Insights into microbiota community dynamics and flavor development mechanism during golden pomfret (Trachinotus ovatus) fermentation based on single-molecule real-time sequencing and molecular networking analysis
Yueqi Wng,Qin Chen,Hun Xing,Dongxio Sun-Wterhouse,c,Shengjun Chen,Yongqing Zho,Liho Li,Ynyn Wu,*
a Key Laboratory of Aquatic Product Processing,Ministry of Agriculture and Rural Affairs of The People’s Republic of China,National R&D Center for Aquatic Product Processing,South China Fisheries Research Institute,Chinese Academy of Fishery Sciences,Guangzhou 510300,China
b Co-Innovation Center of Jiangsu Marine Bio-industry Technology,Jiangsu Ocean University,Lianyungang 222005,China
c School of Chemical Sciences,The University of Auckland,Auckland 92019,New Zealand
d Collaborative Innovation Center of Seafood Deep Processing,Dalian Polytechnic University,Dalian 116034,China
Keywords:Fermented golden pomfret Microbiota community Volatile compound Co-occurrence network Metabolic pathway
ABSTRACT Popular fermented golden pomfret (Trachinotus ovatus) is prepared via spontaneous fermentation;however,the mechanisms underlying the regulation of its flavor development remain unclear.This study shows the roles of the complex microbiota and the dynamic changes in microbial community and flavor compounds during fish fermentation.Single-molecule real-time sequencing and molecular networking analysis revealed the correlations among different microbial genera and the relationships between microbial taxa and volatile compounds.Mechanisms underlying flavor development were also elucidated via KEGG based functional annotations. Clostridium,Shewanella,and Staphylococcus were the dominant microbial genera.Forty-nine volatile compounds were detected in the fermented fish samples,with thirteen identified as characteristic volatile compounds (ROAV >1).Volatile profiles resulted from the interactions among the microorganisms and derived enzymes,with the main metabolic pathways being amino acid biosynthesis/metabolism,carbon metabolism,and glycolysis/gluconeogenesis.This study demonstrated the approaches for distinguishing key microbiota associated with volatile compounds and monitoring the industrial production of high-quality fermented fish products.
1. Introduction
Ferment golden pomfret (Trachinotus ovatus,family Bramidae)is a popular traditional fermented food in Asian countries,including Japan,Korea,Thailand,and China.It is traditionally prepared via spontaneous fermentation that involves enzymatic and chemical decomposition and biotransformation of organic substances in raw materials,enabling unique flavor and increased nutritional value of the end product[1].The development of the typical flavor of fermented golden pomfret was reported in close association with the combined action of endogenous enzymes and microorganisms[2].Our research team found that the number and concentration of volatile compounds increased during the early stage,but the amounts of certain volatile compounds decreased during golden pomfret fermentation[1].The microorganisms contained in the fermented golden pomfret can function as “microbial factories” and induce various hydrolysis reactions (e.g.,proteolysis and lipolysis).A range of reactions occurs during golden pomfret fermentation including amino acid breakdown via Strecker degradation,the Maillard reaction between amino acids/amines and reducing sugars,and lipid degradation/oxidation esterification between alcohols and acids[1].Similar to other fermented fish products,the flavor profiles of fermented golden pomfret are determined by the metabolic activities of microorganisms in the fermentation system[3].
Spontaneous fermentation is an age-old practice and induced by naturally occurring microflorae.The microbiota community is closely associated with the fermenting matrix,presenting complex relationships among microorganisms,including amensalism,parasitism,commensalism,mutualism,and competition[3].Therefore,monitoring the interactions among microorganisms is essential to tailoring the flavor profile of fermented fish products.During spontaneous fermentation,the microbial community may change in response to the nature of starting raw material,fermentation process variables and environment.Thus,the main challenges with the spontaneous fermentation process consist of the production of consistent high-quality fermented products,prediction of contamination,and safety risks attributed to undesirable/unexpected microorganisms in fermented foods[3].For the fermented golden pomfret produced via spontaneous fermentation,the challenges include low raw material conversion,sensitivity to environmental changes,and inconsistent quality (e.g.,production of insufficient desirable flavor,unpleasant flavor;and undesirable by-products).To overcome these challenges,improving the predictability of empirical knowledge-based traditional spontaneous fermentation is imperative.Understanding the mechanism underlying the flavor development of the fermented golden pomfret and the correlation between the microbiota involved and volatile compounds is particularly important.
Foodomics has been applied to study various foods and is especially useful for examining the microbial community and metabolic processes in fermented foods[4,5].Microbial community succession and metabolites of fermented foods evaluated using foodomics can provide insights into the mechanisms underlying food fermentation and flavor development[6].Single-molecule real-time(SMRT) sequencing technology has the advantages of long reading length,high accuracy,and uniform genome coverage,which could reveal the structure and succession of the microbial community.This study examines the dynamic changes in volatile organic compounds and microbiota profiles of fermented golden pomfret based on single-molecule real-time (SMRT) sequencing and molecular networking analysis.Moreover,the study intended to elucidate mechanisms underlying flavor development and correlations between the microbiota involved and volatile compounds,thereby to highlight the approaches for improving the spontaneous fermentation process and product quality of fermented fish.
2. Materials and methods
2.1 Preparation of fermented golden pomfret
Fresh adult golden pomfret ((500 ± 50) g;(28 ± 2) cm) were acquired from a dedicated fish farm in the Guangdong Province,China.Fish were euthanized by blunt force to the head before being bled by trained personnel.All fish samples were cleaned,eviscerated,stored at 37 °C for 2 h,covered with table salt (NaCl),and finally placed into fermentation containers.The additive amount of table salt were 25% (m/m).The salted fish samples were stacked in layers inside a fermenter (1 m long,1.5 m wide,and 1 m high),covered with copious amounts of salt.The fermentation was conducted at (30 ± 1) °C with a relative humidity of (45 ± 15)% for 24 days.Fish samples were withdrawn at different fermentation time points from containers of different fermentation batches (20 fish in a container taken on Days 0,6,12,18,and 24;hereafter,termed as D0,D6,D12,D18 and D24,respectively) for analysis.Then,samples with the same fermentation time were mixed on an ultraclean bench,placed in a sterile food-grade plastic bag as an individual analysis sample,and frozen at −20 °C until further analysis.
