Characterization of meiotic chromosome behavior in the autopolyploid Saccharum spontaneum reveals preferential chromosome pairing without distinct DNA sequence variation
Xin Zhng,Zhung Mng,Jinli Hn,Hris Khurshi,Aymn Esh,Robrt Hstrok,Ki Wng,c,
a School of Life Sciences,Nantong University,Nantong 226019,Jiangsu,China
b Plant Cytogenetics and Molecular Biology Group,Institute of Biology,Biotechnology and Environmental Protection,Faculty of Natural Sciences,University of Silesia in Katowice,Katowice 40-032,Poland
c College of Agriculture,Fujian Agriculture and Forestry University,Fuzhou 350002,Fujian,China
d Sugar Crops Research Institute,Agriculture Research Center,Giza 12111,Egypt
e Oilseeds Research Program,National Agricultural Research Centre,Islamabad 44000,Pakistan
Keywords: Autopolyploidy Saccharum spontaneum Meiosis I Chromosome behavior Chromosome pairing
ABSTRACT Autopolyploidy and allopolyploidy may represent an evolutionary advantage and are more common in plants than assumed.However,less attention has been paid to autopolyploidy than to allopolyploidy,and its evolutionary consequences are largely unclear,especially for plants with high ploidy levels.In this study,we developed oligonucleotide (oligo)-based chromosome painting probes to identify individual chromosomes in S.spontaneum.Using fluorescence in situ hybridization(FISH),we investigated chromosome behavior during pachytene,metaphase,anaphase,and telophase of meiosis I(MI)in autotetraploid,autooctoploid,and autodecaploid S.spontaneum clones.All autopolyploid clones showed stable diploidized chromosome behavior;so that homologous chromosomes formed almost exclusively bivalents during MI.Two copies of homologous chromosome 8 with similar sizes in the autotetraploid clone showed preferential pairing with each other with respect to the other copies.However,sequence variation analysis showed no apparent differences among homologs of chromosome 8 and all other chromosomes.We suggest that either the stable diploidized pairing or the preferential pairing between homologous copies of chromosome 8 in the studied autopolyploid sugarcane are accounted for by unknown mechanisms other than DNA sequence similarity.Our results reveal evolutionary consequences of stable meiotic behavior in autopolyploid plants.
1.Introduction
Cultivated sugarcane (Saccharumspp.) is a sugar and biofuel feedstock crop providing 80% of the world’s sugar and 40% of its ethanol.TheSaccharumgenus is composed of two wild species:S.robustumandS.spontaneum,and four groups of formerly cultivated clones:S.barberi,S.sinense,S.edule,andS.officinarum[1].Saccharum spontaneum,the most primitive species [2,3],has been reported to comprise nearly 40 genotypes,whose chromosome formulas range from 2n=5x=40 to 2n=16x=128[1,4-6].Saccharumofficinarum(2n=8x=80)(referred to as noble cane)was proposed[7-10]to have been domesticated fromS.robustum(2n=10x=60,80)and was cultivated worldwide before the end of the nineteenth century for its high sugar content.Saccharum barberiandS.sinenseare interspecific hybrids betweenS.officinarumandS.spontaneum[11].Saccharum edulemay have originated fromS.robustum[12]or could be an interspecific or intergenic hybrid between eitherS.officinarumorS.robustumwith a species in theSaccharumcomplex[13].Interspecific hybridization using noble cane andS.spontaneumprovides a breakthrough in sugarcane breeding by introducing disease resistance,vigor,stubbling,and other traits into noble cane [14].To restore the high sugar content trait,a series of backcrosses with noble cane were conducted after interspecific hybridization [15].This breeding program for generating modern cultivars is known as nobilization breeding.The resulting modern sugarcane cultivars are highly polyploid interspecific hybrids and represent the most genetically complex crop ever studied [16].
