GmNLP7a inhibits soybean nodulation by interacting with GmNIN1a
Xuesong Wu,Yuping Xiong,Jingjing Lu,Mi Yang,Hongtao Ji,Xia Li,Zhijuan Wang
National Key Laboratory of Crop Genetic Improvement,College of Plant Science and Technology,Huazhong Agricultural University,Wuhan 430070,Hubei,China
Keywords: Nodulation Nitrate GmNLP7 Soybean
ABSTRACT Nitrogen (N) is an essential macronutrient for plant growth and productivity.Leguminous plants establish symbiotic relationships with nitrogen-fixing rhizobial bacteria to use atmospheric dinitrogen gas to meet high N demand under low-N conditions.Nodule formation and N fixation are energy-consuming processes and are inhibited by nitrate present in the environment.Previous studies in model leguminous plants characterized NIN-LIKE PROTEIN (NLP) proteins that mediate nitrate control of root nodule symbiosis,but the mechanism by which nitrate regulates soybean root nodules via NLP remains unclear.In the soybean genome we found four homologs of AtNLP7,named GmNLP7a-GmNLP7d.We showed that the expression of GmNLP7s is responsive to nitrate but not to rhizobial infection and localized GmNLP7a to the nucleus.Downregulation of GmNLP7s increased nodule number,and overexpression of GmNLP7a(GmNLP7aOE) reduced nodule number regardless of nitrate availability,suggesting a negative role for GmNLP7s in nodulation.Nitrogenase activity in the GmNLP7aOE line was comparable to that of the wild type,indicating that GmNLP7a does not affect mature nodule activity.Overexpression of GmNLP7a downregulated the expression of GmNIN1a and GmENOD40-1.GmNLP7a interacted with GmNIN1a via the PB1 domain.Our results reveal a new regulator of GmNLP7 in nodulation and a molecular mechanism by which nitrate affects nodule number in soybean.
1.Introduction
As a major component of proteins,nucleotides,hormones,and chlorophyll,N is an essential macronutrient for plant growth and productivity[1-4].As immobile organisms,plants have developed diverse strategies to acquire N in a fluctuating N nutrient environment [5].In contrast to other plants,legumes obtain N in two ways:taking up mineral N from the soil and establishing a symbiotic interaction with nitrogen-fixing bacteria,rhizobia.Leguminous plants form nodules on their roots to accommodate rhizobia that biosynthesize nitrogenase to fix atmospheric N,which benefits the host plants.
The establishment of legume-rhizobia symbiosis begins when plants encounter a N deficiency.Legume roots secrete flavonoid molecules into the rhizosphere to induce the synthesis of lipochitin oligosaccharide signaling molecules,Nod factors (NFs),from compatible rhizobia[6].NFs are perceived by LysM receptors located in the plasma membrane of root epidermal cells(e.g.,NF Receptors 1 and 5 inLotus japonicus,NF Perception [NFP] inMedicagotruncatula,and NFR1/5 in soybean)[7-11]and activate a signaling cascade(the NF signaling pathway)to trigger nodule formation.InM.truncatula,the downstream events of the NF signaling pathway include the accumulation of calcium concentrations (called ‘‘calcium spiking”)in the nuclei of infected root hair cells and the activation of nuclear calcium-calmodulin kinase MtDMI3/CCaMK[12-14].CCaMK then phosphorylates the transcription factors MtIPD3 and LjCYCLOPS inM.truncatulaandL.japonicus,respectively[15,16].LjCYCLOPS transactivates theNINandERN1genes by binding to the CYC-box sequence in the promoter ofNINandERN1[16,17].MtIPD3 and LjCYCLOPS can also stimulate the GRAS family transcription factors MtNSP1/LjNSP1 and MtNSP2/LjNSP2 to activateNINexpression[18-20].NIN is an RWP-RK family protein that functions in the process of infection threads and nodule primordia[21-23].NIN positively regulates nodulation by directly targeting multiple genes:LjNPL[24],NUCLEAR FACTOR-Y[25],MtENOD11,and cytokinin receptorMtCRE1[26,27].NIN also activates autoregulation of nodulation (AON) inLotusand soybean by upregulating the expression ofCLE-RS1/2andRIC1/2,respectively [28,29].In turn,NIN is tightly regulated at the transcriptional and protein levels.The expression level ofNINin the pericycle is controlled by a remote upstream region in its promoter [30],while its DNA binding activity is negatively regulated by NNC1,the AP2 transcription factor in soybean,to attenuate nodulation [29].
