Inhibitory effect of procyanidin B2 and tannin acid on cholesterol esterase and their synergistic effect with orlistat
Xiangxin Li,Yijing Pu,Haitao Jiang,Wenxiao Jiao,Wenjun Peng,Wenli Tian,Weio Jiang,*,Xiaoming Fang*
a Institute of Apicultural Research,Chinese Academy of Agricultural Sciences,Beijing 100093,China
b College of Food Science and Nutritional Engineering,China Agricultural University,Beijing 100083,China
c College of Food Science and Engineering,Qilu University of Technology (Shandong Academy of Sciences),Jinan 250353,China
Keywords:Cholesterol esterase Polyphenols Interaction mechanisms Molecular dynamic simulation Synergistic effect
ABSTRACT This study was aimed to analyze the effect of procyanidin B2 (PC) and tannin acid (TA) on the activities of cholesterol esterase (CEase) and the inhibitory mechanisms of enzymatic activity.The interaction mechanisms were investigated by enzymatic kinetics,multi-spectroscopy methods,thermodynamics analysis,molecular docking,and dynamic simulations.PC and TA could bind with CEase and inhibit the activity of enzyme in a mixed-competitive manner and non-competitive manner,which was verif ied by molecular docking simulations and dynamics simulations.Also,PC and TA showed the synergistic inhibition with orlistat.Fluorescence,UVvis and the thermodynamic analysis revealed that the complexes were formed from CEase and inhibitors by noncovalent interaction.As revealed by the circular dichroism results,both PC and TA decreased enzymatic activities by altering the conformations of CEase.The inhibition of PC and TA on CEase might be one mechanism for its cholesterol-lowering effect.
1. Introduction
Cholesterol,an important lipid and precursor of bile salts,vitamin D and steroid hormones,is one of main component of cell membrane while participates in the regulation of permeability and fluidity of the membrane[1].However,the excessive intake of cholesterol is related to obesity and cardiovascular,thus it is necessary to remove excess cholesterol and reduce its adsorption and translation for decreasing the risk of chronic diseases[2].In the process of cholesterol metabolism,cholesterol esterase (CEase) is a key enzyme related to the absorption and transport of cholesterol micelles to enterocytes,and it is transferred from pancreas to intestinal,which hydrolyze cholesterol ester to cholesterol monomer[3].Existing studies suggested that reducing the catalytic ability of CEase may efficiently downregulate the cholesterol level and thus suppressed generation of related diseases[4,5].
Orlistat is an anti-obesity drug,which is a potential inhibitor of lipase,CEase,gastric,and carboxylic ester lipases and phospholipase A2.These enzymes are related to the hydrolysis of dietary lipids in the gastrointestinal (GI) tract[6].Currently,orlistat as the only specific pancreatic lipase (PL) inhibitor has been approved for clinical use in Europe[7].Orlistat is an effective CEase inhibitor as well,but it might bring about some side effects[8].Therefore,it is necessary to search for natural,safe,and effective CEase inhibitors to prevent the absorbance of cholesterol.
Existing studies have shown that tannins are easy to influence the activity of enzyme due to their complex structure and binding characteristics.Tannins are composed of condensed tannins and hydrolysable tannins,which extensively distributed in plants[9].Condensed tannins,namely procyanidins,consist of favan-3-ol units,covering (epi) catechin,(epi) gallocatechin,and little (epi) afzelechin,which are linked by carbon-carbon bonds[10,11].Hydrolysable tannins(mostly tannin acid (TA)) cover a glucoside.They primarily consist of gallic acid and have been identified in numerous plants.TA covers several reactive groups (e.g.,phenolic -OH group,-OH group,and -COOH group),which are easy to interact with other nutrition contents[12].Previous reports have demonstrated that catechin extracted from green tea might hinder the absorption of lipid by inhibiting the activities of lipase,and the procyanidins of grape seeds could decelerate the digestion of starch by inhibiting the activity ofα-amylase[13].The conformations and activities ofβ-galactosidase were altered by binding with TA,which indicated that a new complex formed between TA and the enzyme[14].It was also reported that phenolic extract of edible plants inhibited the CEase activity by nearly 50%[5].Furthermore,the procyanidins extract was considered to have a cholesterol-lowering effect through blunting the activity of CEase[15].And condensed tannins extracted from banana pulp could inhibit the activities of CEase by changing the structure of enzyme[16].However,the underlying interaction mechanisms of the enzyme and inhibitors (procyanidin B2(PC) or TA) molecules were not very clear.