AllSaccharumspecies are polyploids and exhibit variations in chromosome numbers [6].The genome complexity has hindered progress in genomic research and the application of genomic tools in sugarcane breeding programs.Fluorescencein situhybridization(FISH) has played a key role in deciphering the classification,genome structure,and evolution ofSaccharum.Applying FISH using ribosomal DNA and whole genomic DNA as probes revealed the basic chromosome numbers of several species and the genomic composition of cultivated sugarcane[17-19].FISH using massively synthesized chromosome-specific oligonucleotide probes (oligo-FISH) have allowed accurate karyotyping in sugarcane [20,21].Oligo-FISH has also revealed a wide range of ploidies from 4x(tetraploidy) to 16x(hexadecaploidy),which have different basic chromosome numbers includingx=8 [8],9 [21,22] or 10 [23].
Newly formed polyploids experience a process of compensatory adaptation to tolerate meiotic irregularity,including meiosis pairing chaos [24,25].The autopolyploids can evolve into a cytologically stable state in which the homologous chromosomes can pair predominantly as bivalents[26-31].However,how autopolyploids develop this meiosis stabilization is still largely unknown.A recent study [32] in a maize autopolyploid suggested that subtle sequence differences could lead to an innate pairing preference between homologous chromosomes with the most similar DNA sequences.However,more than such preference based on subtle DNA diversity may be required to prevent pairing between highly similar homologous chromosomes,given that bivalents formed by two parental chromosomes were observed.Thus,whether such diversity or to what extent the DNA sequence variation among homologous chromosomes could lead to the establishment of exclusive bivalent associations in natural or evolved autopolyploids is still unknown.
The high-level diversities in chromosome number and ploidies in sugarcane provide an excellent system for studying autopolyploidization in plants.However,the large number of chromosomes with similar morphologies and high (>70%) proportions of repetitive DNA [33,34] makes the identification of individual chromosomes in sugarcane meiotic cells difficult.To overcome such problems,we developed oligonucleotide (oligo)-based chromosome painting probes for chromosome 2 in a previous study [22].Here,we developed painting probes for other three chromosomes of 1,7,and 8.By FISH using these probes,we investigated chromosome behavior during pachytene,metaphase,anaphase,and telophase of meiosis I (MI) in autotetraploid,autooctoploid,and autodecaploidS.spontaneumclones.
2.Materials and methods
2.1.Plant materials
Saccharum spontaneumclones,Np-X (2n=4x=40),SES208(2n=8x=64),and Yunnan 82-106 (2n=10x=80) were used for FISH.Seeds of Np-X and SES208 were germinated at 22 °C,with a 20 h light/4 h dark photoperiod and a humidity of 65%.
2.2.Chromosome preparation
The method of preparing metaphase chromosomes from root tips was based on a published procedure[35]with some modifications.Root tips were excised and treated with nitrous oxide for 1.5 h to enrich metaphase cells.They were then fixed in fixative solution (3:1 ethanol:acetic acid) and stored at -20 °C.An enzymatic solution with 4% cellulase (w/v) (Yakult Pharmaceutical,Tokyo,Japan) and 2% pectinase (w/v) (Plant Media,Pittsburgh,PA,USA) was used to digest the root tips for 1 h at 37 °C.Metaphase chromosomes were then prepared as previously described[35].
Meiotic chromosome preparations were made as previously described [36].Briefly,young flowers were collected and fixed in a 3:1 ethanol:acetic acid mixture for 24 h.After digestion with enzymatic solution (2% cellulase (w/v) and 1% pectinase (w/v) for 1 h at 37 °C),anthers were transferred to a glass slide with a drop of 60% acetic acid (v/v).The pollen mother cells were dissected from the anther with tweezers and then squashed with a cover glass slide.Chromosome slides were stored at -80 °C for FISH experiments.
2.3.Development of oligo-based chromosome painting probes
Chromosome-painting oligo probes were designed with Chorus2 software (https://github.com/zhangtaolab/Chorus2) as described previously [37].Chromosomes 1,7,and 8 from the assembly ofS.spontaneum(https://www.ncbi.nlm.nih.gov/datahub/genome/GCA_003544955.1/) were divided into 50 bp oligo sequences using the default parameter ‘‘-l 50 -homology 75 -step 5”.After repetitive oligos were filtered out,oligos with a unique genome assembly site were retained for probe synthesis.To develop dual-color barcoding probes,chromosomes 7 and 8 were evenly divided into 10 segments.Oligos localized in each region were mixed as a single pool and synthesized independently.The adjacent oligo pools on each chromosome were then labeled with contrasting colors (green or red) to produce banding patterns.
