The pentatricopeptide repeat protein EMP601 functions in maize seed development by affecting RNA editing of mitochondrial transcript ccmC
Rongrong Chen,Qianhan Wei,Yan Liu,Jiankun Li,Xuemei Du,Yan Chen,Jianhua Wang,,Yunjun Liu,
a The National Forestry and Grassland Administration Engineering Research Center for Germplasm Innovation and Utilization of Warm-Season Turfgrasses,Jiangsu Key Laboratory for the Research and Utilization of Plant Resources,Institute of Botany,Jiangsu Province and Chinese Academy of Sciences (Nanjing Botanical Garden Mem.Sun Yat-Sen), Nanjing 210008,Jiangsu,China
b Institute of Crop Sciences,Chinese Academy of Agricultural Sciences,Beijing 100081,China
c College of Agronomy and Biotechnology,China Agricultural University,Beijing 100094,China
d College of Agronomy,Henan Agricultural University,Zhengzhou 450002,Henan,China
Keywords: Maize Empty pericarp 601 PPR Mitochondrial Ccmc Seed development
ABSTRACT Although several pentatricopeptide repeat(PPR)proteins are involved in post-transcriptional processing of mitochondrial RNA,it is unclear which specific protein is involved in the RNA editing of ccmC in maize(Zea mays).Here we report the identification of the maize empty pericarp 601 (emp601) mutant and the map-based cloning of the Emp601 gene,which encodes an E2-type PPR protein that is targeted to mitochondria.A single-nucleotide deletion in the emp601 mutant caused a frameshift and introduced a premature stop codon into the predicted EMP601.This mutation was associated with reduced accumulation of mitochondrial complex III as well as with inhibition of growth and differentiation of basal endosperm transfer layer cells,leading to final degeneration of the embryo and endosperm.We determine that loss of EMP601 function prevents the C-to-U RNA editing of the mitochondrial transcript ccmC at position 358.EMP601 binds to the ccmC transcript and directly interacts with Multiple organellar RNA editing factor 8 and may be a component of the plant mitochondrial editosome.We conclude that EMP601 functions in RNA editing of mitochondrial ccmC transcripts and influences mitochondrial function and seed development.
1.Introduction
Pentatricopeptide repeat(PPR)proteins function in RNA editing,splicing,stabilization,processing,and translation.Most PPR proteins localize to mitochondria or chloroplasts,and there appears to be minimal genetic redundancy between PPR genes[1].PPR proteins can be classified as members of P or PLS subfamilies.P subfamily PPR proteins contain an N-terminal organellar targeting sequence and classical P motif consisting of a 35-amino acid repeat.PPR proteins from the PLS subfamily also contain an Nterminal organellar targeting sequence as well as P,L,and S motifs and are further divided into PLS,E1,E2,E+,and DYW subgroups depending on the C-terminal domains they harbor[1].P subfamily PPR proteins can stabilize specific RNAs,activate or repress the translation of specific mRNAs,and stimulate RNA cleavage,while PLS subfamily PPR proteins function mainly in the editing of specific RNAs.
RNA editing in mitochondrial genes is a common phenomenon,and most editing events involve the conversion of cytidine (C) to uridine (U) at specific positions in the RNA [2].It has been estimated that more than 400 cytidine residues are changed to uridines in plant mitochondrial transcripts via RNA editing[3],which is required for producing functional mitochondrion-encoded and translated proteins.Many PPR proteins are involved in RNA editing of transcripts encoding components of mitochondrial electron transfer chain complexes.Arabidopsis thalianaSLOW GROWTH 4(SLO4) is involved in editingnad4[4],while maize (Zea mays)DEK40 is required for editing ofnad2andnad5[5],mutations of them reduce the activity of complex I.Mitochondrial editing factor 35 (MEF35) in Arabidopsis and Empty pericarp 21 (EMP21) and PPR2263 in maize function in editingcob1,and their loss of function impairs the assembly of complex III [6-8].Small kernel 4(SMK4) and EMP18 are required for editingcox1andcox2,respectively,and their mutations affect the activity of complex IV in maize [9,10].
In addition to PPR proteins,multiple organellar RNA editing factors (MORFs),organelle RNA recognition motif proteins (ORRMs),and organelle zinc finger motif-containing proteins (OZs) contribute to RNA editing [11].MORFs can interact selectively with PPR proteins and may form a bridge between the PPR proteins and the necessary deaminase in the editosome[12,13].Arabidopsis MEF13 was reported [14] to interact with MORF3 and MORF8 and to function in RNA editing at eight sites in mitochondrial mRNAs.In maize,DEK53 and PPR27 interact with MORF1,while EMP21 interacts with MORF8 [7,15,16].
