Introgression of sharp eyespot resistance from Dasypyrum villosum chromosome 2VL into bread wheat
Ciyun Liu,Wei Guo,Yng Wng,Bisheng Fu,Jroslv Doležel,Ying Liu,Wenling Zhi,Mhmoud Sid,d,István Molnár,d,e,Kteřin Holušová,Ruiqi Zhng,,Jizhong Wu,f,g,
a Institute of Germplasm Resources and Biotechnology/Jiangsu Provincial Key Laboratory of Agrobiology,Jiangsu Academy of Agricultural Sciences,Nanjing 210014,Jiangsu,China
b College of Agronomy/JCIC-MCP/National Key Lab of Crop Genetics and Germplasm Enhancement,Nanjing Agricultural University,Nanjing 210095,Jiangsu,China
c Institute of Experimental Botany,Centre of the Haná Region for Biotechnological and Agricultural Research,Šlechtitelu˚31,CZ-783671 Olomouc,the Czech Republic
d Field Crops Research Institute,Agricultural Research Centre,9 Gamma Street,Giza,12619 Cairo,Egypt
e Agricultural Institute,Centre for Agricultural Research,ELKH,Martonvásár 2462,Hungary
f Jiangsu Co-Innovation Center for Modern Production Technology of Grain Crops,Yangzhou University,Yangzhou 225009,Jiangsu,China
g Zhongshan Biological Breeding Laboratory,Nanjing 210014,Jiangsu,China
Keywords:Triticum aestivum Rhizoctonia cerealis Dasypyrum villosum Flow sorting KASP
ABSTRACT Wheat sharp eyespot,a stem disease caused by the soilborne fungus Rhizoctonia cerealis van der Hoeven,has become a threat to wheat production worldwide.Exploiting resistance resources from wild relatives of wheat is a promising strategy for controlling this disease.In this study,a new wheat-Dasypyrum villosum T2DS·2V#4L translocation line in the background of Chinese Spring (CS) showed stable resistance to R.cerealis.Introgression of the T2DS·2V#4L chromosome into wheat cultivar Aikang 58 by backcrossing produced a marked increase in sharp eyespot resistance in NIL-T2DS·2V#4L in comparison with NILT2DS·2DL,and no detrimental effects of 2V#4L on agronomic traits were observed in the BC2F2,BC2F2:3,and BC2F2:4 generations.Flow-sorted sequencing of 2V#4L yielded 384.3 Mb of assembled sequence,and 8836 genes were predicted of which 6154 had orthologs in at least one of the 2AL,2BL,and 2DL arms of CS,whereas 1549 genes were unique to 2V#4L.About 100,000 SNPs were detected in genes of 2V#4L and 2DL in 10 sequenced bread wheat cultivars.A Kompetitive Allele Specific Polymerase chain reaction and 30 conserved ortholog sequence markers were developed to trace the 2V#4L chromatin in wheat backgrounds.T2DS·2V#4L compensating translocation lines represent novel germplasm with sharp eyespot resistance and the markers will allow rapid detection in breeding programs.
1.Introduction
Sharp eyespot caused byRhizoctonia cerealisvan der Hoeven is a global bread wheat(Triticum aestivumL.)disease causing 10%-42%yield loss [1,2].It occurs on 6.7-9.3 Mha annually in China [3].Chemical control is not effective because the pathogen is a soilborne fungus and the disease usually appears in the basal stem and sheath of the wheat plant.Resistance breeding against sharp eyespot is challenged by a lack of sources of genetic resistance,although moderate to high resistance has been reported in some wheat cultivars,breeding lines,and landraces,such as ARz,AQ 24788-83,CI 12633,Luke,Niavt 14,and Shanhongmai [4,5].To date,more than 30 quantitative trait loci(QTL)have been reported to be located throughout the wheat genome,but a single locus explained less than 30% of phenotypic variation [5].Because no stable or major QTL for sharp eyespot resistance in wheat has become available for marker-assisted selection in breeding,it is desirable to identify new resistance genes or QTL.
The wild relativeDasypyrum villosum(L.) P.Candargy(2n=2x=14,genome VV) is recognized as a rich source of genes for resistance to fungal diseases of wheat such as powdery mildew,eyespot,stem rust,and leaf rust [6,7].At least fiveD.villosumaccessions(with genomes designated V#1 to V#5)have been used to develop wheat-D.villosumintrogression lines [8].Genes for resistance to diseases such as powdery mildew (Pm21[9,10],Pm55[11],Pm62[12],andPm67[13]),stem rust(Sr52[14]),yellow mosaic virus (Wss1[15]),stripe rust (Yr5V[8]),and cereal cyst nematode(CreV[16])have been transferred to wheat as Robertsonian translocations.T6VS·6AL carryingPm21has been widely used in Chinese wheat breeding [17,18].However,responses of wheat-D.villosumintrogression lines to sharp eyespot have not been evaluated.