2.4.Oligo-FISH
The oligo-FISH procedure was conducted as previously described [23].The hybridization mixture consisted of 10 μL of 100%deionized formamide,2 μL of 20×SSC,4 μL of a 50%dextran sulfate solution,and approximately 100 ng of oligo probe.The hybridization mixture was denatured at 90 °C for 5 min before being applied to chromosome preparations.After hybridization,chromosomes were stained with 4′,6-diamidino-2-phenylindole(DAPI) (Vector Laboratories,Newark,CA,USA),and probe signals labeled with tetramethylrhodamine (red) or carboxyfluorescein(green) were directly examined under a fluorescence microscope.FISH images were captured using an Olympus DP80 CCD camera with cellSens Dimension 1.9 software (https://lifescience.evidentscientific.com.cn) and then adjusted and merged using Adobe Photoshop software (Adobe Systems,San Jose,CA,USA).
2.5.Sequence variation analysis
Genomic sequencing data of NP-X (ASM2245720v1) were retrieved from the National Center for Biotechnology Information Assembly (https://www.ncbi.nlm.nih.gov/assembly).The genomic data were used to identify single-nucleotide polymorphisms(SNPs) and insertion-deletion mutations (InDels,1-200 bp).The FASTA-formatted files of chromosomes to be compared were processed with NUCmer of the MUMmer4 suite [38].For presence and absence variation (PAV,>200 bp) analysis,the chromosomes were divided into 150 bp windows with 10 bp steps.The numbers of SNPs and indels and the length of PAVs were calculated using BEDTools (v.2.27.1) [39].Data visualization was performed using ggplot2 (https://ggplot2-book.org/;https://www.r-project.org/).
3.Results
3.1.Development of chromosomes 1-,7-and 8-specific oligo probes in S.spontaneum
The chromosome painting probes of chromosomes 1,7,and 8 were developed in this study.Chromosome 1 was the longest at 123.3 Mb.Chromosomes 7 and 8 were the two shortest at 81.2 Mb and 65.0 Mb,respectively.For each chromosome,chromosome-specific sequences from the full pseudochromosome assembly were identified and used to design oligos.Respectively 100,665,59,980,and 48,610 oligos were developed for chromosomes 1,7,and 8 and presented average densities of 818,740,and 747 oligos per Mb.Distribution analysis showed that the oligos covered most regions in each chromosome (Fig.1).The centromeric region of each chromosome was defined by mapping CENH3 ChIP-seq reads [33].For chromosomes 7 and 8,low oligo density in the defined centromeric regions was found.The defined centromere in the chromosome 1 assembly was located at the end,and low oligo density was observed.Cytological assay showed that chromosome 1 was metacentric with an arm ratio of 1.21 [20],a finding consistent with the weak FISH signal observed in the middle regions of SES208 metaphase chromosomes (Fig.2a).The displacement of the centromere position derived from ChIP-seq mapping may indicate misassembly of the centromere region in chromosome 1,a common occurrence in highly repetitive regions[40,41].

Fig.1.Distribution of oligonucleotides on chromosomes 1,7 and 8 of S.spontaneum SES208.The x axis indicates the position on corresponding chromosomes,and the y axis shows oligo density (oligos per 500 kb window).Cent indicates the centromere regions revealed by CENH3 ChIP-seq.

Fig.2.FISH mapping using chromosome painting probes in mitotic metaphase and meiotic pachytene chromosomes of S.spontaneum SES208,Yunnan 82-106,and Np-X.The painting probes specific to chromosomes 1,2,7,and 8 were hybridized to mitotic metaphase and meiotic pachytene cells in SES208(a-h),Yunnan 82-106(i-p),and Np-X(qx).Arrows indicate the chromosomal copies bearing the signals on their distal fragments.Scale bars,10 μm.