Nodule formation and N fixation are energy-consuming processes and are inhibited by the presence of mineral N,such as nitrate.InArabidopsis,nitrate is sensed by NRT1.1/NFP6.3 and activates the AtNLP6-AtNLP7-centered transcriptional net,leading to the expression of genes involved in the primary nitrate response(PNR)[31-33].AtNLP7 is a nitrate sensor that controls metabolism and plant development in responsive to nitrate change [34].AtNLP6 and AtNLP7 belong to the plant-specific RWP-RK family of proteins that harbor RWP-RK and PB1 (Phox and Bem1)domains.InLotusandMedicago,genetic studies [35-37] have shown that NLPs are involved in nitrate-induced control of root nodule formation.InLotus,a forward genetic screening for genes involved in nitrate-induced control of root nodule symbiosis identifiedNRSYM1,which encodes LjNLP4[36].LjNLP4 mediates multiple aspects of root nodule symbiosis by nitrate,including nodule organogenesis and mature nodule activity.Nitrate triggers the accumulation of LjNLP4 in the nucleus,where it binds to the promoter ofCLE-RS2in a nitrate-dependent manner to induceCLERS2expression [36].InMedicago,a similar mechanism was found[35] by whichMtNLP1controls root nodule symbiosis in response to nitrate.Nitrate stimulates the translocation of MtNLP1 from the cytosol to the nucleus,where MtNLP1 interacts with MtNIN through the PB1 domain and represses the activation of MtNIN onMtCRE1,which is essential for nodule organogenesis.MtNLP1 also binds to the promoter region ofMtCLE35to induce its expression [38].
Soybean (Glycine max) is a crop of economic importance for its oil and vegetable protein.Although great progress has been made in understanding how nitrate controls root nodule symbiosis through NLP proteins in model leguminous plants,whether NLP proteins regulate nitrate-mediated nodulation in soybean was unknown.In this study,we searched the soybean genome and found four homologs of AtNLP7,named GmNLP7a-GmNLP7d.We investigated the expression patterns of the GmNLP7s,and used an RNAi knockdown and overexpression strategies to analyze their function in nodulation under high N conditions.We also used yeast two-hybrid and bimolecular fluorescence complementation (BiFC)assays to test the interaction between GmNLP7a and GmNIN1a.Our results show that GmNLP7s play a negative role in soybean nodulation via interaction with GmNINa.
2.Materials and methods
2.1.Plant growth and rhizobium inoculation
The soybean cultivar Williams 82 (Ws82) was used in our experiments.Bradyrhizobium diazoefficiensUSDA110 was used for rhizobial inoculation and nodulation.Plant growth conditions and rhizobial inoculation procedures were as described previously[39].The seeds were germinated in vermiculite and grown under 16 h/8 h light/dark conditions in a growth room at 25-26 °C and were then inoculated with a rhizobial suspension (OD600=0.08)at 7 days after germination (DAI).For the infection thread assay,plants were inoculated with β-glucuronidase (GUS)-tagged USDA110 and roots were collected and analyzed at 6 DAI.Three independent experiments were performed and more than five plants were analyzed for each experiment.
2.2.Vector construction and plant transformation
For theGmNLP7aoverexpression (GmNLP7aOE) construct,the full-length coding sequence(CDS)ofGmNLP7awas amplified from cDNA of roots and cloned into theSmaI site of the expression vector pTF101 and driven by the cauliflower mosaic virus 35S promoter.For subcellular localization,the full-length CDS ofGmNLP7awas cloned into pocA30 usingBamH I andXbaI and driven by the 35S promoter.For the RNAi construct,a 459-bp fragment of theGmNLP7aCDS was amplified from Ws82 cDNA and cloned into pTCK303 in the sense and antisense orientations usingKpnI/SpeI andBamH I/SacI.For theCas9-GmNLP7a/cconstruct,the small guide RNAs(sgRNAs)were designed using CRISPR-P 2.0 software (https://crispr.hzau.edu.cn/CRISPR2/) [40].The PCBC-DT1T2 vector was used as the template to amplify the fragments with two sgRNAs,and the amplified fragments were then connected to the skeleton vector pKSE401-GFP by the enzymeBsa1[41].The construct of35S:GmNLP7a-GFPwas transformed intoAgrobacterium tumefaciensstrain EHA101 and then was transformed into Ws82 plants using the cotyledon-node method of Flores et al.[42].
2.3.Soybean hairy root transformation and subsequent B.diazoefficiens inoculation
A.rhizogenesstrain K599 was used for hairy root transformation.The transformation was performed as described previously[39,43,44].Young seedlings at 3-4 days after germination were used for hairy root transformation.Transgenic composite plants were transferred to pots containing vermiculite and watered with nitrogen-deficient nutrient solution.The plants were grown for one week to allow rooting and then were inoculated with 30 mL of rhizobia (OD600=0.08) per plant.Nodule numbers were recorded at 28 DAI.Three independent experiments were performed and more than 20 hairy roots were examined for each experiment.
2.4.RNA extraction and quantitative PCR analysis
Total plant RNAs were extracted with TRIpure Reagent (Aidlab Biotechnologies Ltd.,Beijing,China).Genomic DNAs were removed by gDNA Wiper Mix (Vazyme Biotech Co.,Ltd.,Nanjing,Jiangsu,China) and cDNAs were produced using a FastQuant RT Kit(Vazyme Biotech Co.,Ltd.).qRT-PCR was performed using Super-Real PreMix Plus (SYBR Green;Tiangen Biotech Co.,Ltd.,Beijing,China) and gene-specific primers for the genes analyzed.The expression levels of the genes were normalized to that ofELF1b.