Although introgression of novel genes from wild species into wheat can contribute to wheat breeding,linkage drag of undesirable genes and/or cytological instability often limits their use in cultivars [19,20].DNA markers have been widely used to identify homoeologous relationships between alien and wheat chromosomes.This process has been greatly expedited with the recent development of high-throughput DNA genotyping arrays and next-generation sequencing.Chromosome arms 4VS [21],5VS[8],and 6VS [22] have been flow-sorted and sequenced,and their colinearity with homoeologous wheat chromosomes has been characterized using the assembled sequences of alien chromosome regions and corresponding reference sequences of bread wheat.
A set ofT.aestivum-D.villosum91C43 disomic substitution and translocation lines involving chromosomes 1V#4 -7V#4 was developed in Chinese Spring (CS) [23].In disease nurseries,a durum-D.villosum#4 amphiploid (AABBV#4V#4) and a disomic substitution line DS2V#4(2D) showed resistance to sharp eyespot,whereas the CS-D.villosumT2V#4S·2DL·translocation line was susceptible,suggesting the presence of resistance on chromosome arm 2V#4L.The objectives of this study were to (1) develop and cytogenetically confirm a T2DS·2V#4L translocation,(2) confirm sharp eyespot resistance in the T2DS·2V#4L translocation line and evaluate the agronomic performance of a partial backcross derivative of a wheat cultivar carrying the translocation,and (3)characterize the genetic compensability and diversity of 2V#4L by flow-sorted sequencing and comparative genomics.
2.Materials and methods
2.1.T.aestivum-D.villosum materials and molecular cytogenetic analysis
TheT.aestivum-D.villosumdisomic substitution line DS2V#4(2D) (NAU 1808),and theT.aestivum-D.villosumdisomic translocation lines T2V#4S·2DL (NAU2V-3) and T2V#4L·W (NAU2V-8)were first identified with DNA markers and molecular cytogenetic analysis.The respective markersCINAU184andCINAU1025were used to identify the 2V#4S and 2V#4L chromosome arms [12,24].Genomicin situhybridization (GISH) and fluorescencein situhybridization (FISH) were applied to root tip mitotic metaphase squashes using respectivelyD.villosumtotal genomic DNA labeled with fluorescein-12-dUTP (green) and arm-specific Oligo-pAs1 DNA sequences labeled with 6-carboxytetramethylrhodamine(TAM)as probes [13].Hybridization signals of the probes on chromosomes were visualized with an Olympus BX60 fluorescence microscope (Olympus,Tokyo,Japan) and photographed with a SPOT Cooled Color Digital Camera (Olympus).All cytogenetic materials are maintained at the Cytogenetic Institute,Nanjing Agricultural University (CINAU,Nanjing,Jiangsu,China).
2.2.Field experiments
Sharp eyespot inoculation experiments for CS,theT.aestivum-D.villosumdisomic substitution line DS2V#4(2D)(NAU 1808),and theT.aestivum-D.villosumdisomic translocation lines T2DS·2V#4L(NAU2V-8) and T2V#4S·2DL (NAU2V-3) were conducted in both greenhouse and field at Nanjing Agricultural University experimental station in Nanjing,China during the 2012-2017 growing seasons.Inoculation experiments for segregating populations derived from the cross Aikang 58 × NAU2V-8 were conducted in the greenhouse at the Jiangsu Academy of Agricultural Sciences experimental station in Luhe,Jiangsu,China during 2019-2020,2020-2021,and 2021-2022,and in the field in Nanjing,China during 2019-2020 and 2020-2021.For the greenhouse experiment,each line was planted in a single 1-m row containing 50 seeds sown at 2-cm intervals.For field experiments,each line was planted in a 1.5-m row containing 75 seeds sown at 2-cm intervals.The wheat cultivars Niavt 14,Ningmai 9,and Yangmai 15 were used as resistant,moderately resistant,and susceptible controls,respectively.
For agronomic trait assessment,a BC2F2:3population derived from the cross Aikang 58 × NAU2V-8 was grown in the field in both Nanjing and Luhe,China during 2020-2021.A BC2F2:4population from Aikang 58× NAU2V-8 was grown in the field in Luhe,China during 2021-2022.A BC5F2population derived from the cross Nannong 0686×NAU2V-8 was grown in the field in Nanjing,China during 2020-2021.Wheat lines were arranged in a randomized complete block design with three replications.Each line was planted in one 2-m row containing 100 seeds sown at 2-cm intervals.