In the FISH analysis using these probes,clear signals were observed over the studied chromosomes 1,2,7,and 8 in both the mitotic metaphase and pachytene stages inS.spontaneumSES208 (2n=8x=64) (Fig.2a-h),indicating that these chromosome painting probes could be used as reliable markers for chromosome identification in the highly polyploid speciesS.spontaneum.FISH was also conducted inS.spontaneumclones Np-X and Yunnan 82-106 using the four chromosome painting probes.Np-X is a tetraploid (2n=4x=40) and has been proposed[23] as an ancestral genotype ofS.spontaneum.Yunnan 82-106(2n=10x=80) is a decaploid,and one of the two predominant cytotypes inS.spontaneum(octoploid and decaploid) [6,42].As shown in Fig.2i-p,chromosome-painting signals from metaphase and pachytene chromosomes 1,2,7,and 8 were observed in Yunnan 82-106.In the clone Np-X,four chromosomal copies with whole-chromosome painting signals from each probe were observed at metaphase (Fig.2q-x).Another four chromosomal copies bearing the signals on their distal fragments were observed in FISH using chromosomes 2 and 7 probes(Fig.2r,s,arrows).This outcome is consistent with the previous observations that three chromosomes (named chromosomes 2,4,and 9 in NP-X) in thex=10 ancestor genotype clone Np-X fused into the two chromosomes named chromosomes 2 and 7 inx=8 clone SES208 (the probe was designed based on thex=8 clone SES208) [22,23].
3.2.Chromosome pairing in S.spontaneum clones with diverse ploidies
To investigate chromosome pairing,we conducted FISH on pachytene chromosomes using Np-X,SES208,and Yunnan 82-106 with four chromosome painting probes.A total of 1023 pachytene cells painted by these probes were analyzed in the three clones.We observed 975 cells(95.3%)showing diploid-like behavior,with only bivalents observed(Fig.2).All-bivalent cells in Np-X,SES208,and Yunnan 82-106 showed frequencies of 97.3%,93.1%,and 95.0%,respectively,showing a higher percentage in the tetraploid than in the other two clones with greater genome ploidies(Fig.3a).In the comparison among chromosomes,we observed a much lower frequency of all-bivalent cells for chromosome 8(92.0%) than for the other three chromosomes (95.9%-97.0%)(Fig.3b),a finding we attributed to the lower frequencies of allbivalent cells of chromosome 8 in Yunnan 82-106 (89.7%) and SES208 (91.0%),in contrast to the 96.1% frequency observed in the tetraploid clone NP-X.These results revealed that most cells acted as canonical diploid cells,with all chromosomes pairing into bivalents.At the same time,higher ploidies may cause more abnormal chromosome pairing (non-bivalents) inS.spontaneum.
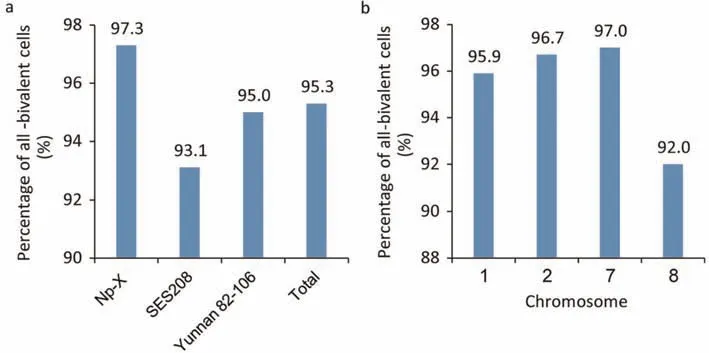
Fig.3.Summary of cells and chromosomes forming bivalents.(a)Percentage of the cells with all bivalents in each clone.(b)Percentage of chromosomes forming bivalents in all three clones.
In addition to bivalent cells,four cells with one bivalent and two univalents were observed in the 369 studied NP-X cells (1.1%)(Fig.4a,b;Table S1).Six (1.6%) cells with a quadrivalent were found(Fig.4c,d;Table S1).In the octoploid clone SES208,15 cells with one quadrivalent and two bivalents(Fig.4e,f;Table S3),three cells with one hexavalent and one bivalent(Fig.4g,h),and one cell with two quadrivalents (Fig.4i,j) were observed among the 277 cells examined (Table S2).In the decaploid clone Yunnan 82-106,three other types of paired cells were observed among the 377 studied cells (Table S3).Eleven cells showed one quadrivalent and three bivalents (Fig.4k,l;Table S3),seven cells showed one hexavalent and two bivalents (Fig.4m,n),and one cell showed one hexavalent and one quadrivalent (Fig.4o,p).These observations of multivalent chromosome pairing are consistent with findings [28,31,43,44] in other autopolyploid plants,indicating an autopolyploid identity or a high level of sequence similarity among the copies of each chromosome studied.