2.2 DNA extraction and SMRT sequencing
An Omniscript Reverse Transcription Kit (Qiagen,Hilden,Germany) was used to extract microbial metagenomic DNA from the fish samples.Nanodrop agarose gel electrophoresis was used to examine the extracted DNA.The full-length bacterial 16S rDNA was amplified using the following primers: 27F_(16S-F)(5’-AGRGTTTGATYNTGGCTCA G-3’) and 1492R_(16S-R)(5’-TASGGHTACCTTGTTASGACTT-3’).The PCR amplification was performed under the following conditions: denaturation at 95 °C for 2 min,followed by 30 cycles at 98 °C for 10 s,annealing at 55 °C for 30 s and 72 °C for 90 s,and extension at 72 °C for 2 min.After the electrophoresis of PCR products,the DNA samples were purified,quantified,and homogenized to create a SMRTbell library,and marker genes were sequenced using a PacBio SMRT sequencing system (Pacific Biosciences,Menlo Park,CA,USA).
2.3 SMRT sequence data processing
The SMRT raw data (subreads) were qualified with minPasses ≥ 5 and minPredictedAccuracy ≥ 0.9 to obtain circular consensus sequencing (CCS) (SMRT Link,Version8.0).Lima v1.7.0 set with default parameters was used to identify the CCS sequences in different samples.The CCS sequences with 1 200–1 650 bp were filtered to generate high-quality (effective) sequences.Finally,the chimeric sequences were removed using UCHIME (version 8.1).Indices,such as operational taxonomic unit richness,α-diversity(ACE,Chao1,Shannon,and Simpson),andβ-diversity,were analyzed using USEARCH (Version 10.0) and QIIME software at BMK Cloud (www.biocloud.net).Linear discriminant analysis(LDA) effect size (LEfSe) was applied to screen the biomarkers.The LDA score measures the degree of influence of the species with significant difference between different groups,and the default LDA score was set to 4.Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) was used to identify microbial functional features.
2.4 Analysis of volatile compounds
The volatile organic compounds in fermented golden pomfret were analyzed using headspace solid-phase microextraction(HS-SPME) coupled with gas chromatography-mass spectrometry(GC-MS) according to a previous method[6]with slight modifications.A minced fish sample (2 g) in a 20-mL headspace vial was subjected to volatile compound enrichment.HS-SPME was performed using a polydimethylsiloxane/divinylbenzene(PDMS/DVB) fiber (65 μm,2 cm;Supelco Analytical,Bellefonte,PA,USA).A magnetic stir bar (IKA,Staufen,Germany) was then embedded in the PDMS/DVB aging extraction head,incubated at 60 °C for 40 min,and introduced into the GC/MS injector port to allow analysis and desorption at 250 °C for 5 min.Continuous extraction with the extraction head was conducted at 270 °C for 10 min to avoid cross-contamination between samples.
The volatile compounds were analyzed using an Agilent 7890B GC-MS system (Agilent,Santa Clara,CA,USA) equipped with a CD-5MS column (30 m × 0.25 mm,0.25 μm;CNW Technologies,Düsseldorf,Germany).Helium was used as the carrier gas to run at a constant flow rate of 1.4 mL/min (constant linear velocity).The initial temperature was set to 35 °C and maintained for 1 min.Subsequently,the temperature was raised to 60 °C at a rate of 5 °C/min and held for 1 min.The temperature was further raised at a rate of 6 °C/min to 140 °C and held for 1 min.Finally,the temperature was increased to 230 °C at a rate of 8 °C/min and held for 5 min.The ion source temperature and the electron ionization energy were set at 200 °C and 70 eV.The mass scan range was 35–350m/z.Each sample was analyzed in triplicate.
2.5 Calculation of relative odor activity value
The volatile components were identified based on the retention indices and via mass spectral similarity match using the NIST database (National Institute of Standards and Technology,Gaithersburg,MD,USA).The contribution of each volatile compound to the overall flavor was evaluated as the relative odor activity value(ROAV) using Eq (1):
WhereCiis the relative concentration of the compound of interest,Cmaxis the relative concentration of the compound with the maximum odor activity value,Tiis the odor threshold of the target compound,andTmaxis the odor threshold of the compound with the maximum odor activity value.
2.6 Statistical analysis
Three biological replicates were used in all analyses.Spearman’s rank correlation analysis was performed to construct networks among the genera with relative abundance >0.05% and elucidate the relationships between microbiota and metabolic pathways.Cluster analysis was performed and hierarchical clustering heatmaps were plotted using R software (v3.1.1,R Foundation for Statistical Computing,Vienna,Austria).The network was constructed and visualized using Cytoscape (V3.7.1) to examine the relationship between the volatile compounds and microbial community.
3. Results and discussion
3.1 Changes in the microbial structure during golden pomfret fermentation
SMRT sequencing generated 351 079 high-quality sequencing reads from all samples.Each sample was covered by an average of 23 405 reads.Among them,239 distinct operational taxonomic units(OTUs) were identified with 97% sequence similarity.As summarized in Table S1,the coverage of all samples was >99%,suggesting that most microbial phylotypes were present in the samples and that the obtained sequence reads were adequate to reveal most microbial communities in the fish samples[7].