One of the characteristics ofc-type cytochromes is that they are covalently bound to their heme cofactor via two thioether bonds between two vinyl side chains from heme and two cysteines from a conserved CXXCH motif.The term ‘‘cytochromecmaturation’’describes the covalent association ofc-type apocytochromes to their heme prosthetic groups [17].In plants,cytochromectransfers electrons from complex III to complex IV in the mitochondrial electron transfer chain,and cytochromec1is a component of complex III.The maturation of plant cytochromecrequires the action of cytochrome maturation (CCM) system I,which comprises nine proteins: CcmA-I,of which CcmA,CcmE,and CcmH are encoded by nuclear genes and the remaining six proteins are encoded in the mitochondrial genome.CcmA,CcmB,CcmC,and CcmD belong to the family of ATP-binding cassette transporters,translocating hemebtogether with CcmE;CcmG and CcmH are grouped in module 1 to maintain apocytochromecin a ligation-competent state.CcmF (CcmF is split into ccmFCand ccmFN(C-terminal and Nterminal part) in several species),CcmH,and CcmI form a membrane-integral complex(CcmFHI)in module 3 and participate in the ligation of apocytochromecwith hemeb[18].Ccmtranscripts are also post-transcriptionally modified to generate mature functional transcripts via RNA editing.In maize,EMP9 and EMP21 are required for editingccmB[7,19];DEK36,EMP7,and PPR27 are necessary for editingccmFN[16,20,21];and EMP21 is responsible for editingccmFC[7].Mutation of any of these genes affects the maturation of cytochromecand the activity of mitochondrial complex III,leading to defective kernel development.CcmC acts in cytochromecmaturation by recruiting heme to CcmE.Arabidopsis RNA PROCESSING FACTOR 3(RPF3)and RPF6 are required for the 5′maturation ofccmCtranscripts[22,23].In rice(Oryza sativa),OPAQUE AND GROWTH RETARDATION 1 (OGR1) is essential for RNA editing ofccmC-458[24],while rice PPR756 affects the RNA editing ofccmC-236 [25].In maize,DEK53 was reported [15] to affect the RNA editing of over 60 targets,including reducing editing efficiency atccmC-446 and increasing editing efficiency atccmC-421,ccmC-436,ccmC-497,ccmC-499,ccmC-568,andccmC-637.However,it is unclear which specific protein is required for RNA editing ofccmCin maize.
In this study,we identified a novel maizeempty pericarp(emp)mutant and cloned its candidate geneEmp601,which encodes an E2-type PPR protein.Loss of EMP601 function impaired the C-to-U RNA editing of the mitochondrialccmC-358,and reduced the accumulation of mitochondrial complex III.These results indicate that EMP601 acts in mitochondrial function and seed development by regulating RNA editing of mitochondrialccmCtranscripts.
2.Materials and methods
2.1.Plant materials and cultivation condition
Theemp601mutant was a spontaneous mutant isolated from the progeny of a hybrid plant derived from a cross between inbred lines Chang 7-2 and G391.The heterozygous (Emp601/emp601)plants were crossed with inbred line B73 to generate F1progenies,and the heterozygous F1were self-pollinated to generate F2.The F2progenies of B73 ×Emp601/emp601were used to perform the following experiments.All plants were grown in the field in Beijing or Hainan province,China.
2.2.Rescue of emp601 mutant embryos
The embryos were isolated from WT or mutant kernels with white pericarps and half-translucent appearance of the F2segregating ears 15 days after pollination (DAP),and cultured on solid Murashige and Skoog (MS) medium containing 20 g L-1sucrose under a 16-h-light/8-h-dark photoperiod at 25 °C.Regenerated seedlings were transferred into soil and grown in a greenhouse under the same photoperiod at 25-28 °C.
2.3.RNA extraction and reverse transcription quantitative PCR (qRTPCR)
Total RNA was isolated from 15-DAP immature embryos and endosperms of maizeemp601and WT kernels using an RNAprep pure plant kit (DP432,Tiangen,Beijing,China).One microgram of RNA was reverse-transcribed into first-strand cDNA using random hexamer primers using a TransScript II One-Step gDNA Removal and cDNA Synthesis SuperMix (AH311,Transgen,Beijing,China).Quantitative PCR (qPCR) was performed with an Applied Biosystems 7300 Real-time PCR System (7300,ABI,Foster city,CA,USA)using TransStart Green qPCR SuperMix (AQ101,Transgen,Beijing,China)according to the manufacturer’s protocol.ZmActinwas used as reference gene.Relative expression levels were calculated by the 2-ΔΔCTmethod [26].Primers are listed in Table S2.
2.4.Light microscopy of cytological sections
WT andemp601mutant kernels were collected from selfpollinated F2heterozygotes at 10,15,and 18 DAP.For preparation of paraffin sections,the kernels were cut along their longitudinal axes and fixed with FAA solution (63% ethanol,2% methanol,and 5% acetic acid,v/v/v).The sections were dehydrated in a graded ethanol series and alcohol benzene,xylene before being infiltrated with and embedded in paraffin.For preparation of resin sections,kernels were cut along their horizontal and longitudinal axes and fixed with 2.5% (w/v) glutaraldehyde.Resin sections were washed in phosphate buffer,fixed in 1% (w/v) osmic acid,and dehydrated in a graded ethanol series.After dehydration,the sections were infiltrated with and embedded in resin.The paraffin sections were cut using a microtome(RM2235,Leica,Frankfurt,Germany)into 8-μm sections,stained with toluidine blue,and observed using an Olympus SZX7 stereomicroscope (SZX7,Olympus,Tokyo,Japan).The resin sections were cut using a Leica EM UC7 ultramicrotome(EM UC7,Leica,Frankfurt,Germany)into 1.4-μm or 70-90-nm sections.The 1.4-μm sections were stained with toluidine blue and observed using an inverted fluorescence microscope (IX71,Olympus,Tokyo,Japan).The 70-90-nm sections were stained with uranyl acetate and lead citrate,collected on copper grids,and observed using a transmission electron microscope(7700,Hitachi,Tokyo,Japan).
2.5.Map-based cloning
The mapping population was developed by using the F2progeny of a B73 ×emp601/Emp601F1hybrid.Preliminary mapping was performed using bulked-segregant RNA-seq (BSR-seq).For finemapping,4308 homozygous mutant kernels from F2ears were genotyped with simple sequence repeat (SSR),single nucleotide polymorphism (SNP),or insertion/deletion (InDel) markers developed within the preliminary mapping interval.TheEmp601locus was delimited to a 248-kb region.Candidate genes in this region were identified in MaizeGDB (https://www.maizegdb.org/) based on the B73 reference genome.These candidate genes in theemp601mutant and WT kernels were amplified by PCR and sequenced to identify mutations.Primers are listed in Table S2.