2.5.Subcellular localization assay
The35S:GFP-GmNLP7aconstruct was expressed in hairy roots.Transformed roots were watered with distilled water 7 days after transfer to pots.The roots were observed with or without 10 mmol L-1KNO3treatment for 30 min using a confocal microscope (SP8,Leica Microsystems,Germany).GFP fluorescence was quantified using SP8 LAS X software and more than 40 cells were quantified for the GFP strength.
2.6.Nitrogenase activity
Nitrogenase activity was measured as described by Wu et al.[45].Nodulated roots of WT andGmNLP7OEplants were introduced into glass bottles,which were then sealed with rubber stoppers.Acetylene(2 mL)was injected into each bottle after the same volume of air was pumped out,and incubated for 2 h at 28 °C.Ethylene in 100 μL of gas from each bottle was measured with a GC-4000A gas chromatograph(East&West Analytical Instruments,Beijing,China).After measurement,the nodules were removed from the root and weighed and the nitrogenase activity was normalized to nodule weight.Three independent experiments were performed and more than five plants were analyzed for each experiment.
2.7.Yeast two-hybrid assay
The CDSs ofGmNLP7aandGmNLP7a-PB1were amplified using soybean cDNAs and the PCR products were cloned into the entry vector pDONR207and were then ligated into pGADT7 (AD).AD and BD constructs were co-transformed into the yeast strainSaccharomyces cerevisiaeAH109 using the Matchmaker GAL4 Two-Hybrid System (Clontech,Mountain View,CA,USA).Yeast cells containing the AD and BD variants were selected on medium lacking Trp,Leu,and His for four days at 30 °C.
2.8.BiFC assay
The coding sequences ofGmNIN1aandGmNLP7awere cloned into pEARLYGAYE201-YFPNand pEARLYGAYE202-YFPC,respectively,using the Gateway reaction system (Invitrogen,Carlsbad,CA,USA).The constructs were transformed intoA.tumefaciensstrain GV3101 for infiltration transformation ofNicotiana benthamianaleaves.YFP fluorescence was observed with a confocal laser scanning microscope (SP8,Leica Microsystems,Germany)after infection 48 h.
2.9.Statistical analysis
The phenotypic and expression data were analyzed with Graph-Pad Prism 8 (GraphPad Software,Inc.,La Jolla,CA,USA).Statistical significance was determined using Two-sided Student’st-tests or ANOVA with Tukey’s post hoc test.
3.Results
3.1.Characterizing GmNLP7 proteins in soybean
To identify NLP members in soybean,we first searched for GmNLPs in the soybean genome and found 14 members.These GmNLPs shared the conserved DNA-binding domain RWP-PK and protein-protein interaction domain PB1,suggesting that their homologs in other legume species might exercise conserved functions.To investigate the evolutionary relationships of NLP amongArabidopsis,soybean,M.truncatulaandL.japonicus,we constructed a phylogenetic tree based on amino acid sequences (Fig.1).GmNLPs were divided into four subfamilies based on protein similarity withArabidopsisNLPs,similar to those reported by Fu et al.[46].Given the important roles of these proteins in nitrate signaling and nodulation regulation under high nitrate,we named them GmNLP7a (Glyma.09G137000),GmNLP7b (Glyma.10G234100),GmNLP7c(Glyma.16G182400)and GmNLP7d(Glyma.20G160200).GmNLP7a and GmNLP7c share 91.23% similarity,while GmNLP7b and GmNLP7d share 93.34% similarity.
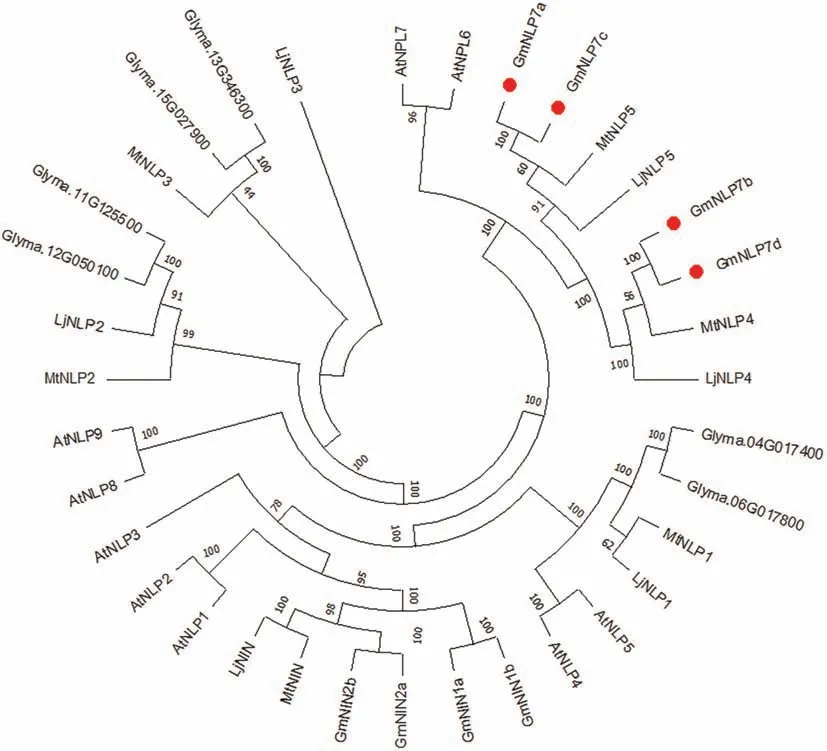
Fig.1.Phylogenetic tree of NIN-like proteins in Arabidopsis,soybean,Lotus,and Medicago.Full-length amino acid sequences were compared and the tree was constructed by the neighbor-joining method.The numbers are bootstrap values from 1000 replicates.
