed KLHDC8A overexpression-induced increase of metastatic capacities of the glioma cells,and the miRNA-128-3p inhibitor obviously attenuated the inhibitory effect of siKLHDC8A of the cells,suggesting that the interaction between miRNA-128-3p and KLHDC8A defines the malignant behavior of gliomas.
Conclusion
Our results demonstrate that KLHDC8A serves likely as an oncogene to promote progression of gliomas,and upregulation of miRNA-128-3p expression inhibits uncontrolled growth of glioma cells by downregulating KLHDC8A and its downstream effectors,suggesting that the miRNA-128-3p-KLHDC8A axis is a potential prognostic indicator as well as a therapeutic target for developing new strategies for glioma treatment.