The rarefaction curve reflects the number of species against the number of samples,indicating species richness.Fig.S1a shows the microbial richness of the fish samples and indicates that the amount of sequencing data from each sequenced sample was sufficient to reflect the true microbial diversity of the sample.The order of OTUs numbers was D0 >>D6 >D18 >D24 >D12.Shannon index measures biodiversity,reflecting species richness and evenness in a specific community.In this study,the Shannon index curves (Fig.S1b) revealed high species abundance/richness,indicating that the amount of sequencing data was adequate.At different fermentation time points resembled,the order of the Shannon index remained similar: D0 >>D18 >D6 >D12 >D24,indicating the highest and lowest microbial diversity in the fish samples at D0 and D24,respectively.Rank abundance curves measure the relative species abundance and community complexity.In this study (Fig.S1c),the changing patterns of the fish samples after fermentation were very similar,which differed greatly from those of the D0 samples (before fermentation).Unfermented fish samples (D0 group) had the highest levels of microbial richness and diversity,suggesting that the relative abundance of the microbial community changed greatly as the fermentation proceeded.The high salt content of the fish samples and their anaerobic environment might facilitate inhibition of the growth of exogenous microorganisms,thereby decreasing the microbial richness and diversity of the fermented samples[8].The microbial activities promoted the accumulation of various metabolites,including nutrients that can be utilized by microorganisms for their growth and other activities.Therefore,the observed fluctuation in microbial richness and diversity during fish fermentation.A principal coordinates analysis (PCoA;also known as multidimensional scaling) allows for the visualization of similarities or dissimilarities among data.The three-dimensional (3D) PCoA score plot shows the proximities between the fish samples obtained at different fermentation time points in three dimensions.
In this study,all the samples were spread by sites and separated into fermentation time-depending clusters in the PCoA plot(Fig.S1d),indicating the differences in microbial communities in the fermentedgolden pomfret samples obtained at different fermentation stages.The locations of the triplicate samples at each fermentation time point in the PCoA plot also varied,although the replicate samples still belonged to the same cluster.
Alpha diversity metrics are useful for evaluating the structure of an ecological community including its richness (i.e.,number of taxonomic groups) and/or evenness (i.e.,distribution and abundance of the different groups).Alpha diversity indices such as Chao,ACE,Shannon,and Simpson indices,are commonly used for this purpose,with Chao and ACE indices measuring community richness while Shannon and Simpson indices reflecting community diversity.In this study,the indices for the D0,D6,D12,D18,and D24 samples differed (Table S1),indicating the dynamic changes in the microbial community structure at different fermentation stages.
3.2 Microbial taxonomic profiles during golden pomfret fermentation
The microbial taxonomic profiles of the samples at different fermentation time points were obtained at the phylum and genus levels based on the sequences generated by SMRT 16S rRNA sequencing.In total,11 phyla and 129 genera were identified in all fish samples.Microbial community dynamics during fermentation is shown in Fig.1.The abundance of each bacterial species fluctuated over the fermentation process (Fig.1a).Firmicutes,Proteobacteria,Bacteroidetes,and Actinobacteria were identified as the dominant bacterial phyla (Fig.1b).These phyla were also found as dominant species inSuan zuo yu(a traditional Chinese fermented fish product) during fermentation[9].As for the raw materials,the relative abundance of Proteobacteria,Firmicutes,and Bacteroidetes was 44.28%,30.58%,and 16.26%,respectively.As the fermentation proceeded,the abundance of Firmicutes increased considerably in the early fermentation stage,while the abundance of the other phyla experienced downward trends during this period.Firmicutes was also the predominant phylum (approximately 90%) in the late fermentation stage.A similar result was previously reported on microbial community succession during Suanyu fermentation[8].As for the abundance of Proteobacteria,a decrease from 44.28% to 7.81%was found,which contributed to the changes in volatile organic compounds.Species of the Proteobacteria phylum are known to play a role in nutrient cycling thereby contributing to the degradation of amino acids,organic acids,and small peptides[10].

Fig.1 (a) Changes in the microbial structure at different levels during fermentation;bacterial community composition of fermented golden pomfret of different fermentation time points at the (b) phylum and (c) genus levels.
At the genus level,Enhydrobacter,Acinetobacter,andClostridiumpredominated at the beginning of fermentation (Fig.1c),which might occur in the raw fish material.Acinetobacteris commonly found in the North-Atlantic cod epidermis[11].As the fermentation of golden pomfret progressed,the abundance of most genera decreased rapidly,except forClostridium.After six days of fermentation,the abundance ofShewanellaandStreptococcussignificantly increased.On Day 6,these two species became the dominant bacteria.The relative abundance ofShewanellaandStreptococcuswere 19.9% and 1.9%,respectively.Shewanellahave been previously reported in various fresh water and marine fish species,and can generate proteases and nucleotidases,which could affect the profiles and availability of peptides and amino acids,and nucleotides,respectively,during fermentation[12].Proteins and nucleotides are broken down and converted to various flavor compounds,including free amino acids,inosine monophosphate,inosine,and hypoxanthine.Streptococcuswithin the orderLactobacillales(lactic acid bacteria),is known as a flavor contributor and able to produce fatty acids and esters[13].Streptococcustend to develop biogenic amines such as histamine when storing fish at elevated temperatures[14].In this study,the abundance ofStaphylococcusincreased sharply and reached a maximum (13.51%) after 12 days of fermentation.In addition,the abundance ofVibriodecreased gradually as the fermentation process progressed.There was significant number ofClostridiumspecies at the end of golden pomfret fermentation.Due to the nature of raw fish samples and the spontaneous fermentation,Clostridiumspecies are often found in fermented fish products.
3.3 Microbial taxon differentiation during golden pomfret fermentation
A LEfSe analysis allows for the assessment of microbial differences at different taxonomic levels.In this study,the LEfSe analysis was used to identify species-specific biomarkers and determine whether any taxa were differentially abundant between groups in fermented golden pomfret at various species classification levels.The microbial biomarkers in the fermentedgolden pomfret samples at the phylotype level are presented in a representative cladogram of the predominant microbiome (Fig.2a).The phyla,Proteobacteriaand Bacteroidetes,were identified as the key phylotypes in D0 samples,while Firmicutes wasthe key phylotype in the Day 24 sample.Firmicutes and Proteobacteria are often found in other salted fermented fish products.At the family level,Shewanellaceae was identified as a biomarker for the D6 fermentation period.Staphylococcaceae and Caloramatoraceaewere the biomarkers for Day 12 fermentation.Clostridiaceae and Vibrionaceae were the biomarkers for the last day of fermentation (D24).