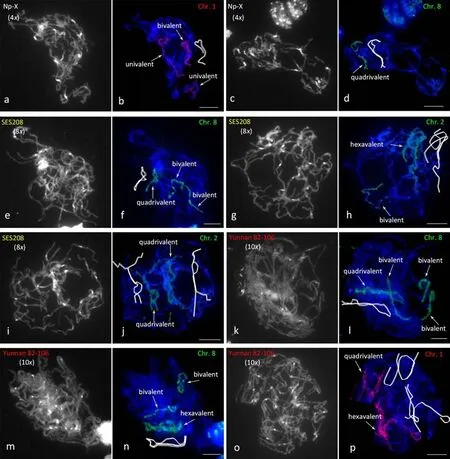
Fig.4.Pairing configurations of pachytene cells with univalent or multivalent associations.(b,d,f,h,j,l,n,and p)FISH signals detected in cells with univalent or multivalent associations.(a,c,e,g,i,k,m,and o) Pachytene chromosomes counterstained with DAPI (gray fluorescence).White lines indicate the configurations of chromosome associations.Scale bars,10 μm.
3.3.Chromosome behaviors at metaphase I,anaphase I,and telophase I
To further investigate chromosome behaviors during MI,we conducted FISH using metaphase I,anaphase I,and telophase I cells.Respectively 100,97,and 96 cells derived from metaphase I,anaphase I and telophase I were identified and examined(Table S4).The chromosomes in most cells displayed normal behaviors in all studied clones (≥96.6%) (Fig.S1;Table S4).Only eight cells were identified with abnormal chromosome behaviors(Table S4).Among them,four cells were found to have lagging chromosomes among the 97 anaphase I cells studied (Fig.5a-f).At least one cell with lagging chromosomes was found in each of the three clones (Table S4).In contrast,only one cell with an anaphase chromosome bridge from Yunnan 82-106 (Fig.5g,h) and two cells with a misaligned metaphase I chromosome from SES208 (Fig.5i,j) were found.One anaphase cell in NP-X with uneven segregation of chromosome 1 was observed (Fig.5k,l).Thus,most cells ofS.spontaneumclones with high ploidies behaved similarly to those of the diploids at MI.

Fig.5.Abnormal chromosome behaviors at anaphase I and metaphase I in S.spontaneum SES208,Yunnan 82-106,and Np-X.(a-f) Chromosome painting in Np-X (a,b),SES208 (c,d),and Yunnan 82-106 (e,f) showing lagging chromosomes.(g,h) Chromosome painting in Yunnan 82-106 showing an anaphase chromosome bridge.(i,j)Chromosome painting of metaphase I cells in SES208 showing a misaligned chromosome 2.(k,l)Images showing uneven segregation of chromosome 1 in Np-X.Scale bars,10 μm.
3.4.Structure comparison of homologous chromosomes in NP-X
In speculating how diploid-like pairing occurred in the autopolyploid species,we hypothesized that homologous copies with similar sizes have higher structure or sequence similarity than any other pairs of homologous copies,which leads to their consistent preferential pairing as well as that found previously[32].As a test,we compared the structures and sequences of the homologous chromosomes.We selected the autotetraploid NP-X for this analysis because of its lower ploidy and the availability of genome assemblies of the complete set of 40 chromosomes.Single-color chromosome painting probes cannot detect intrachromosomal rearrangements,such as inversions,duplications,and deletions occurring within a single chromosome.To overcome this limitation,we designed a dual-color barcoding probe for chromosomes 7 and 8.For chromosome 7,the 81.3 Mb of the chromosomal assembly were divided into 10 segments (~8 Mb each,named C7-1 to C7-10).Each segment consisted of approximately 6200 oligos,except C7-5 (32-40 Mb) (Table S5).The region 32-40 Mb contained the centromere/pericentromere,which is saturated with repetitive DNA.Thus,only 3532 oligos were produced.Similarly,10 segments were obtained for chromosome 8 (C8-1 to C8-10),each containing approximately 4800 oligos.For probe preparation,adjacent probes were labeled with contrasting colors(green or red) to produce a banding pattern to distinguish any probe from its neighboring probe (Fig.6).We observed consistent orders between the FISH signals and their locations on the genome assembly for all bivalents of chromosomes 7 and 8,suggesting no structural variation among these four homologous chromosomes of chromosomes 7 and 8 in NP-X.