2.6.Complementation test of the emp601 mutant
For functional complementation tests,a 1689-bp coding sequence region ofEmp601was cloned into the pCAMBIA3301 vector between theBglII andBstEII sites under the control of the CaMV 35S promoter.This construct was transformed intoAgrobacterium tumefaciensstrain EHA105.Embryos from the maize hybrid HiII were transformed as previously described [27].Transgenic maize lines with a single T-DNA insertion were crossed to heterozygousemp601/Emp601plants to evaluate rescue of the mutant phenotype.The individualemp601/Emp601;T/-plants were selfpollinated to produce an F2population.The segregation ratio of F2segregating ears of this population was calculated to determine theEmp601gene.Ears fromemp601/emp601;T/-plants andEmp601/Emp601;-/-plants were used to evaluate rescue of the mutant phenotype.Primers are listed in Table S2.
2.7.Subcellular localization of EMP601
A 1214-bp coding sequence fragment encoding the N terminus of EMP601 was cloned into the pGWC vector and then into the pEarleyGate 101 vector using the Gateway cloning system [28].The resulting vector was transformed into Arabidopsis by the floral dip method [29].The root hairs of three independent transgenic lines grown on MS medium for 7 days were stained with Mito-Tracker Red (M7512,Invitrogen,Carlsbad,CA,USA) and observed under a Zeiss LSM700 confocal laser-scanning microscope(LSM700,Zeiss,Oberkochen,Germany).Primers are listed in Table S2.
2.8.Analysis of mitochondrial RNA editing
Total RNA was isolated from 15-DAP immature embryos and endosperms of maizeemp601and WT kernels as described above.The full-length coding sequences of 35 mitochondrial transcripts inemp601and WT kernels at 15 DAP were amplified and the resulting PCR products were sequenced as described previously[30].The WT andemp601sequences were aligned to determine the status of RNA editing in the 35 mitochondrial transcripts.The unique unedited site identified inccmCin theemp601mutant was further confirmed in 20 and 25-DAPemp601and WT kernels.Primers are listed in Table S2.
2.9.Blue native-PAGE (BN-PAGE) and NADH dehydrogenase activity assay
A sample of 15 g of immature kernels without pericarps was ground to powder with a mortar and pestle in liquid nitrogen before isolation of mitochondria as described previously[31] with some modifications.The fraction enriched in mitochondria was solubilized using a NativePAGE sample prep kit (BN2008,Invitrogen,CA,USA) according to the manufacturer’s instructions.The solubilized mitochondria were loaded onto a 3% to 13% polyacrylamide separation gel and run at 150 V for 5 h.The gel was then stained with Coomassie Brilliant Blue.An in-gel NADH dehydrogenase activity assay was performed as previously described [31].
2.10.Immunoblotting
Mitochondrial proteins were separated on a 12% (w/v) polyacrylamide,0.1% (w/v) SDS gel and transferred to a nitrocellulose membrane using a semi-dry blotter (Bio-Rad).The immunoblot signal was visualized with a Super Signal ELISA Pico chemiluminescent substrate kit (37069,Thermo Fisher Scientific,Waltham,MA,USA) according to the manufacturer’s instructions.Antibodies against cytochromec1(AS08 343A,Agrisera,Vännäs,Sweden)and AOX1/2 (AS04 054,Agrisera,Vännäs,Sweden) were diluted 1/1000 and HRP-conjugated rabbit anti-goat IgG(D110117,Sangon Biotech) was diluted 1/8000.
2.11.RNA electrophoretic mobility shift assay (REMSA)
The coding sequence ofEmp601was amplified and cloned into the pMAL vector to produce and purify the recombinant protein(MBP-EMP601).RNA probes were synthesized and labeled with 5′-biotin by Tsingke Biotechnology Co.,Ltd.(Beijing,China).To authenticate the predicted binding site between EMP601 andccmCtranscripts,REMSA was performed using a LightShift Chemiluminescent RNA EMSA (REMSA) Kit (Thermo Fisher Scientific,USA).The recombinant protein and RNA probes were prepared in 20-μL reactions containing 2 μL 10× binding buffer,2 μL 50% (v/v)glycerol,and 0.2 μL tRNA (10 mg mL-1).Each binding reaction was incubated at room temperature for 20-30 min.The reaction mixture was then electrophoresed on a 5% polyacrylamide gel and transferred to a nylon membrane at 400 mA (~35 V) for 30 min.The signal was detected after blocking and detection incubations using LightShift Chemiluminescent RNA EMSA Kit (20158,Thermo Fisher Scientific,USA) according to the manufacturer’s instructions.
2.12.Yeast two-hybrid assay
The full-length coding sequences ofEmp601andZmMORF8were individually cloned into the GAL4 DNA-binding domain vector (pGBKT7-BD).The full-length coding sequences ofZmMORF1,
ZmMORF3,ZmMORF8,ZmORRM,ZmORRM2-1,ZmORRM2-2,ZmORRM3-1,ZmORRM3-2,ZmORRM4,andZmORRM5were individually cloned into the GAL4 activation domain vector(PGADT7-AD).Several combinations of BD and AD fusion constructs were cotransformed into the yeast strain Y2HGold (630498,Clontech,Mountain View,USA).The combination of pGBKT7-53 (human P53)and pGADT7-T(T-antigen)was used as positive control.Yeast manipulations were performed according to the instructions of the Frozen-EZ Yeast Transformation II Kit (MKbio).All interactions were tested on synthetic defined (SD) medium lacking Leu and Trp (double dropout,DDO) and SD medium lacking Ade,His,Leu,and Trp(quadruple dropout,QDO)for 4 days at 30°C as previously described [32].