2.3.Evaluation of sharp eyespot resistance
Rhizoctonia cerealisanastomosis group GAG-1 is the dominant strain in Jiangsu province in China,and R0301 is a highly virulent isolate of CAG-1.A wheat kernel inoculation method [4] was used for inoculations.R0301 stock was activated on fresh potato dextrose agar (PDA) plates at 25 °C.Wheat kernels were soaked in sterile water for 24 h and autoclaved at 120 °C for 1 h in culture bags.Small pieces of PDA with R0301 mycelia were added to the culture bags and cultured in an incubator at 25 °C.At Zadoks growth stage (GS) 25 [25],R0301-colonized wheat kernels were inoculated in wheat rows with shallow soil covering.Soil moisture was controlled by sprinkling water before disease scoring at GS75.Fifty stems were scored for each line,and disease severity was scored on the 0-5 scale system[26].Disease index(DI)was calculated by the following equation [5]:
whereiis the infection type andnis the number of stems showing each infection type.
Wheat reactions toR.cerealiswere categorized into resistant(DI ≤ DIresistantcontrol),moderately resistant (DIresistantcontrol<DI ≤ DImoderatelyresistantcontrol),moderately susceptible(DImoderatelyresistantcontrol<DI ≤DIsusceptiblecontrol),and susceptible(DI >DIsusceptiblecontrol).
2.4.Assessment of agronomic traits
For wheat lines,time of heading was recorded when 50%of the plants per plot had reached spike emergence (GS55),and days to heading from seedling emergence stage (GS10) was calculated.Plant height,spike number,spike length,spikelet number per spike,and grain number per spike were recorded at maturity(GS92).Thousand-grain weight was measured after harvest.
2.5.Flow cytometry,DNA sequencing,gene annotation,and SNP calling
Synchronized meristem root tips of the T2DS·2V#4L line were used to prepare suspensions of intact mitotic chromosomes following Vrána et al.[27] and Kubaláková et al.[28].The chromosomes in suspension were fluorescently labelled by fluorescencein situhybridization in suspension (FISHIS) using oligonucleotides 5′-FITC-GAA7-FITC-3′(Sigma-Aldrich,St.Louis,MO,USA) and counterstained with DAPI (4´,6-diamidino 2-phenylindole) following Giorgi et al.[29].Flow cytometric analysis and sorting were performed on a FACSAria II SORP flow cytometer and sorter (Becton Dickinson Immunocytometry Systems,San José,CA,USA).Chromosome samples were separated at 1500-2000 particles per second and T2DS·2V#4L chromosomes were sorted in 15-20/second into a batch of 114,000 chromosomes.The purity of the sorted fractions was determined by FISH with probes for GAA microsatellite and the pSc119.2 DNA repeat on chromosomes sorted onto a microscope slide.
DNA of the sorted T2DS·2V#4L chromosomes was purified following Šimková et al.[30].A total of 13.32 ng DNA was fragmented in 20 μL using a Bioruptor Plus (Diagenode,Denville,NJ,USA) five times for 30 s at the high setting.Sheared DNA was used to prepare a sequencing library using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs,Ipswich,MA,USA) with mean fragment size 900 bp.The library was sequenced on an Illumina NovaSeq 6000 (Illumina Inc.,San Diego,CA,USA) to produce 2× 150-nt paired-end reads to reach 100% coverage.De novoassembly of the Illumina paired-end reads was performed with Meraculous [31].The T2DS·2V#4L scaffolds were used in a BLAST search against the IWGSC RefSeq v1.0 database [32] to remove 2DS sequences and generate the 2V#4L scaffolds.
Protein-coding genes were annotated by bothab initioprediction method with Augustus-3.3.3[33]and homology-based prediction method with GeMoMa-1.6.4 [34].Protein sequences of annotated genes from the long arms of homoeologous group 2 in the 2V#4L assembly (2V#4L),T.aestivum(2AL,2BL,and 2DL)[32],Hordeum vulgare(2HL) [35],Secale cereale(2RL) [36],andThinopyrum elongatum(2EL) [37] were used for gene orthology analysis.The longest protein for each annotated gene was extracted and merged,and orthologous clusters across all merged protein sequences were detected with OrthoFinder v2.3.5 [38]using default parameters.Single-copy genes among 2V#4L,2AL,2BL,2DL,2HL,2RL,and 2EL were extracted,aligned with MAFFT v7.427 [39] in each gene cluster,and linked together by species.A phylogenetic tree was constructed with RAxML v8.2.12 [40]using the PROTGAMMAJTT model with 1000 bootstrap replications.
Annotated gene sequences from 2V#4L were aligned to the 2DL reference genomes of the bread wheat cultivars CS,Arina LrFor,CDC Landmark,CDC Stanley,Jagger,Julius,LongReach Lancer,Mace,Norin 61,and SY Mattis [41] with bwa mem v0.7.17-r1188 [42].SNP calling in 2V#4L and 2DL was performed with bcftools (1.9-170-gd7bb95b) [43].Genes in windows of 10 Mb on CS 2DL were also aligned to both the 2V#4L assembly and other 2DL arms of the above cultivars to call SNP variants.
