SPP-extractor: Automatic phenotype extraction for densely grown soybean plants
Wn Zhou,Yijie Chen,Weiho Li,Cong Zhng,Yjun Xiong,Wei Zhn,Ln Hung,,Jun Wng,,Lijun Qiu
a College of Computer Science,Yangtze University,Jingzhou 434023,Hubei,China
b College of Agriculture,Yangtze University,Jingzhou 434025,Hubei,China
c National Key Facility for Gene Resources and Genetic Improvement/Institute of Crop Sciences,Chinese Academy of Agricultural Sciences,Beijing 100081,China
d MARA Key Laboratory of Sustainable Crop Production in the Middle Reaches of the Yangtze River (Co-construction by Ministry and Province),College of Agriculture,Yangtze University,Jingzhou 434025,Hubei,China
Keywords: Soybean phenotype Branch length Computer vision A* algorithm Phenotype acquisition
ABSTRACT Automatic collecting of phenotypic information from plants has become a trend in breeding and smart agriculture.Targeting mature soybean plants at the harvesting stage,which are dense and overlapping,we have proposed the SPP-extractor (soybean plant phenotype extractor) algorithm to acquire phenotypic traits.First,to address the mutual occultation of pods,we augmented the standard YOLOv5s model for target detection with an additional attention mechanism.The resulting model could accurately identify pods and stems and could count the entire pod set of a plant in a single scan.Second,considering that mature branches are usually bent and covered with pods,we designed a branch recognition and measurement module combining image processing,target detection,semantic segmentation,and heuristic search.Experimental results on real plants showed that SPP-extractor achieved respective R2 scores of 0.93-0.99 for four phenotypic traits,based on regression on manual measurements.
1.Introduction
Soybeans are food,feed,and industrial crops rich in protein and oil [1,2].Improving soybean yield and quality is a focus of breeding.Selection on phenotypic traits,e.g.,the number of pods per plant[3],plant height[4,5],number of effective branches is necessary for identifying superior plants.Previously,such phenotypes were obtained manually,an operation that was labor intensive[6],error-prone,slow,and subjective [7,8].Recording phenotypic traits in an automatic manner allows collecting genetic diversity data rapidly and cheaply,increasing the efficiency of breeding programs that select for these traits.
Recent advances in computer vision and deep learning techniques have powered research in and application of automatic phenotype extraction in agronomy.Examples are to identify the morphology and color of soybean seeds [9] and pods [10-13];to acquire bean grain lengths and widths [14],pod lengths,widths and areas[15,16];and to count pods and leaves on growing plants for yield prediction [17,18].For extracting pod-associated phenotype traits of mature plants,only a few methods can process a plant without taking the pods off first.Yang et al.[19] adapted a model trained with off-plant pods to on-plant pod identification,and Ning et al.[20]identified on-plant pods using target detection.Identifying pods on a mature plant is challenging because pods can be densely packed and overlap other pods and stems,confounding phenotype extraction.
The lengths,distribution and branching traits of the main stem and other branches are target breeding traits of soybean [21-23].On extracting stem-associated phenotype traits from mature plants,Li et al.[24] measured the length of stem nodes and the main stem of mature soybean plants using instance segmentation,with pods and branches first detached from the main stem.Ning et al.[20] designed an ant colony algorithm to calculate plant height.Guo et al.[25] combined target detection and directed search to identify number of branches,branching angles,main stem length,and stem curvature in a mature plant.Guo et al.[26]identified stem nodes and branches by rotational target detection combined with a regional search algorithm to reconstruct the plant skeleton.Most methods identify stems by connecting the stem nodes,which requires straight plant shapes.However,many soybean plants may have irregularly curved,heavily shaded and widely distributed stems and branches,secondary branch structures,and may be densely covered with pods.Such plants pose challenges for the automatic and accurate extraction of stemrelated phenotype traits: they could overlap and mask a stem region,and they could mislead length calculation when two or more pods connect and form a spurious stem when placed on the board.Also,in order to capture as much phenotype information as possible,we would like to keep the pods on the plant so that their distributions can be recorded and measured.This choice must account for overlap among pods and stems.
The objective of this study was to design an algorithm for extracting pod and stem traits from densely grown mature plants without requiring removal of the pods.To address overlap among pods and stems,we proposed to augment a standard neural network model for target detection with an attention mechanism and to combine a semantic segmentation neural network model with a directed search algorithm.