3.2.GmNLP7s are induced by high nitrate but not by rhizobial infection
We first tested the expression levels ofGmNLP7sin tissues in inoculated plants.The results showed that allGmNLP7swere ubiquitously expressed in all tissues tested and were expressed most highly in the leaf and at lower levels in the nodule,root,and stem(Fig.S1).To test whetherGmNLP7sare responsive to rhizobial infection,we measured the expression ofGmNLP7sin soybean roots inoculated withB.diazoefficiensUSDA110 or water as a mock control.GmNLP7sexpression remained unchanged in the early rhizobial infection stage (Fig.S2).To test whetherGmNLP7sare induced by high nitrate,we planted soybean in vermiculite watered with or without 15 mmol L-1KNO3treatment.GmNLP7sexpression was much higher in roots treated with KNO3compared with those without KNO3treatment (Fig.2A),indicating thatGmNLP7scan be induced by high nitrate.

Fig.2. GmNLP7s are responsive to nitrate.(A)Expression assay of GmNLP7s under three nitrate treatments.Values are means±SE from three replicates.GmELF1b was used as an internal control and the transcript level of GmNLP7s under 0 mmol L-1 KNO3 were set as ‘‘1”.Asterisks indicate significant differences compared to the expression of GmNLP7s under 0 mmol L-1 KNO3 treatment.**, P <0.01;***, P <0.001 (Two-sided Student’s t-test).(B) Subcellular localization of GmNLP7a-GFP. 35S:GFP-GmNLP7a was expressed in the hairy roots and the roots were treated with 0 mmol L-1 KNO3 or 10 mmol L-1 KNO3 for 1 h before observation with confocal microscope.Scale bars,25 μm.(C) Quantification assay of GFP signals.Values are means ± SD (n=55 for 0 mmol L-1 KNO3 and 42 for 10 mmol L-1 KNO3 treatments).**, P <0.01;***, P <0.001;ns,no significance (Two-sided Student’s t-test).
To determine the subcellular localization of GmNLP7s,we used GmNLP7a as an example to construct35S:GmNLP7a-GFPand transiently expressed it in hairy roots.The hairy roots were treated with water or KNO3for 1 h before observation.GFP signals were observed in the nucleus in the absence of KNO3.In the presence of KNO3,GFP signal was increased in both nucleus and cytoplasm,but the GFP ratio of nucleus to cytoplasm was not altered(Fig.2B,C).This result suggested that GmNLP7a is located in the nucleus regardless of nitrate availability.
3.3.Downregulation of GmNLP7s increased nodule number
To investigateGmNLP7sfunction in regulating nodulation,we first downregulatedGmNLP7sand evaluated the nodule phenotype in hairy roots by RNA interference(RNAi).We constructed an RNAi vector designed to repress allGmNLP7sfrom a 459-bp sequence ofGmNLP7a,which shares 80% identity withGmNLP7s.We transformed the vector into soybean hairy roots to observe the nodulation phenotype of the composite transgenic plants,using the empty vector as a control.We treated the composite transgenic plants with 0 or 5 mmol L-1KNO3.qPCR assays showed that the average expression levels of the fourGmNLP7sgenes inGmNLP7sknockdown lines were significantly reduced compared with the vector control(Fig.3A,B).The average number of nodules in hairy roots expressing the empty control treated with 5 mmol L-1KNO3was significantly reduced compared with that of the 0 mmol L-1KNO3treatment group,suggesting an inhibitory effect on nitrate nodulation (Fig.3C,D).The average nodule number of theRNAi-GmNLP7slines was significantly increased compared with the empty vector control under both 0 and 5 mmol L-1KNO3conditions.These results indicate thatGmNLP7sfunction as negative regulators of nodulation with or without nitrate treatment.