2.6.Development of KASP and PCR markers
The SNP variation between a pair of orthologous genes of 2V#4L and 2DL was converted to a Kompetitive Allele Specific Polymerase chain reaction (KASP) [44] marker.KASP primers were designed with Primer3 (https://primer3.wi.mit.edu) and synthesized by Tsingke Biological Technology Company (Nanjing,Jiangsu,China).Reactions were performed in a Hydrocycler-16 Water Bath Thermocycler(LGC Genomics,Hoddesdon,Herts,UK),and fluorescence was detected with a PHERAstar high-end microplate reader (LGC Genomics).
Conserved ortholog sequence(COS)markers were developed by comparing the sequences of genes on 2V#4L with their conserved orthologs on 2DL of CS.Primers were designed with DNAMAN(Lynnon Biosoft,Quebec,Canada) and synthesized by the Tsingke Biological Technology Company.PCR reactions were performed on a thermocycler (Eppendorf,Hamburg,Germany),and products were separated in 8%polyacrylamide gels(40%acrylamide bisacrylamide) [45].
2.7.Statistical analysis
Statistical analyses were performed with SAS 9.4 (SAS Institute Inc.,Cary,NC,USA).Multiple comparisons for DI among CS,Niavt 14,Yangmai 15,DS2V#4(2D),T2DS·2V#4L,and T2V#4S·2DL were performed using Duncan’s test with PROC ANOVA in SAS 9.4.Comparison for DI and agronomic traits between wheat lines with and without homozygous T2DS·2V#4L chromosomes was conducted using Student’sttest with PROC TTEST in SAS 9.4.
3.Results
3.1.Identification of the T2DS·2V#4L translocation
The translocation line NAU2V-8 was previously [24] identified as 2VL·W.For identification of the translocated wheat chromosome arm in NAU2V-8,mitotic analysis following pAs1-Oligo-FISH showed that the translocated chromosome was T2DS·2V#4L(Fig.1A).NAU2V-8 showed pairing of 21 bivalents at meiotic metaphase I in pollen mother cells,among which one ring bivalent was formed by two T2DS·2V#4L translocation chromosomes (Fig.S1).PCR markersCINAU1025andCINAU184provided further confirmation:the 2V#4L-specific fragment amplified byCINAU1025primers was present in NAU2V-8,whereas the 2DL-specific fragment was absent;the 2 V#4S-specific fragment amplified by theCINAU184primers was absent in NAU2V-8,whereas the 2DS-specific fragment was present (Fig.1B).
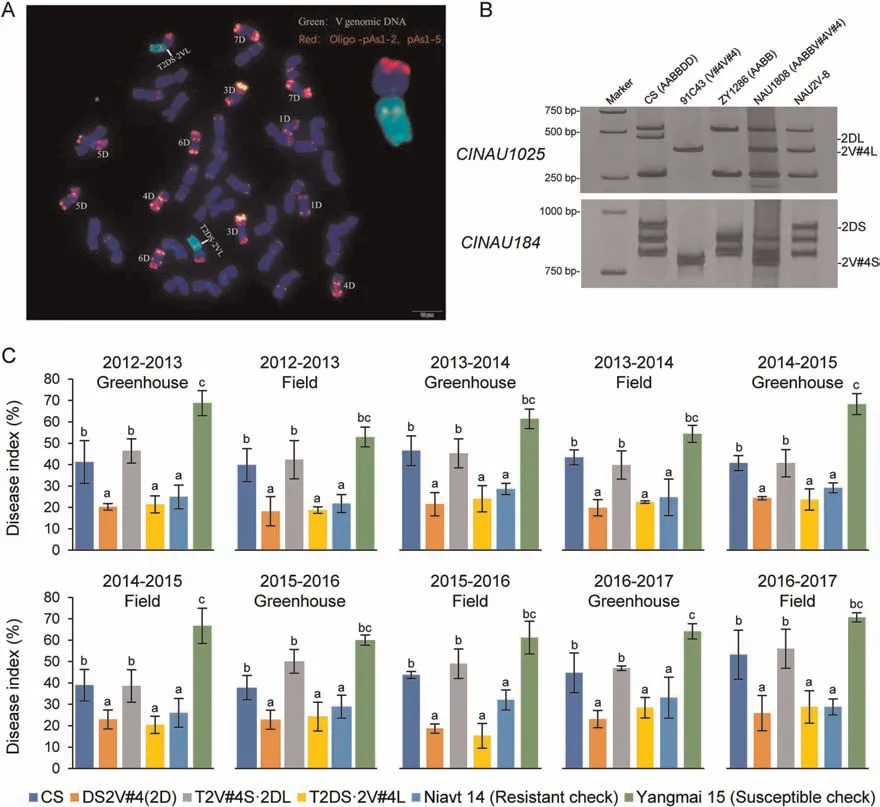
Fig.1.Molecular cytogenetic analysis and sharp eyespot response of T2DS·2V#4L translocation line NAU2V-8.(A)GISH and Oligo-pAs1-FISH patterns indicating that NAU2V-8 has a pair of T2DS·2V#4L translocation chromosomes.D.villosum genomic DNA was labeled with fluorescein-12-dUTP(green),Oligo-pAs1 was labeled with TAM(red),and wheat chromosomes were counterstained with DAPI (blue).(B) Electrophoretic patterns of 2V#4-specific markers.2V#4L amplified by CINAU1025 primers was present,whereas the 2DL-specfic fragment was absent;the 2V#4S-specific fragment amplified by CINAU184 primer was absent,whereas the 2DS-specific fragment was present.(C)Sharp eyespot disease indices for CS,Niavt 14(resistant control),Yangmai 15(susceptible control),the T.aestivum-D.villosum disomic substitution line DS2V#4(2D),and T.aestivum-D.villosum disomic translocation lines T2DS·2V#4L (NAU2V-8) and T2V#4S·2DL in the greenhouse and field during the 2013-2017 growing seasons.CS,Chinese Spring;91C43,accession of D.villosum;ZY1286,durum wheat;NAU 1808,T.durum cv.1286-D.villosum 91C43 amphiploid.Different letters indicate differences at P <0.05.