2.13.Luciferase complementation imaging assays
The coding sequences ofEmp601andZmMORF8without stop codons were cloned into p1300-35s-cLUC and p1300-35s-nLUC vectors using a ClonExpress Ultra One Step Cloning Kit (C112,Vazyme Biotech,Nanjing,Jiangsu,China).The LCI assay was performed on 4-week-oldNicotiana benthamianaleaves transiently infiltrated with various combinations ofAgrobacteriumstrain GV3101 harboring various vectors.After incubation for 48 h under a 16-h-light/8-h-dark photoperiod,the leaves were infiltrated with 1 mmol L-1luciferin(Promega).The resulting luciferase signal was acquired with an automatic chemiluminescence image analysis system (Tanon 5200).
3.Results
3.1.Phenotype of the emp601 mutant
Self-pollinated progenies ofemp601/Emp601heterozygous plants showed a 3:1 segregation ratio between wild-type (WT)andempkernels (Fig.1;Table S1),indicating that theempphenotype is controlled by a single recessive nuclear gene.emp601mutant kernels were easily distinguished from WT kernels at 12 days after pollination (DAP),based on their white pericarps and half-translucent appearance (Fig.1A).Matureemp601mutant kernels were not viable and failed to germinate.We attempted to rescue immatureemp601embryos at 15 DAP by transferring them to Murashige and Skoog (MS) medium.Although we obtained a few successfully growingemp601seedlings,they showed dramatically delayed growth compared to WT seedlings from similarly rescued embryos and never reached reproductive maturity(Fig.S1).

Fig.1.Representative phenotype of maize emp601 kernels(A,B)F2 ears segregating 3:1 for wild-type(WT)and emp601 mutant kernels.(A)Ear at 15 DAP.(B)Mature ear.(CJ) Longitudinal sections of WT and emp601 kernels.(C-E) WT kernels at 10 (C),15 (D),and 18 (E) DAP;(F-H) emp601 kernels at 10 (F),15 (G),and 18 (H) DAP.(I,J)Magnifications of(E)and(H),respectively.SC,scutellum;COL,coleoptile;SAM,shoot apical meristem;LP,leaf primordia;RAM,root apical meristem.Scale bars,0.5 mm.(KP)Development of the basal endosperm transfer cell layer(BETL).The transfer cells are indicated by the red arrows.(K,M,O)Sections of WT kernels at 10(K),15(M),and 18(O) DAP;(L,N,P) sections of emp601 kernels at 10 (L),15 (N),and 18 (P) DAP.Scale bars,50 μm.
To determine at which stage kernel development arrested in theemp601mutant,we examined WT andemp601kernels from the same F2segregating ear using paraffin sections and microscopy.In WT kernels at 10 DAP,the endosperm had began to fill the pericarp and the embryos had reached the coleoptilar stage and began to differentiate(Fig.1C).By contrast,inemp601kernels,the endosperm was reduced and the embryo had reached only the transition stage (Fig.1F).At 15 DAP,WT endosperm filled the pericarp,and the embryos continued to differentiate,having established the scutellum,coleoptiles,leaf primordia,root apical meristem,and shoot apical meristem (Fig.1D).However,emp601kernels were wrinkled and their endosperm did not fill the pericarp,resulting in a large empty space between the endosperm and pericarp.The embryos grew larger but remained at the transition stage(Fig.1G).At 18 DAP,WT embryos were larger than that at the earlier stages with differentiated scutellum,coleoptile,leaf primordium,shoot apical meristem,and root apical meristem(Fig.1E,I),whereasemp601embryos had reached the coleoptilar stage and appeared to differentiate (Fig.1H,J).At maturity,emp601kernels were composed mostly of pericarp (Fig.1B),suggesting that the embryo and endosperm had degenerated during the later stage of kernel development.
In addition to the arrested embryo and endosperm,we observed that the BETL region in theemp601kernels was severely affected.At 10 DAP,the BETL region of WT kernels consisted of relatively large and elongated cells with extensive cell-wall ingrowths (Fig.1K),whereas the BETL region of theemp601kernels was almost absent(Fig.1L).At 15 DAP,there were more cell wall ingrowths in the BETL region of WT kernels (Fig.1M).By contrast,BETL cells ofemp601kernels were small and square and showed almost no cell-wall ingrowths(Fig.1N).At 18 DAP,cell-wall ingrowths in the BETL cells ofemp601kernels had differentiated but much less than those of the WT (Fig.1O,P).Thus,loss of EMP601 function inhibited the growth and differentiation of BETL cells.
3.2.Map-based cloning of Emp601
We maintained theemp601mutation inemp601/Emp601heterozygous plants by successive self-pollination.To generate a mapping population,we crossed a heterozygous plant (emp601/Emp601) to the inbred line B73.F1plants with theemp601/Emp601genotype were self-pollinated to generate a segregating F2population.At 15 DAP,we collected kernels showing theempphenotype for mapping by employing bulked segregant RNA-seq(BSR-seq),which located theEmp601gene on the long arm of chromosome 6 (Fig.S2).