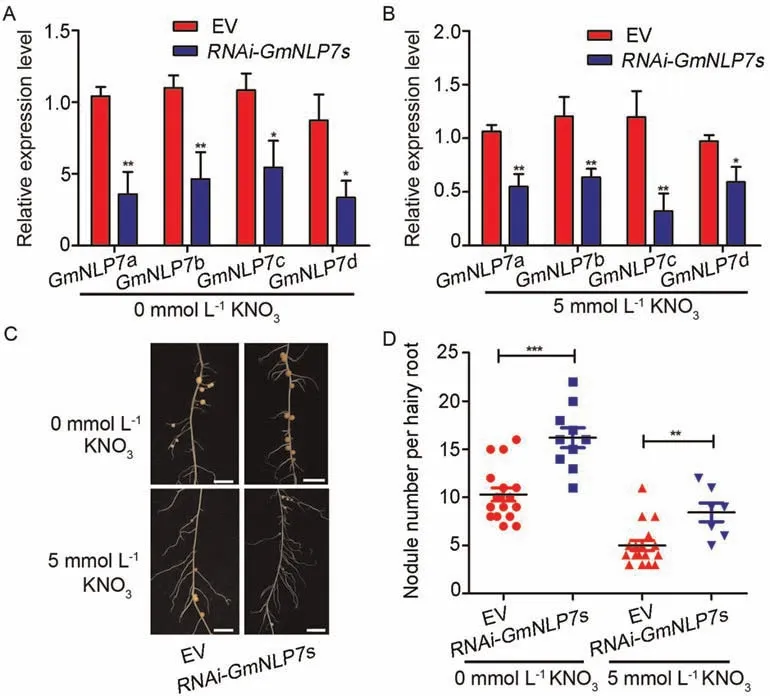
Fig.3.Downregulation of GmNLP7s increased nodulation under high-and low-nitrate conditions.(A) Expression assay of GmNLP7s in hairy roots expressing empty vector(EV)and RNAi-GmNLP7s under 0 mmol L-1 KNO3 condition.Values are means±SE from three replicates.The transcript amounts in each sample were normalized to those of GmELF1b.Asterisks indicate significant differences compared to the expression of GmNLP7s in hairy roots expressing empty vector.*,P <0.05;**,P <0.01(Student’s t-test).(B)Expression assay of GmNLP7s in hairy roots expressing EV and RNAi-GmNLP7s under 5 mmol L-1 KNO3 condition.Values are means±SE from three replicates.The transcript amounts in each sample were normalized to those of GmELF1b.Asterisks indicate significant differences compared to the empty vector.*, P <0.05;**, P <0.01(Student’s ttest).(C)The nodule phenotype of hairy roots expressing EV and RNAi-GmNLP7s under 0 and 5 mmol L-1 KNO3 treatments.Scale bars,2 cm.(D)The nodule number of hairy roots expressing EV and RNAi-GmNLP7s under 0 and 5 mmol L-1 KNO3 treatments.Values are means±SD(n ≥7).Asterisks indicate significant differences compared to empty vector.**, P <0.01;***, P <0.001 (Student’s t-test).Three independent experiments were performed with similar results.
GmNLP7aandGmNLP7care induced by nitrate more strongly thanGmNLP7bandGmNLP7d,and we speculated that these two genes negatively regulate nodulation under nitrate conditions.To test this speculation,we simultaneously knocked outGmNLP7aandGmNLP7cin the hairy root and estimated the nodule number.GmNLP7aandGmNLP7cknockout increased nodule number with or without nitrate treatment (Fig.S3),suggesting thatGmNLP7aandGmNLP7care regulators that repress nodulation.
3.4.Overexpression of GmNLP7a reduced nodule number but not nitrogenase activity
To confirm GmNLP7s function in nodulation,we used GmNLP7a as an example to generate transgenic composite plants overexpressingGmNLP7aunder the control of the 35S promoter and estimated the nodule number.The composite transgenic plants were treated with 0 or 5 mmol L-1KNO3.qPCR assays showed that the average expression levels ofGmNLP7ainGmNLP7aoverexpressing lines were significantly increased compared with those of the vector control (Fig.4A,B).The average nodule number of hairy roots overexpressingGmNLP7alines was significantly reduced compared with the empty vector control under both 0 and 5 mmol L-1KNO3conditions (Fig.4C,D),indicating that GmNLP7a inhibits nodulation with or without nitrate treatment.

Fig.4.Overexpression of GmNLP7s inhibits nodulation under high-and low-nitrate conditions.(A) Expression assay of GmNLP7a in hairy roots expressing EV and 35S:GmNLP7s under 0 mmol L-1 KNO3 condition.GmELF1b was used as an internal control.Values are means ± SE from three replicates.The transcript amounts in each sample were normalized to those of GmELF1b.(B)The expression assay of GmNLP7a in hairy roots expressing EV and 35S:GmNLP7s under 5 mmol L-1 KNO3 condition.GmELF1b was used as an internal control.Values are means ± SE from three replicates.The transcript amounts in each sample were normalized to those of GmELF1b. (C) The nodule phenotype of hairy roots expressing EV and 35S:GmNLP7s under 0 and 5 mmol L-1 KNO3 treatments.Scale bars,2 cm.(D)The nodule number of hairy roots expressing EV and 35S:GmNLP7s under 0 and 5 mmol L-1 KNO3 treatments.Values are means±SD(n ≥7).Asterisks represent statistically significant differences compared to the empty control.*, P <0.05;**, P <0.01 (Student’s t-test).Three independent experiments were performed with similar results.