3.2.Introgression of sharp eyespot resistance from T2DS·2V#4L into wheat cultivar Aikang 58
Greenhouse and field tests during 2012-2017 showed that the substitution line DS2V#4(2D) was resistant to sharp eyespot(mean DI=21.1%compared to CS at DI=41.7%).This finding indicated that the resistance was conferred by chromosome 2V#4.The translocation line T2DS·2V#4L was also resistant (mean DI 22.0%)whereas the T2V#4S·2DL line was susceptible (mean DI 44.1%).Thus,the resistance gene(s) was located on chromosome arm 2V#4L (Fig.1C).
The translocation chromosome was introduced into Aikang 58 by backcrossing and plants with or without the T2DS·2V#4L chromosomes in segregation generations were inoculated withR.cerealis(Fig.S2).In BC2F2,plants disomic for T2DS·2V#4L showed disease indices reduced by respectively 17.4% and 46.7% in the greenhouse and field environments compared to those without the translocation chromosome(Fig.2A).Disomic BC2F2plants with similar plant height and flowering time were selected for selfing to produce the near-isogenic lines NIL-T2DS·2V#4L and NILT2DS·2DL.In BC2F2:3,disomic T2DS·2V#4L lines displayed less necrosis due to sharp eyespot at the flowering and medium milk stages (Fig.2B,C),and mean DI (39.7%-51.7%) was significantly lower than for lines without T2DS·2V#4L (58.9%-85.8%).In BC2F2:4,disomic T2DS·2V#4L lines showed mean DI 27.6% lower than those without T2DS·2V#4L.Thus,disomic Aikang 58 derivatives with T2DS·2V#4L showed less sharp eyespot disease than their disomic counterparts.

Fig.2.Sharp eyespot resistance and agronomic traits of plants with or without the T2DS·2V#4L translocation chromosomes in F2,F2:3,and F2:4 generations of second backcross(BC2 Aikang 58)populations.(A)Sharp eyespot disease indices of plants with or without the T2DS·2V#4L translocation chromosome in greenhouse and field tests in segregation generations.(B,C)Sharp eyespot symptoms on the stems of F2:3 lines with or without T2DS·2V#4L translocation chromosomes at the flowering(B)and medium milk(C)stages.(D-J)Trait comparisons of lines with or without the T2DS·2V#4L translocation chromosome in segregation populations.(D)Plant height,(E)days to heading,(F) spike number,(G) spike length,(H) spikelet number,(I) grain number per spike,and (J) 1000-grain weight.Asterisks indicate differences at P <0.05 .All plants were homozygous.
3.3.Evaluation of agronomic traits in Aikang 58 and Nannong 0686 derivatives
Of the 52 BC2F1plants in Aikang 58 background,20 carried the translocation and 32 lacked it.Chi-squared tests indicated that the segregation followed a 1:1 ratio(χ2=2.76 <=3.84,P=0.096).Among the heterozygous BC1F1-derived BC1F2plants,13 were homozygous,and 36 were heterozygous for presence of the translocation and 20 lacked it,indicating normal inheritance (=1.5 5 <=3.84,P=0.461) (Table S1).
Assessment of agronomic traits in the BC2F2:3and BC2F2:4populations in Aikang 58 background grown under field conditions during 2020-2021 and 2021-2022 indicated no major differences between lines with and without T2DS·2V#4L in heading date,plant height,grain number per spike,or thousand-grain weight (Figs.2,S3).Spikelet number per spike of the homozygous translocation lines was significantly higher than those without T2DS·2V#4L at Nanjing,but there was no significant difference at Luhe (Fig.2H).Spike number and spike length of the homozygous T2DS·2V#4L translocation lines were significantly increased relative to those without T2DS·2V#4L (Figs.2F,G,S3).Thus,the introgressed 2V#4L not only compensated for loss of the 2DL chromosome arm but likely increased spike number and length.