Fig.2 (a) Cladogram and (b) linear discriminant analysis (LDA) score distribution histogram for the unique bacteria at the phylotype level and based on differences in abundance.The lengths of the histogram represent the LDA scores.
The microbial biomarkers were also screened based on linear discriminant analysis (LDA) scores (Fig.2b).Acinetobacter,EnhydrobacterandWolbachiawere the microbial biomarkers of the unfermented samples (D0) at the genus level.Specifically,the genusShewanellaof the family Shewanellaceae within the order Alteromonadales could be the microbial biomarker for Day 6 samples.In D12 samples,the abundance of the Bacilli species was the most different biomarker,along withStaphylococcusandClostridiumas contributors.Bacilli can generate a range of enzymes including proteases and amylases,which greatly influenced the flavor development and accelerate the fermentation process for fermented golden pomfret.In particular,Clostridium_sensu_stricto_4 andVibriowere likely microbial biomarkers at the genus level for the late stage of golden pomfret fermentation.However,the fish samples collected on Day 18 are absent in Fig.2a and 2b,indicating that the D18 group did not have significantly different relative abundance compared with the other groups.
These results confirm the significant effect of fermentation time on microbial species.The spontaneous fermentation of golden pomfret allowed various microorganisms to metabolize substrates.These microorganisms and derived enzymes could jointly act on the initial raw fish material (at the early stage of fermentation) and/or the degraded fish tissues/substances (at the latter stages of fermentation).These activities changed as the fermentation proceeded.The differences in the abundance of the abovementioned microbial species over the fermentation process can be used to identify biomarkers for the quality control of fermented golden pomfret at different fermentation stages.
3.4 Identification of volatile compounds during golden pomfret fermentation
Based on the results of HS-SPME-GCMS,a total of fortynine volatile compounds were identified in fermented golden pomfret samples.According to the detected proportions of different types of volatile compounds (Fig.3a),aromatic compounds,nitrogen-containing compounds,and acids were the major volatile compounds.The heatmap (Fig.3b) revealed the clusters of individual volatile organic compounds in the fermented golden pomfret after fermentation for different periods.The differences in the type and amount of the volatile compounds resulted from the differences in catabolic reactions initiated by the involved microorganisms and derived enzymes at different stages of fermentation.Compared with the samples obtained after fermentation for different periods,D0 samples contained more volatile organic compounds (with alcohols and esters having higher contents).Alcohols and esters accounted for 3.85% and 8.27%,respectively.Methyl butyrate,propyl 2-methylbutanoate,2-butanol were only found in D0 samples.Ethyl butyrate,butanoic acid,butyl 2-methylbutanoate,1-propanol were the main volatile compounds in D0 samples.

Fig.3 (a) Proportions of different types of volatile compounds and (b) heatmap of the volatile organic compounds in the fermented golden pomfret samples obtained at different fermentation time points.
Alcohols likely resulted from the reduction of aldehydes,whereas esters were generated from microbial esterification of acids and alcohols.These compounds contributed to the characteristic flavor of fermented golden pomfret.As for alcohols,1-octen-3-ol content(also known as mushroom alcohol),had the highest content on Day 12 of fermentation.This compound,with a mushroom-like odor,is an important flavor component for fermented fish products[15]derived from the autoxidation of linoleic acid or oxidation of arachidonic acid catalyzed by 12-lipoxygenase[16].Alcohols are also involved in the development of the unique flavor of fermented mandarin fish.Unsaturated alcohols such as 1-penten-3-ol and 1-octen-3-ol provide a metallic smell and can significantly affect the flavor of fermented golden pomfret[17].Ethanol was detected only in samples during the middle fermentation stage (i.e.,D12 and D18 fish samples).Conversely,esters can impart fruity and sweet flavors into fermented fish and help mask those that are fatty notes[18].During fermentation,esters are mainly formed from alcohols and organic acids via nonenzymatic esterification and enzymatic esterification by microorganisms[19].Moreover,reversible alkyl condensation reactions could occur owing to the actions of carboxylesterase and triacylglycerol lipase from microorganisms[20].In this study,ethyl esters,including ethyl propionate and ethyl butyrate,were abundant in the fermented golden pomfret due to the esterification of ethanol and acids.Salt can promote bacterial esterification,thereby enhancing the formation of ester compounds[21].
Aldehydes with low thresholds are known as significant contributors to pleasant the green grass-/cheese-like flavor and fat aroma,thus,they were anticipated to significantly affect the flavor of fermented golden pomfret.In general,aldehydes are mainly from the oxidation/degradation of lipids and the metabolism of amino acids (e.g.,via the Strecker degradation of amino acids or the decomposition of hydroperoxides and peroxyl radicals)[22].In this study,four aldehydes were identified in the fish samples.Benzaldehyde,3-methylbutanal,and hexanal reached their peak levels on Day 12,and decreased gradually in abundance during the last stage of fermentation.Among these aldehydes,hexanal as a straightchain aldehyde provides a green grass-like aroma and is derived primarily from fatty acids (especially unsaturated fatty acids) via the hydrolysis by lipoxygenase[23].3-Methylbutanal was previously reported as a key aroma-associated compound with a malty-and nutlike flavor that could come from leucine through Strecker degradation and biosynthetic pathway[24].In this study,2-methylbutanal was only detected in D18 samples.Benzaldehyde resulting from Strecker degradation or the oxidative degradation of linolenic acid[25]can endow fermented fish with almond and nutty sweet aroma.In general,branched-chain aldehydes and short chain aldehydes originate primarily from branched chain amino acids,whereas straight-chain aldehydes originate from fatty acids.Short-chain aldehydes are derived from amino acids via Strecker degradation or microbial metabolism[6].Similar to aldehydes,ketones are produced due to the oxidation or degradation of unsaturated fatty acids and the breakdown of amino acids via Strecker degradation during fermentation[26].The relative percentage of ketones was the lowest among the different types of volatile compounds,and ketones were only found in D6 and D12 samples.