2.Materials and methods
2.1.Data acquisition and annotation
The study employed 37 populations created from one maternal parent(Zhongdou 41)and 37 paternal parents,planted in June and harvested in October in the experimental field of Yangtze University in Jingzhou,Hubei,China.The range of maturity periods was 82-127 d.Fifteen plants of each population were double-sown 10 cm apart in one 1.5-m row,with 1 m between rows.Five plants were selected from each row for imaging.Plants were photographed at harvest,using a digital camera,which was fixed right above the plant and in the middle of the data-acquisition device.One image was taken per plant with a resolution of 3456 × 5184 pixels and saved in JPG format.
Preprocessing of the plants included removing the leaves and petioles,and removing the plant roots to the cotyledonary node.Plants were placed flat on a black base plate,with the main stems kept as vertical as possible.The plant branches were spread for minimum overlap.Preparing and imaging one plant took from two to seven minutes,and 2950 plants were imaged.Figure S1 shows the structure of the data-acquisition device and an example image.After image acquisition,pods and effective branches were manually counted,and the plant height and the length of each branch were measured with a measuring tape.An effective branch[27] should have more than one internode and more than one mature pod.Manual measurement process took 5-10 min per plant.Table S1 shows the descriptive statistics of the four phenotype traits studied.These manually measured values served as the standard for the algorithm.
Two datasets from the original images were annotated and produced,one for the target detection subtask and the other for the semantic segmentation subtask.The purpose of the first was to identify pods and stems from the input image and obtain their coordinates and that of the second was to extract the stem regions.Images were annotated with the LabelImg [28] and the LabelMe[29]software.Table S2 lists the guidelines used for the annotation of plant parts and Fig.S2 shows an annotated example image from each subtask.
2.2.Main methods
SPP-extractor’s purpose is automatic extraction of pod-and stem-associated phenotype traits from densely grown soybean plants.To this end,we decomposed the task into three sub-tasks and designed three modules accordingly: pod and stem region identification,stem and branch extraction,and branch-length calculation.Fig.1 illustrates the overall structure of SPP-extractor and displays each module in a different color.Pod and stem regions were first identified to provide the basis for subsequent extraction.Then stem and branches were identified for extracting stemassociated phenotype traits.
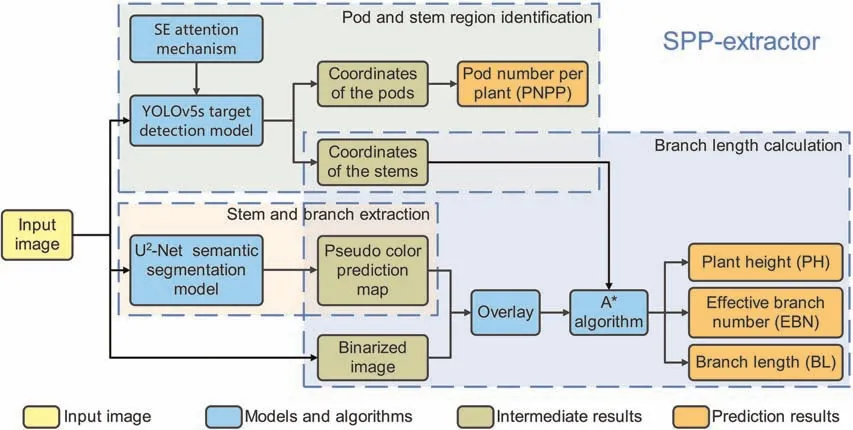
Fig.1.Structure of SPP-extractor,with three major modules: for pod and stem region identification,stem and branch extraction,and branch-length calculation.
2.2.1.Pod and stem region identification
We employed target detection[30]for the pod and stem region identification task.Target detection identifies instances of a specified object type from an input image,that is,pods and stems in our case.The high overlap among pods and stems poses challenges to standard target detection models.We solved this problem in three ways.
First,instead of labeling an intact pod,we elected to label only the tip region of a pod as an alternative indicator.This allows a pod that is obscured to be recognized,provided part of it is visible.Fig.S2 A shows an annotated example.Each rectangle formed by four green points labels a target: either a pod or a stem.Similarly,given that pods were kept on the plant and could obscure stem regions,when stems were labeled,only the unobscured parts were labeled.