We frequently observed length differences in NP-X FISH between the two bivalents of chromosome 8 (Fig.6b).This observation was confirmed by the average sizes of long (25.3 ± 5.4 μm)and short bivalents (22.9 ± 4.5 μm) from 41 pachytene cells with well-spread chromosomes in NP-X (P=3.85 × 10-7,two-way ANOVA).We inferred that the four homologous copies of chromosome 8,named Chr.8A-8D,are not completely identical and that the pairing likely occurs between homologous copies of similar size.In the current genomic assembly,Chr.8A (44.7 Mb) and 8C(47.8 Mb) are the short copies and Chr.8B (53.4 Mb) and 8D(54.1 Mb) are the long copies.The size ratio between long and short homologous copies was 1.16,similar to the ratio of the bivalent lengths (1.11,25.3 μm/22.9 μm).
In a further FISH assay using barcoding probes of chromosomes 7 and 8 in SES208 and Yunnan 82-106,consistent signal orders were observed in all probes between the two clones (Fig.6c-f).However,probes C7-9 and C7-10 showed a signal order in reverse of their locations in the genome assembly (Fig.6c,e).This finding suggests incorrect alignment around the regions C7-9(64-72 Mb)and C7-10 (72-81.3 Mb).
3.5.DNA sequence variation of homologous chromosomes in NP-X
Using the genomic sequencing data of autotetraploid NP-X,we examined the sequence similarity between the homologous copies of chromosome 8.We found 613,505 SNPs between chromosomes 8A and 8C (Table S6),633,254 SNPs between Chr.8A and 8B,and 624,244 SNPs between Chr.8A and 8D.There were similar distributions for the SNPs (SNPs per 500 kb) along chromosome 8 between each chromosome pair(Fig.7).This result was confirmed with InDels and PAVs between each chromosome pair (Fig.7).However,the InDel and PAV variation levels of chromosome pair 8A/8C were not as low as that of 8A/8B and Chr.8A/8D(Table S6).Analogous analysis of the remaining nine chromosomes showed no apparent differences for any homologous pair (Fig.S2;Table S6,).We conclude that the diploid-like pairing for each chromosome or the specific pairing of Chr.8A/8C and Chr.8B/8D is unlikely to have been due to their high structural similarity.
3.6.Seed germination and fertility of S.spontaneum clones
Given that normal segregation of MI chromosomes in all studied polyploid clones was observed,we expected the clones to show normal reproductive development and seed germination.To confirm this expectation,we collected mature spikes from Np-X and SES208 (no fertility and seed germination data were available for Yunnan 82-106,as only one spike developed and was used for chromosome analysis).By examining individual spikelets,we collected only 11 seeds from 3000 spikelets from Np-X (0.4%) and 329 seeds from 10,000 spikelets (3.29%) from SES208.We conducted seed germination assays to check the fertility of the collected seeds.However,no seedlings emerged from the 11 Np-X seeds.In contrast,39 of the 329 SES208 seeds were viable,leading to a germination rate of 11.9%.These results suggest that most spikelets were sterile in both Np-X and SES208.