Acids significantly affect the overall aroma of fermented golden pomfret owing to their variety and unique acidity.In this study,butanoic acid had the highest relative abundance in the fish samples,followed by acetic acid,pentanoic acid,2-methyl butyric acid,and 3-methylbutanoic acid.The level of butanoic acid,originating from carbohydrate metabolism,increased from 0 to 12 days,and subsequently decreased.Butanoic acid at a lower concentration provides a desirable acid flavor but an unpleasant rancid butter odor at a high level.Acetic acid was generated mainly from microbial metabolites,while 3-methylbutanoic acid resulted primarily from the oxidation of 3-methylbutanal in the presence of aldehyde dehydrogenase.The content of 2-methyl butyric acid exhibited an upward trend within the first 12 days.The production of organic acids during fermentation also led to an acidic environment,which enhanced the inhibition of the growth of many spoilage-causing microorganisms.In this study,the total alcohol,aldehyde,and acid contents peaked on Day 12,whereas the ester level increased mainly during the late-stage fermentation (e.g.,Day 18).
Hydrocarbons originated from the oxidation of fatty acids,in particular,decarboxylation of higher non-volatile fatty acids,make little contribution to flavor development due to their high-threshold characteristics[27].In this study,the fish samples contained a significant amount of indole,which might have contributed to the distinct flavor of fermented fish products[28].Styrene is an aromatic compound with a plastic-like smell that was found in the fish samples of this study,which was likely derived from sugars during fermentation[29].In total,seven aromatic compounds were found in the fermented fish samples,possibly due to the catabolism of aromatic amino acids[17].D18 samples had the highest amount of aromatic compounds,with phenol accounting for the largest proportion.Meanwhile,toluene was only detected on Day 18,which might result from the decomposition of phenylalanine or esters.Toluene levels are correlated with the degree of fish oxidation[30].Sulfur-containing compounds were also present in significant amounts in fermentedgolden pomfret samples,with dimethyl disulfide being the most abundant,while a relatively high level of dimethyl trisulfide was found.These sulfur-containing compounds provide garlic-and cooked cabbage-like flavors[31]and were likely derived from sulfur-containing amino acids viathe Strecker degradation,followed by oxidation[32].Methanethiol may be generated from methionine due to the actions of amino acid lyases[33].As for the nitrogen-containing compounds,2,6-dimethylpyrazine was detected during the early stage of golden pomfret fermentation(Day 0–6).Nitrogen-containing compounds with a roasted coffee and peanut flavor might result from the Maillard reaction and/or thermal decomposition of amino acids[22].The highest content of trimethylamine was detected in D18 samples.Trimethylamine-Noxide might be transformed into trimethylamine during microbial metabolism,which causes a strong fishy odor[34].
3.5 Contributions of the characteristic volatile compounds to the overall flavor during golden pomfret fermentation
The contribution of a volatile compound to the overall flavor of fermented golden pomfret was measured using ROAV.According to ROAV,the characteristic volatile compounds in fermented fish samples were determined.In general,volatile compounds with ROAVs between 0.1 and 1 are considered contributors to the overall flavor and those with ROAV ≥ 1 are important for the specific aroma of a particular product.In this study,thirteen characteristic volatile compounds with ROAV >1 were identified in the fermented golden pomfret samples (Table S2).These compounds were alcohols,aldehydes,acids,esters,sulfur-containing compounds,and nitrogencontaining compounds.Dimethyl trisulfide with a garlic-and cooked cabbage-like flavor was the dominant contributor to the overall aroma profile of the samples,thus,its ROAV was set as 100.Trimethylamine presents a fishy odor and is usually used as an indicator of the freshness of fish,and in this study,had a ROAV in the range of 36-96.In addition to dimethyl trisulfide,trimethylamine had a very high ROAV (96.09) on Day 0,indicating the highest intensity of fishy odor in the D0 samples.Dimethyl disulfide also had a quite high ROAV (70.31) on Day 0,indicating a strong presence of a garlic-like or decaying fish odor.In addition to dimethyl trisulfide,ethyl butyrate showed very high ROAV (96.16) on Day 6,suggesting that the pineapple-like or distinct nail polish remover smell was dominant in the golden pomfret after 6 days of fermentation.Trimethylamine (ROAV 89.27;fishy odor),dimethylpyrazine (ROAV 88.97;roast and nutty odor),and dimethyl disulfide (ROAV 65.83;a garlic-like or decaying fish odor) were also significant contributors to the odor of D6 samples.Moreover,dimethyl trisulfide,dimethyl disulfide,3-hexanone,and trimethylamine had relatively high ROAVs(52.81,49.54 and 35.99,respectively),indicating that a garlic-like or decaying fish odor,ethereal,grape or wine-like odor,and fishy odor made similar contributions to the overall flavor of D12 samples.On Day 18,trimethylamine had a remarkably higher ROAV (88.09) than other volatile compounds (except for dimethyl trisulfide).Therefore,the D18 samples mainly had a fishy odor.On Day 24,dimethyl disulfide(ROAV 48.31;garlic-like or decaying fish odor) and trimethylamine(ROAV 36.81;fishy odor),along with garlic-and cooked cabbage-like flavors (dimethyl trisulfide),were the key flavors.
3.6 Construction of the co-occurrence networks during golden pomfret fermentation
Microbial interactions influence the metabolic activities of the microbial community thereby affecting the production of metabolites and flavor development.Co-occurrence and co-exclusion relationships were established to evaluate relationships between the microbiota community and volatile flavors.A Spearman’s rank correlation analysis was conducted to determine the relationships among the dominant microbial genera.Co-occurrence networks were established to reveal the actual interactions among the different microbial genera and identify the key genera.As shown in Fig.S2,50 genera were included in the microbial network of fermented golden pomfret.Streptococcuswas found in positive correlation withHydrogenophagaand Veillonellaceae.Clostridiumwas negatively correlated withEnterobacter,Aeromonas,andChryseobacterium,whereas it had a positive correlation withPsychrobacter.Shewanellawas negatively correlated withVibrioandSoonwooa.Staphylococcusexhibited a negative correlation withBrevundimonas,Serratia,andAcinetobacterbut had a positive correlation withHydrogenophaga.