Second,we elected to use the YOLOv5s neural network [31] as the base model for target detection.The YOLOv5 model is the fifth generation of the YOLO (You Only Look Once) series algorithms[32] and has achieved accuracy improvements.YOLOv5 uses non-maximum suppression (NMS) [33] and the CIoU bounding box regression loss function [34] to improve classification and positioning accuracy.Its detection speed is also satisfactory.YOLOv5 consists of four parts: Input,Backbone,Neck,and Head.The Input module takes an original input image and performs data augmentation by Mosaic processing[35]to increase data diversity.The Backbone module,composed mainly of the Conv,C3 and SPPF submodules,extracts image features.The C3 submodule uses a residual feature structure,which consists of Conv and BottleNeck.SPPF (spatial pyramid pooling fast) uses three serial MaxPooling calculations with kernel size of 5,which reduces the number of repetitive calculations and increases the speed of the network compared to SPP’s three parallel MaxPooling calculations with kernel sizes of 5,9,and 13.The Neck module uses FPN (feature pyramid network) [36] and PAN (pyramid attention network) [37]structures to enhance the fusion effect of features and increase the detection accuracy of objects at different scales.The Head module makes predictions and outputs three scales of feature map to detect objects of various sizes.
Finally,to increase the model’s predictability in both dense and sparse regions,we added an SE (the squeeze-and-excitation network [38]) attention mechanism into the YOLOv5s structure,resulting in the YOLOv5s-S model.In contrast to the conventional convolutional pooling process that assigns equal weights to each channel of the feature map,the SE attention mechanism learns the importance of each channel together with the task at hand,and weights each channel according to its relevance to the end task.Figure S3 illustrates this process: each channel of the feature map X from the previous layer starts with equal importance and is weighted differently after the squeeze and the excitation process,where the different colors indicate the different weights.The learning process then focuses on patterns in channels with larger weights.The SE mechanism is usually integrated into the C3 submodule,forming the SeC3 submodule.Figure S4 describes the structure of the proposed YOLOv5s-S model.
Given an annotated image,YOLOv5s-S is trained to recognize pod and stem regions in the image,and outputs their coordinates.Pod number per plant(PNPP)is then counted in a single scan of the plant.Another output is the coordinates of the predicted stem regions.These regions are required for identifying the main stem and the branches,and they form the basis for subsequent extraction of effective branch number (EBN),plant height (PH) and branch length (BL).
2.2.2.Stem and branch extraction
To solve the challenges of overlapping pods on accurate stem and branch extraction,SPP-extractor employs the U2-Net [39]semantic segmentation model.Given an input image,semantic segmentation classifies each pixel in the image into one of several semantic object categories: stem,pod,and background.U2-Net is the most advanced version of the classic U-Net structure [40].It adopts an encoder-decoder architecture.The encoder module extracts features from the input image,increases the perceptual field by acquiring local features of the image,and performs picture-level classification.The decoder module up-samples by fusing features from several different scales.The overall network structure forms a letter U.U2-Net attempts to capture contextual information at different scales by nesting two-layers of the Ushaped structure.
Given an input image,the stem and branch extraction submodule identifies the main stem and all the branches on the plant,including the short secondary branches that grow from a branch,and annotates them in bright green(as shown in Fig.2c).Using this image,SPP-extractor then identifies the main stem and the effective branches,measures the plant height and branch lengths,and discards the undesired secondary branches.