4.Discussion
Both autopolyploidy and allopolyploidy may represent an evolutionary advantage and are more common in plants than assumed[31,45-47].However,autopolyploidy has received less attention than allopolyploidy,and its evolutionary outcome and consequences in natural populations are still unclear.Indeed,studies[30,31,45] in some autopolyploid plants revealed that their chromosomes commonly exhibited diploid behaviour in MI,with homologs forming exclusively or almost exclusively bivalents.However,these studies focused mainly on plant autotetraploids or autotriploids[25,31,46,48],which are the lowest ploidies among autopolyploids.Because associating bivalents/multivalents with individual chromosomes is difficult,the exact meiotic behavior of each chromosome in autopolyploids,especially those with higher ploidy levels,is largely unclear.The finding that all studied autopolyploids formed almost exclusively bivalents in MI regardless of ploidy inSaccharumspecies reveals two novel features.First,two shorter copies of four homologous chromosomes 8 in autotetraploid NP-X may preferentially pair at the pachytene stage.Such preferential pairing between specific pairs of homologs in autopolyploid sugarcane may account for the stable MI behaviors observed.With the ability to distinguish each homologous chromosome,whether such pairing occurs for other chromosomes or in other autopolyploid sugarcanes could be tested by developing and applying a haplotype-specific chromosome painting probe[49] to sugarcane chromosomes.
Second,given that substantial problems in gamete formation can be observed when polyploidization arises,it is believed[30,31,50] that the fertility barrier must have been rapidly bypassed to produce fertile and successful lineages for naturally stable polyploids.But we observed extremely low seed fertility despite regular chromosome behavior at MI.We can exclude the possibility that the environment influenced seed fertility,because the same phenomenon was also observed for NP-X seeds collected from Yunnan province,which is near the original collection location of this plant [23].Although we do not know the molecular basis of this seed sterility,these findings suggest that stable MI is insufficient for achieving plant fertility,at least in autopolyploid sugarcane.
A fundamental question behind this study is whether or to what extent the DNA sequence variation among homologous chromosomes could contribute to establishing exclusive bivalent associations in naturally evolved autopolyploids.Because the four copies of each chromosome in NP-X showed a highly similar extent of sequence variation among them,we suggest that mechanisms other than DNA sequence similarity lead to stable diploidized bivalent pairing.In fact,previous studies[51-53]showed that a stable bivalent association could be obtained in newly formed autopolyploids by selection for fertility over multiple generations.Moreover,the majority of chromosomes formed bivalents in a modern cultivar although chromosomal abnormalities were frequently observed[54].These findings suggest that the DNA sequence similarity is not a decisive factor in forming stable diploidized bivalent pairing in autopolyploids.Genetic mechanisms can enforce pairing between homologous chromosomes in some allopolyploids.In allohexaploid wheat,a 2.5 Mb-long interstitial region of chromosome 5B,known as thePh1locus,is required for suppressing homoeologous chromosome pairing,thus assuring a sequencesimilarity-based preferential pairing [55].Whether a genetic system responsible for the rapid development and maintenance of stable bivalent associations after polyploidization is present in autopolyploid plants remains an open question.
CRediT authorship contribution statement
Xin Zhang:Software,Data curation,Writing -original draft.Zhuang Meng:Data curation and investigation.Jinlei Han,Haris Khurshid,andAyman Esh:Data curation and Formal analysis.Robert Hasterok:Conceptualization and Writing -review &editing.Kai Wang:Conceptualization,Writing-review&editing,and Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the National Field Genebank of Sugarcane Germplasm of China for supplying us withS.spontaneumplants.This work was funded by the Startup Foundation from Nantong University (03083074) and partially supported by the National Natural Science Foundation of China (31771862),Special Funds for Technology Innovation of Fujian Agriculture and Forestry University(KFA20001A),and the Research Program of Guangxi Key Laboratory for Sugarcane Biology (GXKLSCB-20190203).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.02.008.
- The Crop Journal的其它文章
- Reversible protein phosphorylation,a central signaling hub to regulate carbohydrate metabolic networks
- Genetic and environmental control of rice tillering
- High-throughput phenotyping of plant leaf morphological,physiological,and biochemical traits on multiple scales using optical sensing
- The R2R3-MYB transcription factor GaPC controls petal coloration in cotton
- The photosensory function of Zmphot1 differs from that of Atphot1 due to the C-terminus of Zmphot1 during phototropic response
- Disruption of LEAF LESION MIMIC 4 affects ABA synthesis and ROS accumulation in rice