For the fine-mapping ofEmp601,we genotyped 4308 kernels with theempphenotype,narrowing the candidate interval to a 248-kb region flanked by the SNP markers M5 and M6,between 115,494,237 and 115,742,129 bp on chromosome 6 in the B73 reference genome (RefGen V4 at https://www.maizegdb.org/).This region contained six putative genes: Zm00001d037199,Zm00001d037200,Zm00001d037201,Zm00001d037202,Zm00001d037203,and Zm00001d037204 (Fig.2A).To identifyEmp601,we amplified the coding regions of each gene and subjected them to direct sequencing.Alignment with the WT sequences revealed a 1-bp deletion in Zm00001d037203 in theemp601mutant,causing a frameshift and a premature stop codon(Fig.2B),whereas the sequences of the other genes did not differ between WT andemp601.We assigned Zm00001d037203 as the candidateEmp601gene.It encodes a PPR protein with fourteen PPR motifs (Fig.2C).

Fig.2.Map-based cloning of Emp601 and complementation test.(A)The Emp601 gene was mapped to a 248-kb region on chromosome 6 containing six putative genes.(B)One gene contained a 1-bp deletion in the emp601 mutant,leading to the introduction of a premature stop codon.(C) Schematic diagrams of EMP601 and the predicted truncated EMP601 protein produced by the emp601 mutant.(D) Complementation test of the emp601 mutant.Heterozygous emp601/Emp601 plants were pollinated with three independent transgenic maize lines harboring a transgene expressing the WT copy of the candidate gene.F2 ears that were heterozygous for both the emp601 mutation and the transgene showed a 15:1 segregation ratio between WT and mutant kernels.Scale bars,1 cm.
To confirm that Zm00001d037203 wasEmp601,we performed a transgenic complementary assay.Segregating ears ofemp601/Emp601;T/-showed a segregation ratio of 15:1 (245:20,χ2=0.56;368:23,χ2=0.04;442:32,χ2=0.13) between WT andempkernels (Fig.2D).These results indicate that the transgene can rescue theempphenotype and confirm that the observed mutation in Zm00001d037203 is responsible for theemp601mutant phenotype.
3.3.Emp601 is ubiquitously expressed and encodes an E2-type PPR protein that is targeted to mitochondria
Emp601encodes a PRR protein from the E2-type subgroup with 14 PPR motifs,an E1 motif (amino acids [aa] 484-519) and an E2 motif (aa 520-553) at its C terminus,and a signal peptide (aa 1-10) at its N terminus (Fig.2C) [33,34].Most PPR proteins are targeted predominantly to plastids or mitochondria[1].In agreement with this rule,a mitochondrial signal peptide was predicted at the N-terminal end of EMP601 by both TargetP (https://www.cbs.dtu.dk/services/TargetP) and PREDOTAR (https://urgi.versailles.inra.fr/Tools/Predotar) (Fig.2C).To determine the subcellular location of EMP601,we cloned the sequence encoding the N-terminal region of EMP601 (aa 1-404) in-frame and upstream of the yellow fluorescent protein(YFP)sequence and transformed the resulting construct into Arabidopsis.We examined YFP fluorescence by confocal microscopy in the root hairs of transgenic seedlings,together with MitoTracker Red staining as a control for mitochondrial localization.We observed co-localization of YFP signals with MitoTracker Red signals(Figs.3A,S3),indicating that EMP601 localizes to mitochondria.We also investigated theEmp601expression pattern in wild-type maize by reverse transcription quantitative PCR (qRTPCR)and found thatEmp601was expressed in all tissues examined(Fig.3B).

Fig.3.Subcellular location of EMP601,expression pattern of Emp601,and mitochondria showing altered morphology in emp601. (A) Subcellular location of EMP601 in mitochondria.Transgenic Arabidopsis lines harboring a 35S:Emp601-YFP transgene were generated,and the YFP fluorescence signal was observed in the root hairs of T1 seedlings.MitoTracker Red was used to label mitochondria.The merged image shows the overlap between YFP and MitoTracker Red signals.Scale bars,20 μm.(B)Relative Emp601 expression in six tissues.The expression level of Emp601 in the root was set to 1,and values are means ± SEM of three independent biological replicates.(C-F)Transmission electron microscopy images of the WT (C,E) and the emp601 mutant (D,F).(C,D) Scale bars,10 μm;(E,F) Scale bars,1 μm.Panels E and F show magnified images of C and D,respectively.Mt,mitochondria;Pb,protein body.
3.4.Loss of EMP601 function affects the formation of mitochondria
The proper formation of the cristae membranes in mitochondria is critical for the function of the electron transport chain(ETC)[35].We accordingly investigated,by examining endosperm cells by transmission electron microscopy,whether the loss of EMP601 function in theemp601mutant compromised mitochondrial ultrastructure.At 15 DAP,WT mitochondria formed dense and normal inner membranes (Fig.3C,E),whereasemp601mitochondria showed a poorly developed membrane system without normal cristae structures (Fig.3D,F).These results suggest that EMP601 function is essential for mitochondrial structure differentiation at an early stage of kernel development.