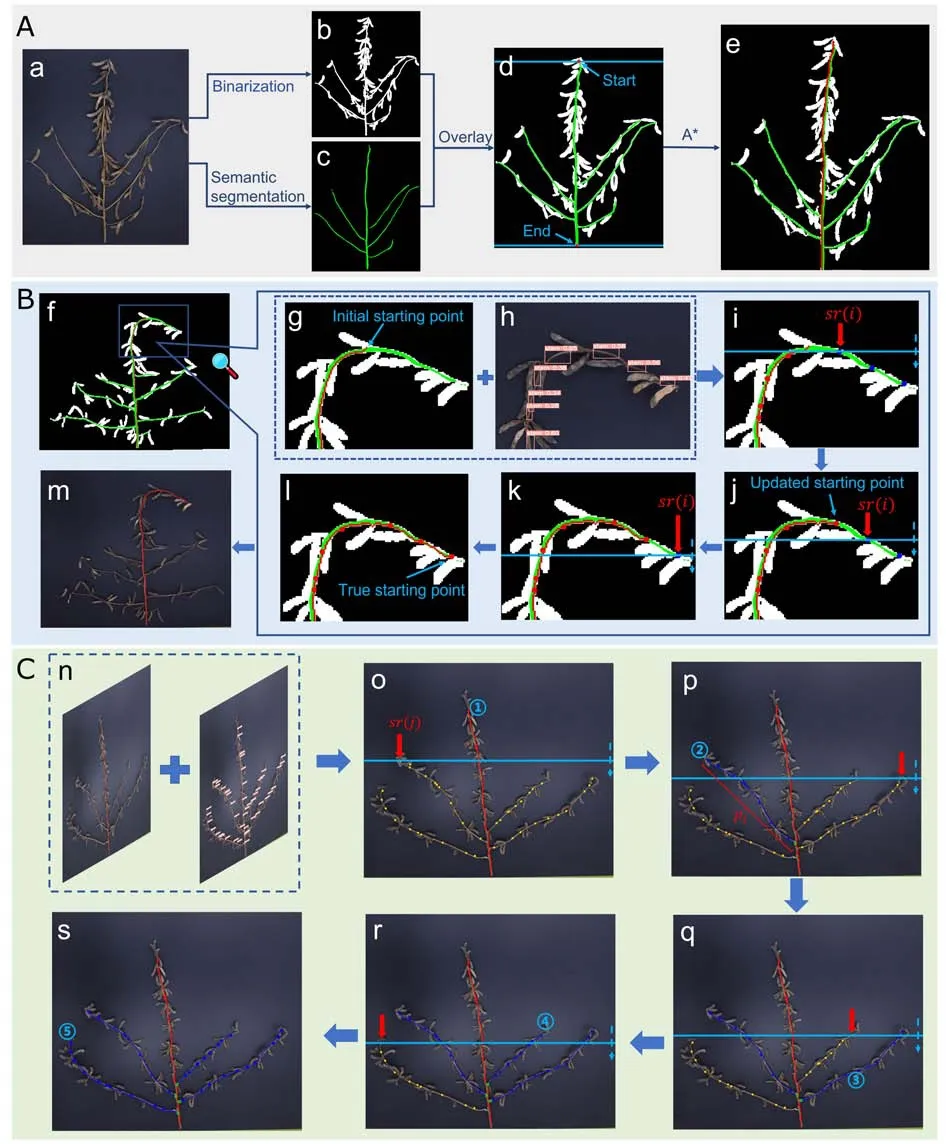
Fig.2.Examples of stem-associated phenotype extraction processes.(A) Map overlay and main stem extraction.(a) The original image.(b) The binarized image.(c) The semantically annotated image.(d) The composite map for path searching.(e) The annotated image with extracted main stem.(B) Bending top treatment.(f) The initial incomplete main stem path.(g)Determining the starting point.(h)Obtaining relevant stem regions.(i-l)The path expanding process.(m)The final complete main stem path.(C) Branch identification and length calculation.(n) The composite map for branch identification.(o-s) Extraction and measurement of each of the four branches.
2.2.3.Route search for stem-associated phenotype trait extraction
Measuring the plant height and the branch lengths is modeled as finding the shortest path between two specific points on the pixel map of an image;for example,between the top of the plant and the cotyledon node on the main stem for plant height calculation,and from the top of the branch to the point where it intersects with the main stem for branch-length calculation.In general,there are two options for generating the pixel map: binarization or semantic segmentation as described above.SPP-extractor combines both binarized and semantically annotated maps to form a composite map for subsequent path finding,as shown in Fig.2A.
Given the pixel map with binarized and annotated branches,we developed a route search algorithm based on A* (A-Star) search[41] to find the shortest end-to-end path through a stem or a branch.The lengths of the stem and branches are measured as respectively PH and BL.EBN was also counted according to the number of paths found along the branches.A* search is one of the most efficient directed-search methods for finding the shortest path in static networks.Compared with other path planning algorithms such as genetic and ant colony algorithms,A*search is efficient and usually achieves satisfactory accuracy on domainspecific problems.At the core of the A* algorithm is its cost function:
Given noden,f(n)denotes the estimated cost from the starting point to the end point via noden,and consists ofg(n)the cost of moving from the starting point to noden,andh(n)the estimated cost from nodento the ending point.The input image is first compressed to one eighth of the original width and height to reduce the size of the raster map required by A* search.Then it is binarized using the OpenCV [42] library,followed by denoising and expansion to remove small particles of noise from the target area and to fill in voids in the image.The input image is read from top to bottom and from left to right and is represented as a twodimensional array A.Each element of A represents a pixel in the image and is assigned 0 if it belongs to the background and 1 if it is part of the plant.The semantically annotated map is represented similarly with a two-dimensional array B,with values 1 and 0 denoting the stem and the background regions respectively.Then the two arrays are added elementwise,resulting in the three values 0,1,and 2.An element with value 0 means that the corresponding pixel is recognized as the background in both maps,and accordingly is set as an unpassable obstacle during search.Elements with values 1 and 2 are possible nodes in the search process and movement costs are set to 1 and 0.1 respectively.In the composite map,the A* algorithm selects the node with the smallestf(n)value as the next node to traverse each time.Because the points on the stems have lower costs than those in the background,the search will be extended largely through points on the stem,i.e.along the stems,until the target point is reached.