As shown in Fig.4a and b,strong correlations were noted between the microbes and volatile organic compounds.Acinetobacterwas positively correlated with 1-octen-3-ol,3-methylbutanal,and ethyl butyrate,indicating a significant contribution ofAcinetobacterto the formation of these three volatile organic compounds.During sufu fermentation,Acinetobacterwas found to act as the dominant genus and showed a positive correlation with flavor formation owing to its capacity to secrete esterolytic enzymes[35].In this study,1-octen-3-ol showed a negative correlation withClostridium,indicating that the formation of 1-octen-3-ol was inhibited by this microbial species.ClostridiumandVibriowere the notable bacteria that positively regulated the production of trimethylamine and indole,whereasStaphylococcusexerted an opposite effect.Trimethylamine and indole are responsible for the typical off-odor of fish.Therefore,Staphylococcuscould inhibit the formation of off-odor and improve the quality of fermented golden pomfret.Staphylococcushas enzymes with lipolytic and proteolytic activities,and in this study,was negatively correlated with hexanal (a typical product of lipid oxidation).This result indicates that lipid oxidation was inhibited byStaphylococcus.In comparison,Vibriowas beneficial to the formation of hexanalduring the current golden pomfret fermentation,probably due to the hydroperoxide lyase encoded byVibrioto catalyze the production of hexanal.Furthermore,Chryseobacterium,Enhydrobacter,andAcinetobacterwere found in close association with the formation of volatile compounds during the golden pomfret fermentation,such as methanethiol,dimethyl disulfide,1-octen-3-ol,and indole.Thus,these microbial genera were important for the formation of characteristic volatile compounds of fermented golden pomfret.As forShewanella,there were no exhibit significant effects on the formation of characteristic volatile compounds during the golden pomfret fermentation.

Fig.4 (a) Correlations among dominant microbial genera and characteristic volatile compounds;(b) network of dominant correlations among microbial genera and volatile organic compounds.The red and blue lines represent the positive and negative correlations,respectively,among the genera and volatile compounds.
3.7 The possible mechanism underlying the formation of flavor compounds
The prediction of the possible functional pathways and genes associated with the metabolism of microbial species based on sequencing data provides deeper insights into the adaptive responses of the microbiota in fermented foods during food fermentation.The relative abundance plots of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and the abundance of dominant microbial genera,based on KEGG database annotation,are shown in Fig.5.Six functional gene groups were categorized at level 1 to infer the different functional pathways.At level 1 (Fig.5a),the relative abundance of the genes for metabolism was the highest,whereas that for organismal systems was the lowest.Considering the KEGG pathways at level 2(Fig.5b),the global and overview maps,carbohydrate metabolism,and amino acid metabolism were the most abundant pathways during the golden pomfret fermentation.Similar results were found for the Chinese yellow wine huangjiu[36].Since proteins,carbohydrates,and lipids are the primary precursors of volatile organic compounds in fish,it was anticipated that carbohydrate metabolism,amino acid metabolism,and lipid metabolism contributed considerably to the organoleptic properties of fermented golden pomfret.Carbohydrates can be broken down byStaphylococcus,which facilitated the development of the distinct aroma of traditionally preserved meat products[4].Furthermore,the formation of alcohols and aldehydes is known to be associated with amino acid and lipid metabolism,and esters result from lipid metabolism.In all fish samples of this study,267 metabolic subsystems were identified within the annotated dataset at KEGG classification level 3 (Fig.5c),with 16 dominant KEGG pathways.The average relative abundance was greater than 1%.Amino acid and amino sugar biosynthesis,nucleotide sugar metabolism,carbon metabolism,and glycolysis/gluconeogenesis are important pathways for flavor development during golden pomfret fermentation[37].The enrichment of the genes associated with the metabolism of carbohydrates,amino acids,and lipids suggests that these biomolecules are particularly important for flavor formation during golden pomfret fermentation.The enrichment in the metabolic pathways among the key bacterial genera is presented in Fig.5d.Relatively high abundances ofChryseobacterium,Staphylococcus,andAcinetobacterwere related to amino acid metabolism,carbohydrate metabolism,and lipid metabolism,respectively.Thus,the differences in metabolic pathways related to the functional genes in microbial communities denote the various biochemical processes responsible for flavor formation (including the breakdown and utilization of proteins,lipids,and carbohydrates in the fish material).