To confirm the effect ofGmNLP7aoverexpression on nodulation,we generated a stable transgenic plant overexpressingGmNLP7aunder the 35S promoter in the Ws82 background (GmNLP7aOE)(Fig.S4).The plants were treated with 0,5,or 15 mmol L-1KNO3,and the nodule and root phenotypes were evaluated.The primary length and fresh weight were comparable between Ws82 andGmNSP7aOEunder three nitrate treatments (Fig.S5).The nodule number decreased with increasing KNO3in both the Ws82 andGmNLP7aOElines (Fig.5A,B),but the nodule number of theGmNLP7aOElines was significantly reduced compared with that of Ws82 under all KNO3conditions(Figs.5A,B,6B,S6A),confirming thatGmNLP7ais a negative regulator of nodulation regardless of nitrate concentrations.
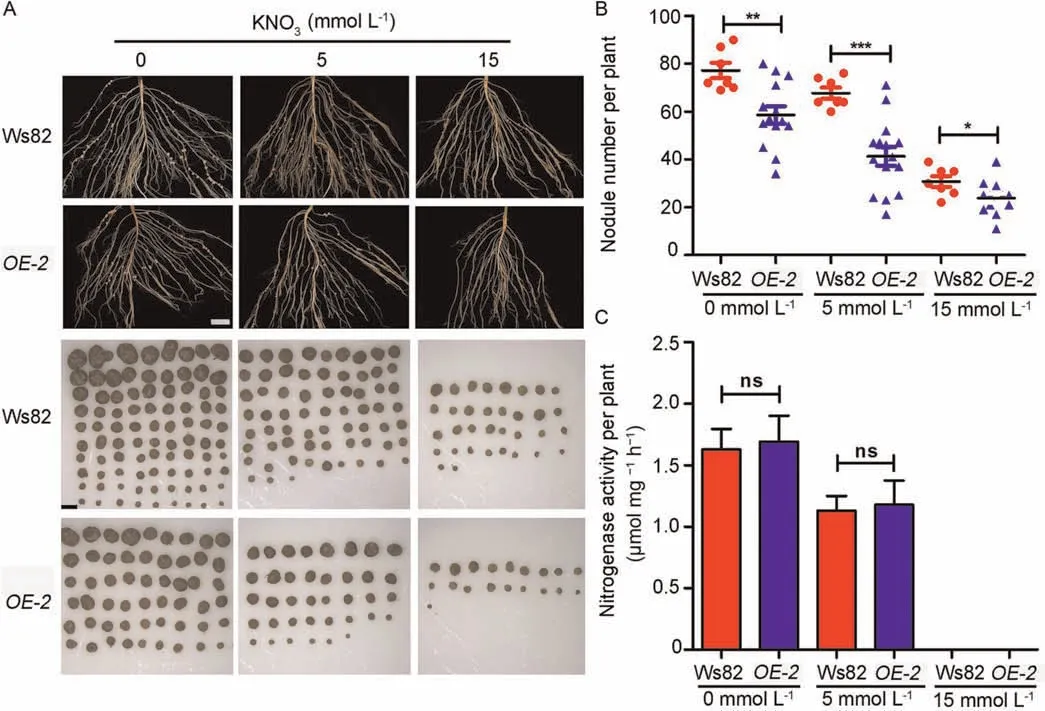
Fig.5.Overexpression of GmNLP7s inhibits nodulation but not nitrogenase activity.(A)The nodule phenotype of Ws82 and GmNLP7aOE-2 line under three nitrate treatments.Bar,3 cm for root and bar,2 mm for nodule.The seeds were germinated under three KNO3 concentrations and seven-day-old seedlings were inoculated with B.diaefficiens strain USDA110;the nodule phenotype was counted at 28 DAI.(B) Nodule numbers of Ws82 and GmNLP7aOE-2 line under three KNO3 treatments at 28 DAI.Values are means ± SD (n ≥7).Asterisks represent statistically significant differences from the wild type.*, P <0.05;**, P <0.01;***, P <0.001 (Two-sided Student’s t-test).Three independent experiments were performed with similar results.(C)The nitrogenase activity of Ws82 and GmNLP7aOE-2 line under three KNO3 treatments at 28 DAI.Values are means ± SD (n ≥7).Three independent experiments were performed with similar results.
To further investigate whetherGmNLP7ais involved in the regulation of nitrogen fixation,we inoculated Ws82 andGmNLP7aOEplants withB.diazoefficiensUSDA110.The plants were grown under three nitrate treatments for 28 days and their nitrogenase activity was measured by acetylene reduction.The nitrogenase activity of Ws82 was significantly reduced after nitrate treatment,but there was no significant difference between Ws82 andGmNLP7aOEplants (Figs.5C,S6C).Thus,GmNLP7a was required for nitrate inhibition of nodule number but not for nitrogenase activity.