With the A* search algorithm and combining previous results from target detection and semantic segmentation,SPP-extractor can then extract the main stem and the other branches,and measure their lengths accordingly.The following two sections describe the methods.
2.2.4.Main stem recognition and length measurement
Soybean plant height (PH) is a phenotypic trait [43],measured as the length from the cotyledonary node to the top growing point of the main stem [44,45].Extracting PH is easier when pods are removed from the plant.For densely grown plants with pods,we first surveyed the plant images in our dataset and found that the semantic segmentation module was well able to detect the highest and lowest points of the stem region.Accordingly,the highest and the lowest within-stem-region points in the semantically-colored binarized image were used as the starting and the ending points,and the A*algorithm described above was used to find the shortest path between them.Fig.2e shows the identified main stem with a red line.
To map the identified path length to actual PH and BL,the magnification ratio was calculated for each photo by dividing the fixed lengthLScaleby the number of pixels involved in itLPixel.Letting PHPixelbe the number of pixels along the identified shortest path,the predicted plant height value in units of cm PHScalewas calculated as follows:
One difficulty in densely grown plants is that the main stem could be bent downward at the top by the weight of the pods concentrated there.The previous algorithm is no longer applicable for such cases because the starting point of the main stem is not necessarily the highest point of all stem regions.Fig.2f shows such an example and Fig.2g shows the initially extracted path.To address this problem,we designed an algorithm MSE (Main Stem Extraction,pseudocode in Fig.3) to compute and extract the complete main stem.MSE has four steps.First,it represents each rectangular frame detected by YOLOv5s-S with five digits:

Fig.3.Pseudocode of the MSE and BIL algorithms.
wherex,ydenote the horizontal and vertical coordinates of the center of the frame andw,hdenote its width and height.The first digit is an indicator with initial value 0.Givennthe number of stem regions identified in an image,the image is then represented with ann×5 matrix.Second,all the stem regions are sorted ascendingly by theirycoordinates.In our samples,xandycoordinates start from the top left corner.Thus,stem regions are sorted from the top of the plant to the bottom.Third,the indicator’s value is changed to 1 for all regions along the main stem path identified by the A* search (i.e.,sr(i)∈pmain):
Then,MSE continues to read down theycoordinate and find the next stem region sr(i)with identifier 0,denoted as sr(i)with red arrows in Fig.2i;sets it as the new starting point(i.e.,the updated starting point in Fig.2j) and finds the path from this point to the main stem’s endpoint.Once the path is found to bypass the current top point of the main stem,i.e.,bypassing the stem region at the top of the current main stem path,sr(i)’s center is set as the new starting point of the main stem.The fourth step is repeated if more such stem regions are found,and the main stem is grown accordingly,until all stem regions are checked.Fig.2(i-l) shows step by step how the stem regions on a bending top(denoted by blue dots)become identified (noted with the red dots).The horizontal line represents the scanning process from the top stem region downwards to the actual plant top.The red arrows indicate the current stem region being checked.
2.2.5.Branch identification and length measurement
Branches are lateral sub-stems that extend from the main stem.BL is defined as the length from the end of a branch to its originating point on the main stem.EBN and BL are phenotype traits selected in plant breeding.In our dataset,a plant can have 0 to as many as 12 branches (as shown in Table S1).Some branches gather around the main stem while others spread out.Some branches also have secondary branches that need to be discarded from the EBN count.
The BIL (Branch Identification and Length measurement) algorithm (pseudocode in Fig.3) is designed specifically for branch identification and length measurement,using the samen×5 matrix as in the MSE algorithm.Using the output of equation (4),BIL scans all the stem regions along the main stem recorded in the matrixIsequentially,for those with an indicator value of 0.Such regions can be the top points of branches,given that all stem regions are sorted ascendingly by theirycoordinates,denoted as sr(j)in Fig.2o.Given such sr(j),BIL finds a pathpjfrom sr(j)to the end of the main stem using the A* search as described previously.Ifpjis found to intersect with the main pathpmain,the intersection point is set as the end point ofpjand the length ofpjis computed accordingly,as shown in Fig.2p.Finally,all stem regions alongpjare updated with an indicator value ofjto indicate that they belong to thejth branch of the plant.The process is repeated until all stem regions have a nonzero indicator value.Fig.2C illustrates the work process of the BIL algorithm.Stem regions with an indicator value of 1 (the main stem),0 (to be determined)and 2 +(the branches) in are shown as red,yellow,and blue dots respectively.Considering that the starting point of the branch is the centroid of the stem frame at the branch top,half of the diagonal of the top stem region is added to the length ofpjas the final branch length.