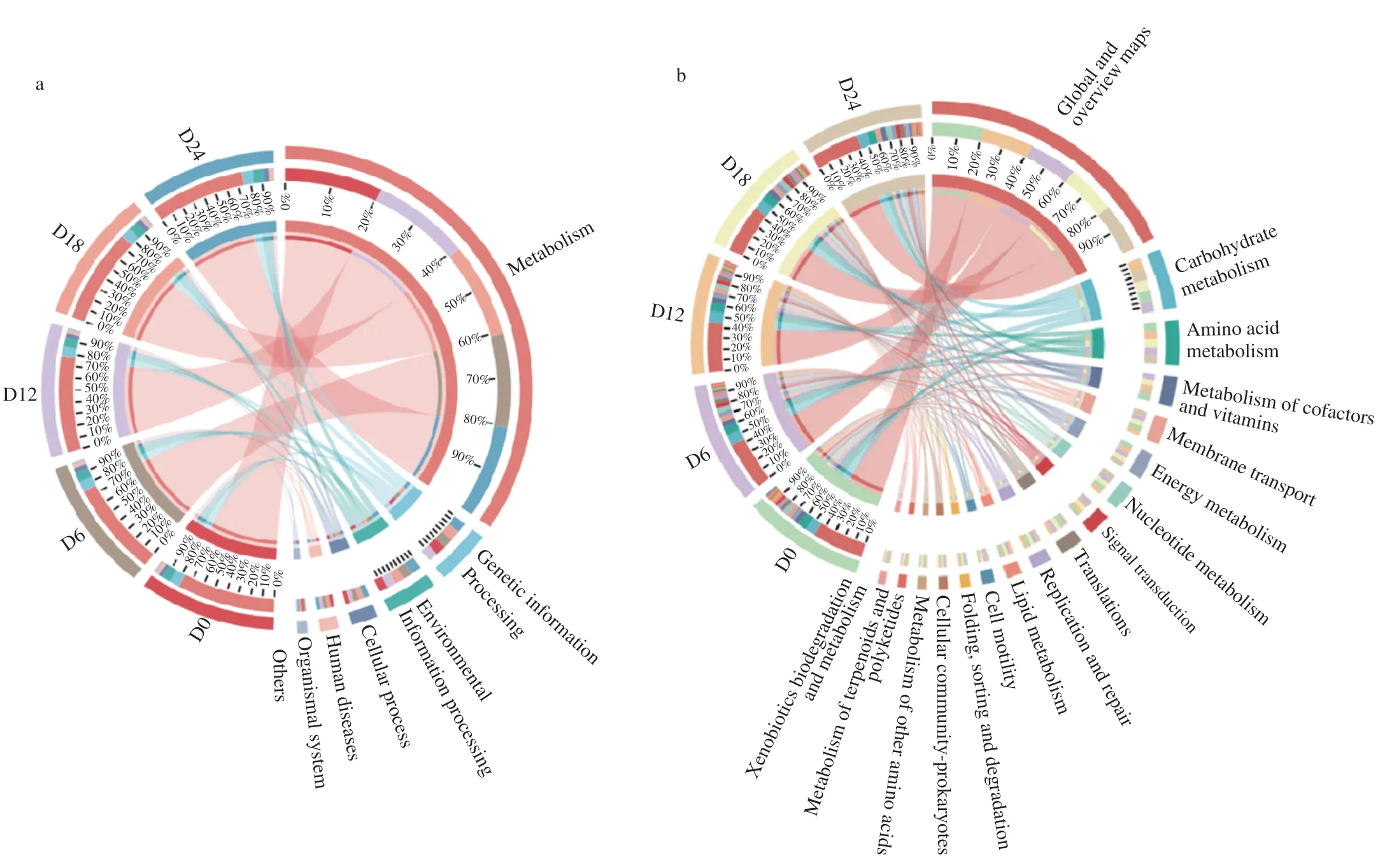
Fig.5 Relative enrichment of microbial Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in samples at (a) level 1,(b) level 2 and (c) level 3;(d) functional gene prediction for dominant microbial genera in fermented golden pomfret by PICRUSt2;(e) heatmap of Spearman’s rank correlations among the dominant microbial genera and KEGG pathways.

Fig.5(Continued)
To understand the interrelationships among the microbial genera and KEGG pathways in the fermentedgolden pomfret samples,a Spearman’s rank correlation analysis was performed (Fig.5e).In total,29 pairs of significant relationships (P<0.05) were obtained from seven dominant microbial genera and 16 KEGG pathways.Among them,16 pairs of positive correlations and 13 pairs of negative correlations were identified.Staphylococcuswas positively correlated with carbon metabolism and antibiotic biosynthesis.This result was consistent with the finding related to the predictive functional gene abundance inStaphylococcus.Acetic acid was derived from the carbohydrate metabolism ofStaphylococcus,involving the key enzymes related to flavor development,such as acetate kinase and pyruvate dehydrogenase.The oxidative decarboxylation of pyruvate generated acetyl-CoA;then,acetyl-CoA reacted with phosphoric acid to form acetyl phosphate;finally,acetyl phosphate converted to acetic acid by acetate kinase[5].Moreover,Clostridiumwas negatively correlated with carbon metabolism and antibiotic biosynthesis but positively correlated with glycolysis/gluconeogenesis,pyruvate metabolism,purine metabolism,and pyrimidine metabolism.Clostridiumcontributed to the production of alcohols,ammonia,trimethylamine,ethyl acetate,ketones,and sulfur compounds during fermentation[11].Chryseobacterium,Acinetobacter,Enhydrobacter,andVibriowere negatively correlated with the processes of glycolysis/gluconeogenesis,pyruvate metabolism,purine metabolism and pyrimidine metabolism.Glucose could be converted to pyruvate via the glycolytic pathway through enzymatic action.Pyruvate might participate in the synthesis of aromatic amino acids or turn into ethanol and acetic acid due to microbial metabolism.As fish have a low glucose content,the proteins and amino acids in the fish samples were converted to glucose by microorganisms via gluconeogenesis.Chryseobacteriumspecies can produce enzymes like proteases and gelatinase,and in this study,were positively correlated with the metabolic pathways such as gluconeogenesis/glycolysis,amino acid metabolism,sulfate reduction,propanoate metabolism,and tricarboxylic acid cycle.During the fermentation of golden pomfret,Chryseobacteriumpresented a high relative abundance for amino acid metabolism.Proteolysis was anticipated to take place as one of the most important processes in fermented golden pomfret due to its high protein content.The breakdown and metabolism of the proteins in fermented golden pomfret resulted from the joint actions of endogenous proteases of fish and the enzymes from microorganisms.In general,endogenous proteases contribute to the hydrolysis of proteins into oligopeptides,whereas microbial enzymes further degrade oligopeptides.Thereafter,amino acids are converted into their correspondingα-keto acids by aminotransferases,and the resultingα-keto acids are converted into aldehydes,alcohols,and esters.In this study,Chryseobacteriumwas also positively correlated with 3-methylbutanal,a malty flavor compound from fatty acids and amino acids such leucineviaStrecker reactions and through deamination followed by decarboxylation[38].Taken together,the above results indicate that the formation of volatile compounds in the fermented golden pomfret was primarily associated withStaphylococcus,Clostridium,andChryseobacteriumvia the glycolysis/gluconeogenesis pathway,pyruvate metabolism,and amino acid biosynthesis and metabolism.