3.5.Complex III is severely affected in the emp601 mutant
To further investigate the consequences of the loss of EMP601 function to the assembly of mitochondrial complexes,we performed BN-PAGE and in-gel NADH dehydrogenase activity assays.Complex III and complex I+III2were less abundant in theemp601mutant,whereas there were no differences in complex I or complex IV between WT andemp601kernels (Fig.4A).The ingel NADH dehydrogenase activity assay confirmed that complex I+III2activity is dramatically reduced inemp601relative to WT,whereas complex I activity was comparable between the two genotypes (Fig.4B).To further confirm the loss of mitochondrial complex III assembly inemp601,we performed immunoblotting using an antibody against cytochromec1.We detected a very weak signal for cytc1in theemp601mutant relative to the WT(Fig.4C),a finding consistent with the disrupted assembly of complex III in mitochondria in theemp601mutant.Complex III and complex I+III2assembled normally in kernels from transgenic complementation lines(Fig.4D,F),and an in-gel NADH dehydrogenase activity assay showed that the activities of complex I and complex I+III2were comparable between kernels from transgenic complementation lines and control nontransgenic kernels(Fig.4E).The presence of the35S:Emp601transgene brought the abundance of cytochromec1in transgenic kernels to levels comparable to those of nontransgenic kernels (Fig.4F).
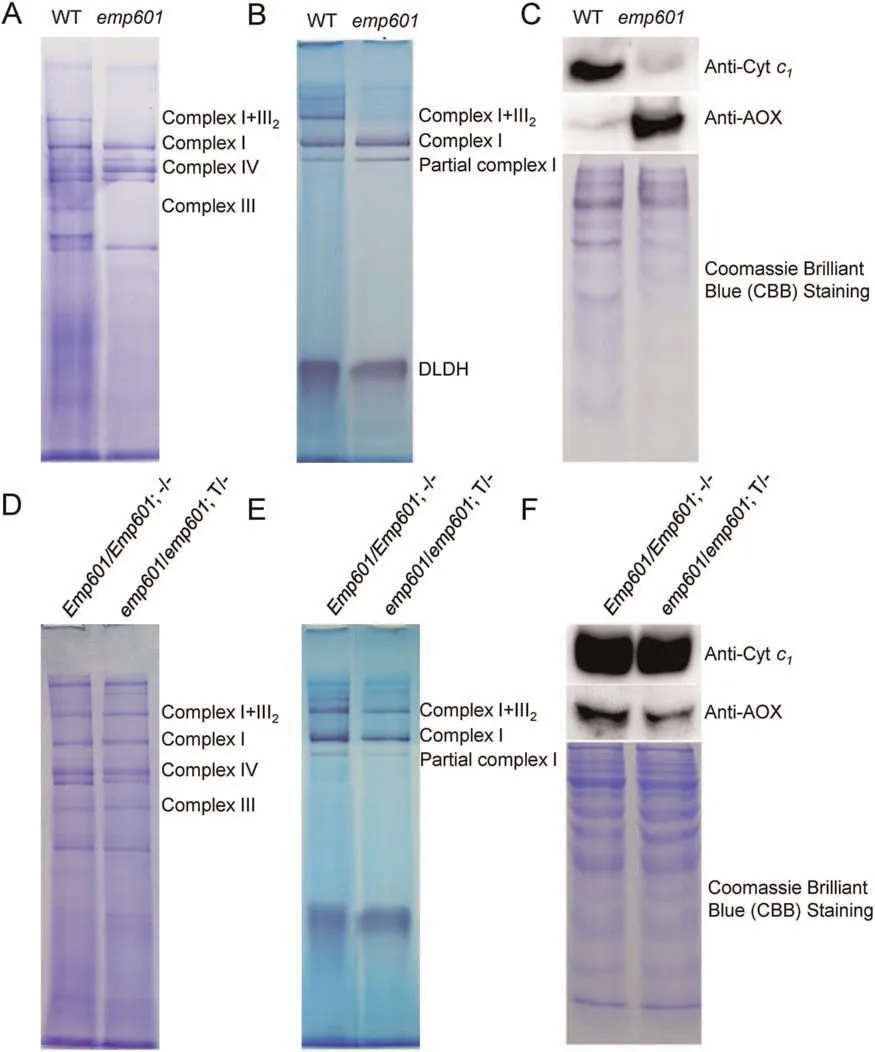
Fig.4.Mutation of Emp601 abolishes the activity of complex III.(A-C) Analysis of mitochondria isolated from WT and emp601 mutant kernels.(D-F) Seeds with the genotypes Emp601/Emp601;-/-and emp601/emp601; T/-from F2 ears of the same transgenic complementation line were grown,and the plants were self-pollinated to generate F3 ears.Mitochondria were isolated from WT-like kernels of F3 ears.(A,D)Blue native polyacrylamide gel stained with Coomassie Brilliant Blue(CBB).(B,E)In-gel NADH dehydrogenase activity assay.Mitochondrial protein complexes were separated on a blue native polyacrylamide gel with dihydrolipoamide dehydrogenase(DLDH)as a loading control,and NADH dehydrogenase activity was evaluated.(C,F)Immunoblot analysis with antibodies against cytochrome c1(subunit of complex III)and AOX.CBB staining of the gel was used as a loading control.
When the mitochondrial respiratory complex is blocked,levels of alternative oxidase (AOX) increase to compensate for malfunction of the ETC [36].Theemp601mutant showed high AOX abundance,whereas WT kernels showed only low levels (Fig.4C).Again,the introduction of the35S::Emp601transgene rescued AOX abundance in kernels from transgenic complementation lines(Fig.4F).These results suggest that the loss ofEmp601function impairs the assembly and activity of complex III,thus substantially impairing respiration inemp601mitochondria,leading to upregulation of the alternative respiratory pathway.