In real plants,a branch can also have one or more small secondary branches.Such branches are usually short (<3 cm in length) inflorescences and thus are not counted as true branches during manual measurement.Yet the BIL algorithm can still identify and measure them.BIL distinguishes a secondary branch if the pathpjfrom sr(j)to the end of the main stem passes the bottom region of a branch instead.To be consistent with the manual measurement criterion,all such branches are discarded.
2.3.Evaluation metrics
For the pod and stem region identification task,the target detection model was evaluated with three metrics: precisionP,recallR,and mean Average Precision mAP@.5.mAP@.5 refers to the mean AP for each category when the ratio of the intersection and the union of the predicted and true outcomes(i.e.,IoU)threshold is 0.5.Definitions of these metrics are shown in equations(5)-(8):
TP,FP,FN,and TN refer respectively to true positive,false positive,false negative,and true negative cases.Nrefers to the total number of images in the test andkis the number of classes.
For the stem and branch extraction task,the semantic segmentation model was evaluated using the mean IoU(mIoU)defined by equation (9),the mean value of IoU over all classes.
For the extraction of PH,BL,EBN and the PNPP features,SPPextractor was evaluated by root mean square error (RMSE),mean absolute error(MAE),and coefficient of determination(R2)metrics against the ground-truth values collected manually by human experts.Equations (10)-(12) show their definitions,wherey,︿yanddenote respectively the true value,the predicted value,and the sample mean.
2.4.Experimental setup
SPP-extractor was implemented and evaluated on a Windows 10 computer with an i5-12600KF CPU,32G RAM,an RTX 3090Ti GPU,24 GB RAM.The software environment included the CUDA 11.6 [46] acceleration package,Python 3.9,Pytorch 1.12.1 [47]for general deep learning functionality,and Baidu PaddlePaddle 2.3.2 framework [48] for building the semantic segmentation model.
SPP-extractor was first trained with 2065 training images and then validated with 590 validation images,when the various hyperparameters involved in training SPP-extractor were optimized.Values that achieved the highest predictive accuracy on the validation images were kept.The validated model was then tested on the testing set to generate experimental results.Tables S3 and S4 list the hyperparameters for training the YOLOv5s and the U2-Net series models.
3.Results
3.1.Overall performance of SPP-extractor
Fig.4 plots SPP-extractor’s predictions of PNPP,PH,EBN,and BL against manually measured values.

Fig.4.Regressions for six phenotypic traits.PNPP,pod number per plant;PH,plant height;EBN,effective branch number;BL,branch length.
On PNPP,SPP-extractor achieved anR2coefficient of 0.93 and an MAE of 6.34 pods per plant.As shown in Fig.4A,SPP-extractor matched well with human performance on plants with an average of about 100 pods per plant.The mean absolute difference between the automatically and the manually counted values was 6.34 pods per plant.On plants with many pods,pods are dense and some are largely hidden by other pods and stems.SPP-extractor was challenged in such scenarios,as indicated by the points deviating from the model line in the upper right corner of Fig.4A.
On PH,SPP-extractor achieved anR2coefficient of 0.99 and an MAE of 0.68,as shown in Fig.4B.The average absolute difference between SPP-extractor’s predicted PH and the manually measured PH over the 295 testing images was only 0.68 cm,less than the 1-cm threshold set for the human experts when they physically measured the plant.
On EBN,SPP-extractor achieved anR2coefficient of 0.98 and an MAE of 0.04,as shown in Fig.4C.Thus,SPP-extractor correctly identified almost all the branches in almost all images.A closer examination showed that on only 13 test images did SPPextractor predict a plus or minus one EBN value.
On BL,SPP-extractor achieved anR2coefficient of 0.98 and an MAE of 0.97,as shown in Fig.4D.In our test set,a plant can have as many as 10 branches and on average each plant has 4.2 branches,resulting in about 1200 branches in total.The average absolute error in predicting all their lengths was 0.97 cm.
3.2.Performance of SPP-extractor on various plant types
To demonstrate SPP-extractor’s capability in handling plants with secondary branches and bending main stem tops,Fig.5 compares the predicted PNPP,PH,EBN and BL values against manually measured values for three representative plant types from our dataset: one with sparse and easy-to-identify branches,one with several secondary branches,and the third with a bending main stem.All identified branches were numbered in ascending order of their heights in the plant.As Fig.5 shows,SPP-extractor was able to recognize all branches,including secondary branches(as shown in Fig.5B) and to calculate PH and BL in all cases.The MAE for BL and PH were 0.48 cm and 0.4 cm over the three plants.