3.5.GmNLP7a inhibits early infection events and NF signaling
Having found that altering the expression ofGmNLP7adramatically affected the nodule number in soybean,we questioned whether GmNLP7a affects early infection and nodule organogenesis.To test this proposition,we first evaluated the infection threads 6 days after inoculation withB.diazoefficiensUSDA110-GUS in theGmNLP7aOEline with three concentrations of nitrate treatments.Nitrate significantly reduced the number of infection threads,and overexpression ofGmNLP7astrongly inhibited infection regardless of nitrate concentrations (Fig.6A,B),suggesting that GmNLP7a inhibits rhizobial infection.

Fig.6.GmNLP7a inhibits early infection events and NF signaling.(A)Infection threads in Ws82 and GmNLP7aOE plants.Plants were planted with 0,5 or 15 mmol L-1 KNO3 treatment and inoculated with B.diazoefficiens USDA110-GUS at 5 days after germination,and the infection threads were analyzed at 6 DAI.The red arrows indicate infection threads.(B) Density of infection threads in different nitrate conditions in (A) (n=6);Significant differences were determined by two-sided Student’s t-test,**, P <0.01;***,P <0.001.Two independent experiments were performed with similar results.(C) The average number of nodule primordia per plant of Ws82 and GmNLP7aOE plants.The plants were planted in 0,5 and 15 mmol L-1 KNO3 and inoculated with B.diazoefficiens USDA110 at 5 days after germination;the nodule primordia were counted at 8 DAI.Values are means ± SD (n=6);Significant differences were determined by two-sided Student’s t-test,***, P <0.001;****, P <0.0001.Three independent experiment were performed with similar results.(D)qRT-PCR analysis of ENOD40-1 and GmNIN1a in roots of Ws82 and GmNLP7aOE plants without(-R)or with(+R)USDA110 treatment at 2 DAI.The transcript amounts in each sample were normalized to those of ELF1b.The expression levels shown are means ± SE from three biological replicates.
We then tested whether GmNLP7a affects nodule organogenesis.We compared the numbers of nodule primordia between Ws82 andGmNLP7aOEplants with three nitrate treatments.The result was similar to that for infection threads.Nitrate reduced nodule primordia number in Ws82,and overexpression ofGmNLP7areduced nodule primordia number regardless of nitrate(Fig.6B),suggesting that GmNLP7a inhibits nodule organogenesis.
Rhizobial infection activates NF signaling,and we next questioned whetherGmNLP7aaffects the NF signaling pathway.To address this question,we examined the expression pattern of marker genes of NF signaling,includingGmNIN1aandENOD40-1,in roots at 3 DAI.The expression ofENOD40-1andGmNIN1awas induced by rhizobial infection in the wild type,whereas their induction was inhibited inGmNLP7aOEroots (Fig.6C),suggesting that GmNLP7a downregulatesGmNIN1aandENOD40-1to regulate nodulation.
3.6.GmNLP7a interacts with GmNIN1a in the PB1 domain
NLP interacts with NIN inM.truncatulato regulate nitrate control of root nodule formation [34].Having found that GmNLP7a inhibits nodulation,we wondered whether GmNLP7a interacts with GmNIN1a,the ortholog of LjNIN in soybean [29].To answer this question,we first tested the interaction between GmNLP7a and GmNIN1a in a yeast two-hybrid (Y2H) system.We clonedGmNLP7ainto the pGADT7 vector andGmNIN1ainto the pGBKT7 vector.The AD and BD variants were co-transformed into yeast strain AH109 and selected on medium lacking Leu,Trp and His.Only yeast cells transformed with GmNLP7a-AD and GmNIN1a-BD grew on the selective medium,indicating that GmNLP7a and GmNIN1a interact in the Y2H assay (Fig.7A).The interaction between GmNLP7a and GmNIN1a was confirmed in a BiFC assay by coexpressingGmNIN1a-YFPNandGmNLP7a-YFPCin pavement cells ofN.benthamiana.As shown in Fig.7B,GmNLP7a interacted with GmNIN1a in the nuclei of transformed cells,in agreement with their subcellular localization.These results suggest that GmNLP7a may form a complex with GmNIN1a to mediate nodulation.
GmNIN1a and GmNLP7a contain the conserved DNA-binding domain RWP-RK and the protein--protein interacting domain PB1.To test whether PB1 is responsible for their interaction,we divided GmNIN1a into an N-terminal end containing the RWP-RK domain and a C-terminal end containing the PB1 domain and tested their interaction with GmNLP7a in Y2H assay.Only the Cterminal end interacted with GmNLP7a (Fig.7A),indicating that the PB1 domain of GmNIN1a is involved in its interaction with GmNLP7a.We tested the interaction of PB1 of GmNLP7a with GmNIN1a.As shown in Fig.7A,GmNLP7a-PB1 interacted with the full-length and C-terminal end of GmNIN1a.Thus,the PB1 domain of GmNLP7a and GmNIN1a mediates the interaction between GmNLP7a and GmNIN1a.