The pathways associated with the formation of volatile organic compounds during golden pomfret fermentation are summarized in Fig.6.Proteins and lipids are vital precursors of the flavors formed in fermented golden pomfret.The catabolism of leucine,valine,isoleucine,phenylalanine,and methionineled tointermediates such asα-keto acids and aldehyde,and products including hydroxyacids,CoA-esters,esters,alcohols,and other high flavor-impact aldehydes.α-keto acid is a key node compound and can lead to the formation of corresponding aldehydes and ketones.Leucine could be transformed intoα-ketoisocaproic acid due to the catalysis by transaminase before decarboxylation into 3-methylbutanal by decarboxylase[24].Methionine with sulfur atoms might be degraded to methional via Strecker reaction then to methanethiol.The resulting methanethiol could be oxidized further to dimethyl disulfide,which significantly affected the characteristic flavor of fermented golden pomfret[33].Ammonia-like odor might be generated due to the conversion of glutamate toα-ketoglutarate and the degradation of arginine.The alcohol compounds in the fermented fish were derived from the Ehrlich pathway (an amino acid metabolic decomposition pathway involving branched-chain amino acid aminotransferases and producingα-keto acids as the main products from amino acids) and/or the Harris pathway (an anabolic pathway using glycogen as a precursor to formα-keto acid for subsequent decarboxylation to produce alcohol compounds)[36].

Fig.6 Potential pathways involved in the formation of the volatile compounds in fermented golden pomfret.
Lipid metabolism was another main metabolic pathway associated with flavor formation of fermented golden pomfret.The oxidation and degradation of fatty acids yield aldehydes and alcohols[39].Linoleic acid could undergo oxidation to form 13-hydroperoxide,and hexanal was then generated via alkoxy radicalβ-scission[40].Polyunsaturated fatty acids,such as eicosapentaenoic acid and arachidonic acid,could be used to produce different alcohols via the action of lipoxygenase.Acetyl-CoA is the precursor of fatty acid and polyketides biosynthesis[41]and,as an important intermediate,can also enter the tricarboxylic acid cycle via citrate synthase.
The microbial community present in the golden pomfret samples could also initiate carbohydrate transport,degradation,and metabolism.The breakdown and utilization of carbohydrates in the fish material played an important role in flavor formation during golden pomfret fermentation e.g.,carbohydrate metabolism in the fish samples cause an increase in butanoic acid content in the first 12 days of fermentation and a decrease in butanoic acid content after Day 12.Staphylococcuscould produce acetic acid via carbohydrate metabolism during fermentation.Metabolic processes,glycolysis (in which glucose is broken down into pyruvate) and gluconeogenesis (in which glucose is synthesized from pyruvate and/or lactate) are two important pathways involving carbohydrates during the development of fermented golden pomfret flavor[37].
Notably,interconversion occurred among the alcohols,aldehydes,acids,and esters.That is,aldehydes could be reduced to alcohols via the action of alcohol dehydrogenase,and alcohols might be oxidized further to carboxylic acids by aldehyde dehydrogenase.In addition,alcohols and carboxylic acids could undergo reversible esterification owing to the actions of alcohol hydroxyl transferases.The balance among the alcohol,aldehyde,acid,and ester contents is important for the ultimate flavor of fermented golden pomfret.During the fermentation of golden pomfret,the interactions among the microorganisms in the fermentation systems and the dynamic changes of the microbial interactions caused the fluctuations/changes in the concentrations of metabolites.The characteristics of the fermentation system underwent dynamic changes due to the produced metabolites,which in turn,caused the dynamic changes in the microbial community and the rerouting of pathways associated with the formation of volatile organic compounds.
4. Conclusions
This study showed the dynamic changes in microbial community and volatile organic compounds during golden pomfret fermentation.Forty-nine volatile compounds were detected in all the fish samples before and after fermentation,with thirteen identified as characteristic volatile compounds.The correlations among different microbial genera and the relationships between microbial taxa and volatile compounds were revealed.The concentrations of trimethylamine and indole were positively correlated withClostridiumandVibrio.Staphylococcusinhibited lipid oxidation and produced acetic acid via carbohydrate metabolism during the fermentation of golden pomfret.Amino acid metabolism,lipid metabolism and carbohydrate metabolism were the three main metabolic pathways associated with flavor development.The mechanisms related to flavor development were also elucidated based on KEGG functional annotations.Thus,the profiles of volatile compounds resulted from interactions among the microorganisms and derived enzymes.This study demonstrated the research approaches for distinguishing key microbiota associated with volatile compounds and examining their roles in flavor formation during golden pomfret fermentation.The obtained findings can also help select the appropriate flavor-inducing microorganisms for the industrial production of fermented fish products.
Conflicts of interest
Dongxiao Sun-Waterhouse is a senior editor forFood Science and Human Wellnessand was not involved in the editorial review or the decision to publish this article.All authors declare that there are no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (32001733),the Earmarked fund for CARS(CARS-47),Guangxi Natural Science Foundation Program(2021GXNSFAA196023),Guangdong Basic and Applied Basic Research Foundation (2021A1515010833),Young Talent Support Project of Guangzhou Association for Science and Technology(QT20220101142) and the Special Scientific Research Funds for Central Non-profit Institutes,Chinese Academy of Fishery Sciences(2020TD69).
Appendix A.Supplementary data
Supplementary data associated with this article can be found,in the online version,at http://doi.org/10.26599/FSHW.2022.9250008.
- 食品科学与人类健康(英文)的其它文章
- GUIDE FOR AUTHORS
- Targeting gut microbiota in osteoporosis: impact of the microbial based functional food ingredients
- Weizmannia coagulans: an ideal probiotic for gut health
- Natural sources,refined extraction,biosynthesis,metabolism,and bioactivities of dietary polymethoxyflavones (PMFs)
- A review of salivary composition changes induced by fasting and its impact on health
- Minerals in edible insects: a review of content and potential for sustainable sourcing