Fig.5.Comparison of SPP-extractor’s predictions and manually measured values on three plant types.(A) With few branches.(B) With secondary branches.(C) With a bending top.PNPP,pod number per plant;PH,plant height;EBN,effective branch number;BL,branch length.
3.3.Comparison results of multiple target detection models
As stated previously,to solve the occlusion problem in densely grown plants,SPP-extractor improves the YOLOv5s model by adding the SE attention module.To fairly evaluate its effectiveness,we compared it with three counterparts: the standalone YOLOv5s model,the YOLOv5s model with four CBAM attention units(YOLOv5s-C),and the YOLOv5s-S model with four SE attention units.Table S5 shows that YOLOv5s-S outperformed the other models in identifying pods and stems.Adding the SE attention helps the training process to focus on the most relevant features and improve the model’s predictability in hard-to-distinguish cases.The mAP values of YOLOv5s-S model were respectively 0.93 and 0.733 on pod and stem regions,yielding increases of 1.4%and 4.7%over the YOLOv5s baseline.Fig.6 visually and quantitatively compares YOLOv5s and YOLOv5s-S for detecting pods and stems in a very densely grown plant.As it shows,YOLOv5s-S’ count of 147 pods was comparable to the count of 151 made by the human labelers who counted pods from the static image,whereas YOLOv5s’ count was only 140.

Fig.6.Comparison of PNPP counts.(A) The original image.(B) Pods detected by YOLOv5s.(C) Pods detected by SPP-extractor.
3.4.Comparison results of binarization and semantic segmentation
SPP-Extractor combines binarization and semantic segmentation for route search.Both components are required for accurate main stem and branch identification and their length calculation.Fig.7 compares branch extraction results using the binarized image only(Fig.7A)and when the semantically annotated regions are added on top (Fig.7B).Without semantic segmentation,as shown in Fig.7A,routing would follow any connected region,even when they are formed by overlapping pods,resulting in inaccurate PH and BL.In fact,if semantic segmentation were not used,on the test set,the overallR2coefficient for PH calculation would drop from 0.99 to 0.93,and the MAE would increase from 0.68 cm to 2.73 cm,as shown in Fig.4E.For many plants,the predicted heights deviate greatly from their true heights.This result showed that adding semantic segmentation could improve the fitness between the planned paths and the real stems and branches.
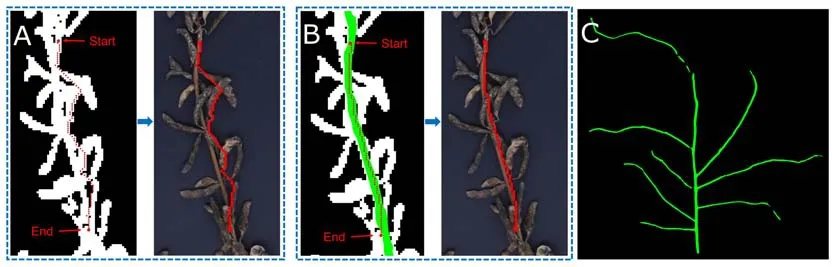
Fig.7.Comparison of three branch length calculation methods.(A)Using binarized images only.(B)Using both binarized and semantically annotated images.(C)Using the semantically annotated image only.
The U-Net and the U2-Net models were the two major options for the semantic segmentation subtask.Quantitatively,U-Net and U2-Net achieved respective mIoU values of 81.35% and 82.55% on our test set.Visually,as shown in Fig.S5,the U2-Net model could recognize more continuous regions on stems and branches.Accordingly,U2-Net was chosen as the semantic segmentation model for subsequent tasks.
If semantic segmentation were used alone without the binarized image,the stem regions would be disconnected,such as by being interrupted by pods that were accidentally placed on top of a stem,as can be seen in Fig.7C.This occurence is common when densely grown plants are placed onto a limited twodimensional plane.As a result,routing could be either incomplete or even infeasible.Adding the binarized images helps to reconnect these regions and form a plausible route.
3.5.Comparison results of main stem top bending and non-bending conditions
Treatment of bending tops affects mainly PH calculation,especially for plants with many pods concentrated at the top.Fig.4F plots the PH values calculated without the treatment against manually measured values.It is obvious that many of the plants in our dataset do have downward-bending main stems,as indicated by the points deviating from the regression line.For these images,the predicted PH values were all smaller than the true values.The bending-top treatment effectively solved this problem.As shown in Fig.4B,the deviating points in Fig.4F now match much better with the true PH values.In fact,the overallR2and MAE values improved from 0.87 and 2.07 cm when not using the treatment to 0.99 and 0.68 cm when the treatment was added.