To further investigate its potential in breeding,the T2DS·2V#4L translocation chromosome was introduced into the wheat cultivar Nannong 0686.In the BC5F1generation,13 plants carried the T2DS·2V#4L translocation and 16 lacked it(χ2=0.31 <=3.84,-P=0.577).Among 54 BC5F2plants,14 were homozygous,27 were heterozygous for the T2DS·2V#4L chromosome,and 13 lacked it(=0.22 <=3.84,P=0.982),indicating normal inheritance(Table S1).Agronomic traits of the BC5F2population grown under field condition during 2020-2021 indicated no major differences between plants with and without T2DS·2V#4L in maximum tiller number,plant height,effective spike number,spikelet number,grain number per spike,or thousand grain weight,whereas the spike length of homozygous T2DS·2V#4L translocation plants was significantly increased relative to those without T2DS·2V#4L (Table S2).
3.4.Flow-sorting,sequencing,and comparative genome analysis of 2V#4L
To dissect the genetic relationship between 2V#4L ofD.villosumand 2DL ofT.aestivum,chromosome T2DS·2V#4L was purified by flow sorting from the translocation line NAU2V-8 (Fig.S4).The purity of sorted T2DS·2V#4L chromosomes was 97.1%.Sequencing produced 86 Gb paired-end reads (each read being 150 bp,PE90)andde novoassembly generated T2DS·2V#4L scaffolds.Sequence alignment of T2DS·2V#4L and 2D from IWGSC RefSeq v1.0 revealed similarity greater than 97%.After removal of 2DS-anchored scaffolds,the locations of the remaining 40,075 scaffolds were predicted to be 2V#4L with total assembly length of 384.30 Mb(Table 1).The maximum and minimum scaffold lengths were 172.56 and 1.00 kb,respectively,with an N50 length (minimum length of scaffolds representing 50% of the assembly) of 20.72 kb.The GC content of the 2V#4L assembly was 46.74%,repetitive sequences accounted for 83.68% (321.57 Mb) of the genomic content of 2V#4L,and retrotransposons accounted for 71.20% and transposons for 8.64%.

Table 1Statistics of the 2V#4L chromosome assembly.
A total of 8836 genes were predicted and 7886(89.2%)had clear functional annotations.Gene family analysis indicated that 6154 genes inD.villosum2V#4L had orthologs in 2AL,2BL,and 2DL of CS (Fig.3A);513 and 1549 gene families and genes were unique to 2V#4L.Among 46 annotated nucleotide binding site-leucine rich repeat (NBS-LRR) genes,15 were specific to 2V#4L.A total of 667single-copy orthologous genes were identified among homoeologous group 2 long arms,namelyD.villosum2V#4L andT.aestivum2AL,2BL,and 2DL,Th.elongatum2EL,H.vulgare2HL,andS.cereale2RL(Fig.3B).A phylogenetic tree built using the single-copy genes indicated that 2V#4L was more closely related to 2RL and 2HL than to the wheat chromosome arms (Fig.S5A).Alignment of all annotated genes on arm 2DL showed close synteny between 2V#4L and 2DL,and that 2V#4L compensated well for 2DL (Fig.S5B).
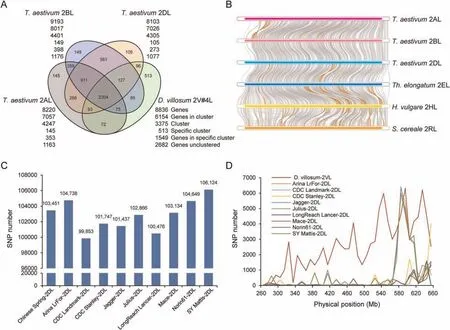
Fig.3.Genomic sequence analysis of chromosome arm 2V#4L of D.villosum.(A)Gene family clustering of 2V#4L of D.villosum,2AL,2BL,and 2DL of T.aestivum.The numbers below each species name are the total number of genes analyzed,genes in clusters,gene clusters,specific clusters,specific genes,and non-clustered genes.Numbers in different sections of the diagram indicate numbers of clusters.(B) Syntenic regions of single-copy orthologous genes among the long arms of homoeologous group 2 chromosomes from Triticeae species.An inverted fragment is indicated in brown color.(C)SNP number in genes on 2V#4L compared with homologous regions on sequenced 2DL of 10 bread wheat cultivars.(D) Distribution of chromosome-wide SNP in genes in 2V#4L and 2DL from nine bread wheat cultivars in comparison with homologous regions of Chinese Spring.