4.Discussion
Nodulation is an energy-expensive process and is inhibited strongly in the presence of nitrate,leading to decreased nodule number,reduced mature nodule activity,and accelerated nodule senescence [47].In the present study,we found that GmNLP7s function in nitrate-mediated control of nodulation in soybean.
AtNLP6 and AtNLP7 are central transcription factors that govern nitrate-inducible gene expression [48].Nitrate triggers AtNLP6/7 accumulation in the nucleus by phosphorylation modification,where it activates genes involved in PNR via its conserved RWPRK domain in the presence of nitrate [45,49].Owing to multiple genome duplications,there are four GmNLP7s in soybean that contain the conserved DNA-binding domain RWP-RK and the proteinprotein interaction domain PB1.GmNLP7s share a high level of amino acid sequence identity with AtNLP7,suggesting that they play similar roles in regulating genes involved in nitrate uptake and assimilation.In distinction from AtNLP7,GmNLP7a is located in both the nucleus and cytoplasm in the absence of nitrate,like LjNLP1 [37].The finding thatGmNLP7sexpression could also be induced in the presence of nitrate suggests thatGmNLP7sfunction under high nitrate conditions.
Previous study[47]has highlighted a role for nitrate in the inhibition of nodulation.Genetic and molecular biology data indicate that NLP proteins function in nitrate-induced control of nodulation in leguminous plants [35-37].However,there are no such reports on the involvement of NLP proteins in nodulation in soybean.We showed thatGmNLP7swere expressed in soybean roots and nodules.Taking into account thatGmNLP7sexpression is induced by nitrate,we speculate that GmNLP7s function in N control of nodulation.Indeed,GmNLP7sdownregulation increased nodule number andGmNLP7aoverexpression reduced nodule number under nitrate and nitrate-free conditions.We found that GmNLP7a is located in the nucleus even under nitrate-free conditions,a location that may contribute to its role in decreasing nodule number under nitrate-free conditions.The finding that GmNLP7a does not affect the nitrogenase activity of nodules indicates that GmNLP7a regulates specifically nodule organogenesis and not mature nodule activity.Given that there are many more GmNLPs in the soybean genome,we speculate that other GmNLP members are responsible for mature nodule activity under high-nitrogen conditions.
NIN was first identified inL.japonicusand functions in the coordination of rhizobial infection,nodule organogenesis,and mature nodule activity [21].NIN has a conserved RWP-RK domain and can bind to NIN binding sites (NBSs)in the promoter of its targets to induce genes required for rhizobial infection and nodule organogenesis [28].Chromatin immunoprecipitation-PCR analysis showed [50] that LjNIN targeted NREs in the promoters of nitrate-inducible genes,includingLjNIR1andLjNRT2.1,suggesting that NLP and NIN may have the same targets.Recently it was reported [34] that MtNLP1 inMedicagointeracts with MtNIN to inhibit MtNIN activation of genes required for nodulation by the PB1 domain.GmNLP7s contain a conserved PB1 domain and we speculate that GmNLP7s interact with NIN.We found that GmNLP7a interacts with GmNIN1a,an ortholog of NIN in soybean[29,48].The interaction between GmNLP7a and GmNIN1a is mediated by a PB1 domain.Overexpression ofGmNLP7areduced the expression ofGmENOD40-1andGmNIN1a.The mechanism of depression ofGmENOD40-1andGmNIN1aby GmNLP7a is still unclear.One possibility is that GmNLP7a binds to the promoters ofGmENOD40-1andGmNIN1aand depresses their expression.Another possibility is that the GmNLP7a interaction with GmNIN1a suppresses its ability to activateGmENOD40-1.We propose the following model for GmNLP7s inhibition of nodulation.Under low nitrate condition,GmNLP7aexpression is low;thus,GmNIN1aexpression can be induced by NF to initiate rhizobial infection and nodule formation.In the presence of nitrate,GmNLP7genes are induced in response to nitrate signals.GmNLP7s inhibitGmNIN1aexpression or form heterodimers with GmNIN1a to suppress its activation of nodulation gene expression,leading to inhibition of nodulation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2018YFD10009000).
CRediT authorship contribution statement
Xuesong Wu:Data curation,Formal analysis.Yuping Xiong:Data curation,Formal analysis.Jingjing Lu:Data curation,Formal analysis.Mi Yang:Data curation,Formal analysis.Hongtao Ji:Formal analysis,Funding acquisition.Xia Li:Conceptualization.Zhijuan Wang:Conceptualization,Writing -original draft,Writing -review &editing.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.03.016.
- The Crop Journal的其它文章
- Reversible protein phosphorylation,a central signaling hub to regulate carbohydrate metabolic networks
- Genetic and environmental control of rice tillering
- High-throughput phenotyping of plant leaf morphological,physiological,and biochemical traits on multiple scales using optical sensing
- The R2R3-MYB transcription factor GaPC controls petal coloration in cotton
- The photosensory function of Zmphot1 differs from that of Atphot1 due to the C-terminus of Zmphot1 during phototropic response
- Disruption of LEAF LESION MIMIC 4 affects ABA synthesis and ROS accumulation in rice