3.6.EMP601 is required for C-to-U RNA editing of ccmC-358
That E-PPR proteins are involved in RNA editing[1]suggested a potential functional target for EMP601.Analysis of mitochondrial RNA editing showed that C-to-U RNA editing of theccmC-358 site was entirely abolished inemp601,causing an amino acid change from tryptophan (W) to arginine (Fig.5A) in the highly conserved tryptophan-rich motif WGXXWXWD of CcmC (Fig.5B).This conserved residue functions in the interaction between CcmC and heme to form holo-CcmE during cytochromematuration [37,38].In the transgenic maize complementation lines (emp601/emp601;T/-),the editing of theccmC-358 site was restored from C to U(Fig.5A).We also observed lower editing efficiencies at the sitesccmFN-137,ccmFN-190,ccmFN-743,ccmFN-790,andccmFN-824 in theemp601mutant,wherea the editing efficiencies at these five sites were comparable to those of the WT in the complementation transgenic plants (Fig.S4).

Fig.5.EMP601 functions in RNA editing of ccmC transcripts at position 358 by binding to ccmC transcripts.(A)Sequencing chromatograms for ccmC from the WT,the emp601 mutant,and a complementation line,and the corresponding amino acid changes are shown.(B)Multiple sequence alignment of the conserved domain of ccmC protein from 16 plant species.The WWD domain of ccmC is indicated by the red line,and the edited site is indicated by an asterisk.(C)Proposed PPR recognition code of EMP601 for RNA sequences.The matching target nucleotides in ccmC are indicated in orange and the editing site is indicated in red.(D)RNA EMSAs indicating that EMP601 directly binds ccmC transcripts.
3.7.EMP601 directly binds ccmC transcripts
Based on the proposed code for recognition of RNA sequences by PRR proteins [39,40],we predicted the binding sites of EMP601.Nine predicted codes matched sequences upstream of theccmC-358 editing site (Fig.5C).To assess whether EMP601 bound to the -17 to -3 bp upstream of theccmC-358 editing site,we performed an RNA electrophoretic mobility shift assay(REMSA).To this end,we purified recombinant EMP601-MBP(maltose-binding protein)fromE.coliBL21 and chemically synthesized a 15-bp biotinylated RNA probe (-17 to -3 bp upstream of theccmC-358 editing site).We observed a clear shift when the biotinylated RNA was incubated with recombinant EMP601-MBP,indicating that EMP601 binds toccmCtranscripts at the predicted binding site(Fig.5D).Moreover,ccmCtranscripts were more abundant inemp601kernels than in WT and transgenic complementation kernels,as shown by qRT-PCR (Fig.S5).The transcript levels ofccmB,ccmFC,ccmFN,andcob,whose encoded proteins also participate in the assembly of complex III,were also higher inemp601relative to the WT,and all butccmFCaccumulated to similar levels in WT and complementation transgenic kernels (Fig.S5).
3.8.EMP601 interacts with ZmMORF8
MORFs participate in C-to-U RNA editing in plant mitochondria[11].The maize genome encodes six putative MORF orthologs,of which three (ZmMORF1 [Zm00001d024674],ZmMORF3[Zm00001d026307],and ZmMORF8 [Zm00001d048291]) localize to mitochondria [7].We tested the potential for interaction between EMP601 and the three mitochondrion-targeted ZmMORFs in a yeast two-hybrid assay.EMP601 interacted with ZmMORF8 but not with ZmMORF1 or ZmMORF3 (Fig.6A).We confirmed the interaction between EMP601 and ZmMORF8 by performing a luciferase complementation imaging (LCI) assay (Fig.6B).These results suggested that EMP601 and ZmMORF8 might form an editosome involved in the RNA editing ofccmC-358.

Fig.6.EMP601 interacts with ZmMORF8.(A) Yeast two-hybrid assay showing interaction between EMP601 and ZmMORF8.EMP601 did not interact with other MORF or ORRM proteins.(B) Luciferase complementation imaging (LCI) assay confirming the interaction between EMP601 and ZmMORF8.
Organelle RNA recognition motif proteins (ORRMs) are also involved in RNA editing,and the maize genome encodes seven mitochondrion-targeted ORRMSs: ZmORRM2-1 [Zm00001 d024675],ZmORRM2-2 [Zm00001d006566],ZmORRM3-1[Zm00001d041750],ZmORRM3-2 [Zm00001d023734],ZmORRM4[Zm00001d022065],ZmORRM5 [Zm00001d012052],and ZmORRM [Zm00001d041949] [15].A yeast two-hybrid assay showed that EMP601 did not interact with any of these editing factors(Fig.6A).Given that Arabidopsis ORRMs have been reported to interact with MORF8,we asked whether the same might hold true in maize.In another yeast two-hybrid assay,ZmMORF8 interacted with ZmORRM3-1,ZmORRM3-2,and ZmORRM4(Fig.S6),suggesting that EMP601,ZmMORF8,and a subset of ZmORRMs might form an editosome to editccmC-358.
4.Discussion
4.1.EMP601 is essential for C-to-U editing of the ccmC-358 site
PPR proteins from the PLS subfamily mainly function in RNA editing of specific RNAs [1].We identified EMP601,an E2-type PPR protein of the PLS subfamily that is essential for the C-to-U editing of theccmC-358 site.Although several PLS members in maize have been reported to be involved in the RNA editing[5,7,8,9,10,15,16,19,20,21,30],but none was reported to specifically function in the RNA editing ofccmC.Maize Dek53 was shown to affect more than 60 RNA editing sites,including decreased efficiency at theccmC-446 site and increased efficiency at theccmC-421,ccmC-436,ccmC-497,ccmC-499,ccmC-568,andccmC-637 sites in thedek53mutant relative to the WT [15].In contrast to these changes in editing efficiency ofccmCindek53,loss of EMP601 function completely abolished the editing ofccmC-358 in theemp601mutant,indicating that EMP601 is essential for RNA editing ofccmC.