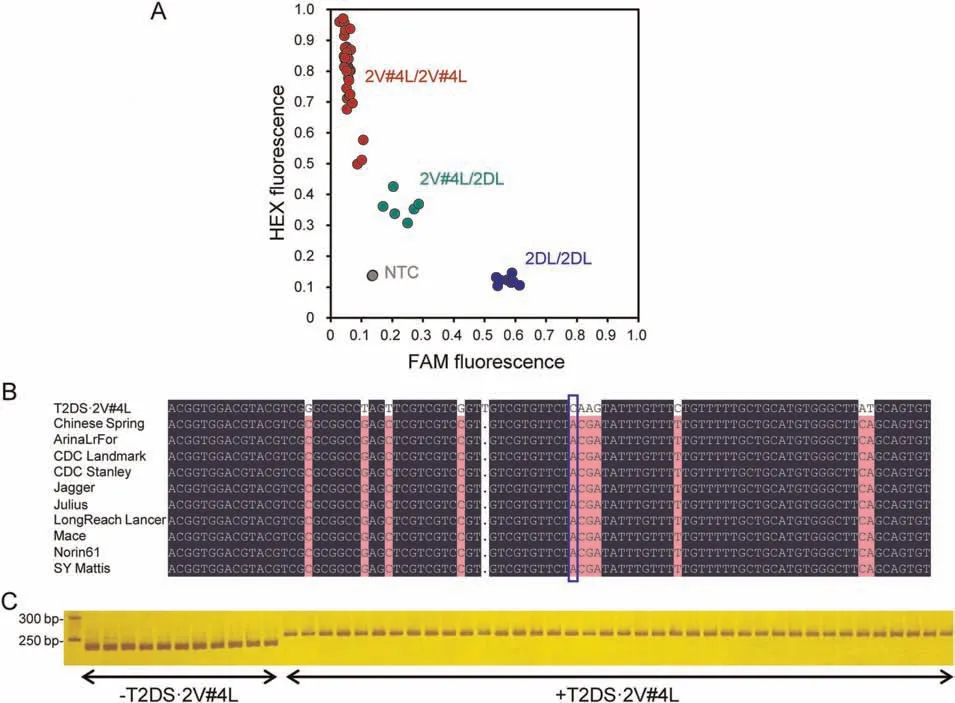
Fig.4.2V#4L-specific Kompetitive Allele-Specific PCR(KASP)and conserved ortholog sequence(COS)markers for the detection of T2DS·2V#4L chromosome in bread wheat background.(A)Scatter plots of 2V#4L-specfic KASP assay showing clustering within the Aikang 58×NAU2V-8 BC2F2:4 population.(B)Position of the SNP used to design the KASP primers.(C) Co-dominant COS marker W1818 distinguishing plants with or without T2DS·2V#4L in the Aikang 58 × NAU2V-8 BC2F2:4 population.NTC,non-template control.
There were 99,853-106,124 SNPs between 2V#4L and 2DL among 10 wheat cultivars (Fig.3C).The number of SNPs was less than 1000 in window sizes of 10 Mb on 2DL between most cultivars and CS,but significantly more between 2V#4L and CS 2DL in most genomic regions (Fig.3D).Thus,the substitution of 2DL by 2V#4L broadened genetic diversity.
A KASP marker based on the SNP variation in a pair of orthologous genes between 2V#4L and 2DL detected the T2DS·2V#4L chromosome and could distinguish homozygosity or heterozygosity for the translocation in multiple genetic backgrounds (Fig.4A,B;Table S3).Thirty COS markers were also developed based on the sequences of orthologous genes in 2V#4L and 2DL from CS,and could be used to detect the T2DS·2V#4L chromosome in wheat backgrounds (Figs.4C,S6;Table S4).
4.Discussion
Sharp eyespot has become a widespread disease and few immune or highly resistant wheat accessions have been identified[5].Identification and introgression of resistance genes from wild relatives is a promising strategy for control.In the present study,the cytological status of the wheat-D.villosumRobertsonian translocation line NAU2V-8 in CS was verified as T2DS·2V#4L by molecular marker assays,GISH and FISH analysis.NAU2V-8 showed stable resistance toR.cerealisat the adult growth stage.To validate the resistance in a different wheat background,NAU2V-8 was backcrossed to cultivar Aikang 58 with selection for 2V#4L using markers and BC2F2,BC2F2:3,and BC2F2:4materials were tested for response to sharp eyespot.Plants with homozygous T2DS·2V#4L chromosomes showed a 30% mean reduction in disease index compared with those without T2DS·2V#4L across the three generations.The T2DS·2V#4L translocation line represented a novel germplasm with sharp eyespot resistance.