4.Discussion
4.1.SPP-extractor can be an effective adjunct to manual measurement
SPP-extractor’s estimates are consistent with manual measurements.On most phenotypes,SPP-extractor’s mean absolute error fell within the allowed error range for human data collectors.On PNPP,PH,EBN,and BL,SPP-extractor’s MAE values were 6.34,0.68,0.04,and 0.97,whereas the error ranges set for humans were 6,1,1,and 1.
Only on PNPP,SPP-extractor’s MAE value 6.34 exceeded the allowable range for manual error.Manual counting was performed in a three-dimensional space,whereas SPP-extractor works on a two-dimensional plane.In fact,densely grown plants have made the task of counting PNPP from static images challenging for humans as well.When we asked the data labelers to count the PNPP while they label the images,their counting results achieved a coefficient of 0.956 with the ground truth,not much greater than SPP-extractor’s coefficient of 0.93.This indicates that the task itself is difficult: accurately identifying all pods in two-dimensional images.
SPP-extractor’s estimates are competitive with related methods.On PH,SPP-extractor achieved anR2of 0.99 and was comparable with those that used instance segmentation (with anR2of 0.98) [29] or used stem node identification and reconnection(with anR2of 0.99) [30].On EBN,in a comparison with a study[31] that identified branches using rotated target detection (with an MAE of 0.03),SPP-extractor offers two advantages.First,it does not require additional labeling of the branches: only the stem regions are annotated.Second,it can identify secondary branches without specifically annotating them.Fig.5B shows such an example: a plant with two secondary branches identified.None of the major branch and the secondary branch needs to be annotated specifically.On BL,no directly comparable results from calculating branch lengths of soybean plants have been reported.
What makes SPP-extractor appealing is its efficiency.On average,inference times for performing target detection and semantic segmentation on a single image were respectively 0.06 s and 1.34 s.Subsequent extraction of all the phenotype features required only 2.8 s.This was much faster than the manual measurement process,which took 5 to 10 min per image when preparing this dataset.The automated extraction process also requires no manual intervention,reducing labor costs.
4.2.Challenges for future improvement
SPP-extractor encountered difficulties in certain cases.When two branches accidentally intersect,correctly identifying the branches is challenging.When a single plant has many pods,e.g.,more than 200,as shown in Fig.4A,accurate pod counting is challenging.Using static images may not be sufficient in these cases.SPP-extractor’s capability to handle these scenarios awaits improvement.
5.Conclusions
We have proposed SPP-extractor,a unified algorithm for automatic extraction of four phenotypic features from soybean plants at harvest stage.SPP-extractor accommodates extensive overlap among pods and stems and does not require removal of pods from plants beforehand.To account for mutual overlap among pods and stems,we labeled only pod tips and augmented the YOLOv5s model with the SE attention mechanism for accurate detection.To solve the overlap of pods with stems,we combined target detection,semantic segmentation,image binarization,and path search for accurate PH and BL calculation.To accommodate concentrated pods on top of the main stem,we designed an effective search algorithm to measure bending tops.SPP-extractor adopts a unified framework for these subtasks,reducing tedious manual annotation work.Its performance in extracting the four phenotypic traits was comparable,and in many cases superior,to that of human data collectors.Its accuracy and efficiency could make it a helpful tool for automated phenotype feature extraction.
CRediT authorship contribution statement
Wan Zhou:Formal analysis,Visualization,Investigation,Methodology,Writing -original draft.Yijie Chen:Investigation,Data curation,Validation.Weihao Li:Investigation,Methodology.Cong Zhang:Validation.Yajun Xiong:Investigation,Data curation.Wei Zhan:Supervision,Funding acquisition.Lan Huang:Supervision,Methodology,Writing -review &editing.Jun Wang:Supervision,Project administration,Funding acquisition.Lijuan Qiu:Supervision,Project administration,Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the editors,the anonymous reviewers,and Professor James C.Nelson for their valuable comments.This work was supported by the National Natural Science Foundation of China(62276032,32072016) and the Agricultural Science and Technology Innovation Program (ASTIP) of Chinese Academy of Agricultural Sciences.
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.04.012.
- The Crop Journal的其它文章
- Reversible protein phosphorylation,a central signaling hub to regulate carbohydrate metabolic networks
- Genetic and environmental control of rice tillering
- High-throughput phenotyping of plant leaf morphological,physiological,and biochemical traits on multiple scales using optical sensing
- The R2R3-MYB transcription factor GaPC controls petal coloration in cotton
- The photosensory function of Zmphot1 differs from that of Atphot1 due to the C-terminus of Zmphot1 during phototropic response
- Disruption of LEAF LESION MIMIC 4 affects ABA synthesis and ROS accumulation in rice