CcmC functions in the cytochrome maturation system by recruiting heme to CcmE,and CcmC contains a conserved WWD motif in the second periplasmic loop and two histidine residues in the first and third periplasmic loops,which are required for the interaction of CcmC with heme [41].The conserved residues W119,G120,W123 (equivalent to W120 in maize),V124,W125,and D126 within the tryptophan-rich WWD motif ofE.coliCcmC participate in holo-CcmE formation.Single amino acid changes W119A,G120A,W123A,W125I,D126A and deleting V124(ΔV124)inE.coliCcmC inhibited the covalent attachment of heme to CcmE,showing that these residues are critical for CcmC activity[42].Inemp601,the lack of editing atccmC-358 changed the W120 residue typical of the WT protein to R120 within the conserved WWD motif,a change that would be expected to affect cytochromecmaturation and thus the activity of mitochondrial complexes.Indeed,we detected much lower complex III levels and severely impaired mitochondrion formation inemp601kernels.About 17%of complex I was estimated to be involved in the formation of the I+III2supercomplex [43],a finding consistent with the observed loss of supercomplexes I+III2inemp601kernels.As in otherempmutants with defects in RNA editing of some mitochondrial transcripts [19,21,30],the loss of editing ofccmC-358 inemp601caused defective kernel development.
In addition to the complete loss of editing atccmC-358,RNA editing efficiency decreased at severalccmFNsites inemp601.Many PPR proteins are involved in RNA editing at multiple sites,such as in Arabidopsis MEF13[14]or maize EMP5[30]and Dek53[15].PPR proteins recognize and bind to specific RNA sequences following the one PPR repeat:one RNA nucleotide rule,although some PPR proteins can also interact with other proteins or with DNA[44,45].According to the proposed PPR recognition codes [39,40],we identified a nine-nucleotide sequence upstream of the editing siteccmC-358 matching the predicted codes for EMP601,and we provided support for the binding of EMP601 toccmCtranscripts by REMSA.These results suggest that a strong and specific binding of PPR proteins to their target RNA is necessary for RNA editing.How PPR proteins recognize and bind target sites awaits further study.
4.2.EMP601 interacts with MORF8 and may be a component of the plant mitochondrial editosome
MORFs can interact with PPR proteins and may form a bridge between PPR proteins and the deaminase in the editosome[12,13].Arabidopsis MEF13 interacts with MORF3 and MORF8 for RNA editing at eight targets in mitochondrial RNAs [14].Maize Dek53 and PPR27 interact with MORF1,and EMP21 interacts with MORF8 to edit multiple RNA targets [7,15,16].We detected an interaction between EMP601 and ZmMORF8,but not with other ZmMORFs.ZmMORF1 was reported as well as found in this study(Fig.S6) also to interact with ZmMORF8 [16].Likewise,MEF35-MORF1-MORF8 or MEF13-MORF3-MORF8 complexes contribute to RNA editing in Arabidopsis [6,14].Perhaps EMP601 forms a complex editosome with ZmMORF8 and ZmMORF1 to regulate RNA editing ofccmC-358.
Organelle RNA recognition motif proteins (ORRMs) are also components of RNA editosomes [11].Arabidopsis ORRMs interact with MORF8.Our finding that ZmMORF8 interacting with ZmORRM3-1,ZmORRM3-2,and ZmORRM4 (Fig.S6) suggests that EMP601,ZmMORF8,and ZmORRMs may form an editosome to editccmC-358.Based on the results of this and previous studies[6,14],we propose a model for the editing of mitochondrialccmCtranscripts by maize EMP601 (Fig.7).EMP601 binds to theccmCRNA target site,while ZmMORF8 acts as a bridge holding EMP601,ZmORRM3-1,ZmORRM3-2,and ZmORRM4 together to form editosomes for the editing of theccmC-358 target site.Further investigation might reveal whether additional components are included in the editosome(s).

Fig.7.Working model of EMP601 for the editing of ccmC transcripts.EMP601 binds to the RNA target upstream of ccmC-358.EMP601 interacts with MORF8,which acts as a bridge holding EMP601,ZmORRM3-1,ZmORRM3-2,and ZmORRM4 together to form editosomes for the editing of the ccmC-358 target site.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Rongrong Chen:Methodology,Investigation,Data curation,Writing-original draft,Writing-review&editing,Funding acquisition.Qianhan Wei:Methodology,Investigation,Data curation,Writing -original draft.Yan Liu:Investigation.Jiankun Li:Methodology,Data curation.Xuemei Du:Data curation.Yan Chen:Methodology.Jianhua Wang:Conceptualization,Supervision,Data curation,Writing -review &editing.Yunjun Liu:Conceptualization,Supervision,Writing -review &editing,Funding acquisition.
Acknowledgments
This work was supported by the Agricultural Science and Technology Innovation Program of CAAS,the Research Program of Sanya Yazhou Bay Science and Technology City (SKJC-2020-02-005),and the Natural Science Foundation of Jiangsu Province(BK20200288).
Appendix A.Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2023.03.004.
- The Crop Journal的其它文章
- Reversible protein phosphorylation,a central signaling hub to regulate carbohydrate metabolic networks
- Genetic and environmental control of rice tillering
- High-throughput phenotyping of plant leaf morphological,physiological,and biochemical traits on multiple scales using optical sensing
- The R2R3-MYB transcription factor GaPC controls petal coloration in cotton
- The photosensory function of Zmphot1 differs from that of Atphot1 due to the C-terminus of Zmphot1 during phototropic response
- Disruption of LEAF LESION MIMIC 4 affects ABA synthesis and ROS accumulation in rice