Although many transfers of genetic traits by means of chromosome translocation involving wild relatives have been reported,only a few have become widely used in wheat cultivars.Poor genetic background (often in CS),linkage drag,and insufficient compensation for loss of wheat chromatin has limited the exploitation of most translocation lines [19,20].Several genes for agronomic traits including plant height,photoperiod response,and grain weight are located in homoeologous group 2 chromosomes[28,46-48],and homoeologous recombination between alien group 2 chromosomes transferred to wheat has been reported to have detrimental effects on agronomic traits in some translocation lines.A wheat-rye T2BS·2RL translocation line delayed heading and maturity [49].In the present study,among the BC2F2:4lines with homozygous T2DS·2V#4L,three lines carried the samePpd-D1aallele on 2DS as Aikang 58,indicating that exchange and recombination occurred between chromosomes T2DS(CS)·2V#4L and T2DS(Aikang 58)·2DL (Fig.S7).The T2DS·2V#4L translocation line with 2DS from Aikang 58 rather than CS may increase its use in breeding.A successful example for utilization of alien chromatin harboring disease resistance genes from homoeologous group 2 of wheat wild relative in breeding was the wheat-Triticum timopheevitranslocation T2B·2G,in which chromosome 2B harbors a 427-Mb part of 2G bearing the stem rust resistance geneSr36and powdery mildew resistance genePm6[41].Two hard red spring cultivars served as the original sources ofSr36andPm6in wheat breeding programs worldwide [50,51],and in a recent study,the proportion of cultivars carryingSr36was up to 40% in a soft red winter wheat breeding population developed at Purdue University in the USA [52].
With the development of technology,next-generation sequencing provides new approaches for homoeologous chromosome study.Chromosome arms 4VS,5VS,and 6VS ofD.villosumshowed high colinearity with their homoeologous chromosome arms of wheat in flow-sorted sequencing [8,21,22].Similarly,a high colinearity between 2V#4L and 2DL was also observed in the present study.The finding that the NIL-T2DS·2V#4L and NIL-T2DS·2DL near-isogenic lines showed no obvious differences in main agronomic traits suggests that 2V#4L compensated the loss of the 2DL chromosome arm in Aikang 58 background,confirming its potential for breeding.
In breeding,it is desirable to accurately select plants with the target chromatin from large populations in each generation and in multiple genetic backgrounds.With the available sequences of 2V#4L and wheat 10+genome [41],a KASP marker developed based on a SNP between orthologous genes will permit fast and accurate identification of progeny homozygous or heterozygous for the T2DS·2V#4L chromosome in diverse backgrounds when the T2DS·2V#4L introgression is used for wheat improvement.
The characterization of T2DS·2V#4L in Aikang 58 background provides breeders with an alternative genetic resource for developing sharp eyespot-resistant cultivars.It can be pyramided with reported sharp eyespot-resistance QTLs using markers developed in this study to breed cultivars with multiple resistance genes.Although the T2DS·2V#4L translocation line is susceptible to powdery mildew,a T2DS·2V#5L translocation line carries the powdery mildew resistance genePm62[12].It should be possible to produce lines carrying both sharp eyespot and powdery mildew resistance by conventional breeding.The available KASP and PCR markers along with the 2VL sequence will expedite transfer of these resistances to other wheat backgrounds by marker selection.
CRediT authorship contribution statement
Jizhong Wu:Supervision,Conceptualization,Writing -Review&Editing.Ruiqi Zhang:Supervision,Resources,Writing -Review&Editing.Caiyun Liu:Formal analysis,Visualization,Validation,Writing-Original Draft,Writing-Review&Editing.Wei Guo:Data Curation,Visualization,Validation.Yang Wang:Investigation,Formal analysis.Bisheng Fu:Methodology.Jaroslav Doležel:Methodology.Ying Liu:Investigation.Wenling Zhai:Investigation.Mahmoud Said:Investigation.István Molnár:Investigation.Kateřina Holušová:Investigation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Prof.Robert McIntosh,University of Sydney,for reviewing the manuscript.We also thank P.Cápal,Z.Dubská,R.Šperková,and J.Weiserová,Institute of Experimental Botany,Olomouc,for assistance with chromosome flow sorting.This work was supported by the National Key Research and Develpment Program of China(2021YFD1200600),the National Natural Science Foundation of China (31871619,32101703,and 32101800),the Natural Science Foundation of Jiangsu Province (BK20210152),the Jiangsu Seed Industry Revitalization Project (JBGS (2021) 013),the Key Research and Development Program of Jiangsu Province(BE2022346),and Jiangsu Agricultural Science and Technology Innovation Fund of China (CX (20) 3029).Jaroslav Doležel and his team in Olomouc was supported by the European Regional Development Fund (CZ.02.1.01/0.0/0.0/16_019/0000827).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.04.013.
- The Crop Journal的其它文章
- Reversible protein phosphorylation,a central signaling hub to regulate carbohydrate metabolic networks
- Genetic and environmental control of rice tillering
- High-throughput phenotyping of plant leaf morphological,physiological,and biochemical traits on multiple scales using optical sensing
- The R2R3-MYB transcription factor GaPC controls petal coloration in cotton
- The photosensory function of Zmphot1 differs from that of Atphot1 due to the C-terminus of Zmphot1 during phototropic response
- Disruption of LEAF LESION MIMIC 4 affects ABA synthesis and ROS accumulation in rice