Recently,molecular docking and dynamic simulation were used to comprehend the interaction between the small molecule and the target protein for achieving high-through screening from the database,and widely used in elaborating molecular mechanisms and simulating complex structures at molecular levels.In this study,we used above technology combining with spectral and thermodynamic analysis technology to reveal the lowering-cholesterol mechanisms of PC and TA.We supposed that PC and TA,two phenolic compounds easy to obtain from plants and their byproducts,could be used as functional food to improve human health by reducing the absorption and metabolism of cholesterol,thereby preventing cholesterol metabolic disorder-associated diseases or complications.This study may provide a theoretical basis for the further research of PC and TA as a natural anti-hypercholesterolemia supplement.
2. Materials and methods
2.1 Chemicals
CEase (50.8 U/mg solid) was obtained from Yuanye Biology Technology Co.(Shanghai,China).TA (mw=1 701.2,AS ≤ 3‰),PC (mw=578.5,≥ 98%) andp-nitrophenyl butyrate (p-NPB)(≥ 98%) were purchased from Sigma-Aldrich Co.(St.Louis,MO,USA).NaH2PO4,Na2HPO4and NaCl were purchased from Sinopharm Chemical Reagent Factory (Shanghai,China).
2.2 Apparent studies of CEase-inhibitors complexes
The transmittance testing of CEase-inhibitors (PC and TA)mixtures was analyzed as described in previous research[17].CEase solution (25 mg/mL) and inhibitors (PC or TA) solutions (0−0.1 mg/mL)were prepared in buffer solution (0.1 mol/L PBS,pH 7.0).Enzyme and inhibitor solutions were incubated together for 15 min,and the absorbance was determined at 600 nm using a UV-vis spectrophotometer (Thermo Fisher Scientific Co.,Waltham,MA,USA).For this experiment,the transmittance of CEase-inhibitors complexes was calculated using the following equation:
Where,Asampleis the absorbance of samples at 600 nm.The particle size of CEase-inhibitor complexes was determined by dynamic light scattering using a Zetasizer Nano ZS instrument(Malvern Instruments,Malvern,UK) at the same conditions of transmittance analysis.
2.3 Inhibitory effect of PC or TA on CEase activity
According to previous study[4],enzyme solution (2 U/mL) and inhibitors solutions were incubated at volume ratio of 1:1 for 5 min,after which reaction substrate (p-NPB,0.4 mmol/L) and buffer were mixed with the above solution at volume ratio of 1:4 and incubated for 15 min.All of the regents were prepared in buffer,which contained 0.1 mol/L NaH2PO4-Na2HPO4and NaCl at pH 7.0.The absorbance of the reaction solution was analyzed using a UV-vis spectrophotometer at 405 nm.The CEase inhibitory activity was calculated according to the following equation:
WhereAcontrolrepresents the control absorbance,andAsamplewas the sample absorbance.
2.4 CEase inhibition kinetics
Kinetic parameters were calculated according to Lineweaver-Burk equation,and the specific method was similar to the method stated in section 2.3,but the substrate concentrations were 0,0.08,0.16,0.24,0.32,0.40 mmol/L.The CEase solution concentration was 2 U/mL.The Lineweaver-Burk equation was referred to the previous study as follow equation[4]:
Where,V0is the initial reaction speed,Vmaxis the maximum reaction speed,Kmis Michaelis constant,[S] is substrate concentration.
2.5 Combined inhibition measurement
The IC50values of PC,TA,and orlistat were calculated from the inhibition rate curves.Different ratio of PC/TA (0,1/8 IC50,1/4 IC50,1/2 IC50,and IC50) and different ratio of orlistat (0,1/8 IC50,1/4 IC50,1/2 IC50,and IC50) were incubated in the same conditions.According to the above method,the combined inhibition capacity on CEase were carried out.The obtained data was analyzed by the CompuSyn program to evaluate the effects (synergy,addition,or antagonism) of the association[18].
2.6 Fluorescence and UV-vis spectroscopy measurement
Binding mechanisms of CEase with inhibitors (PC and TA) were investigated by measuring the fluorescence emission spectra at 310 K using a fluorescence spectrometer according to a previous report with some modifications[19].Specifically,different concentrations of PC aqueous (0−0.17 mmol/L) and different concentrations of TA aqueous(0−5.88 μmol/L) were added to CEase solutions and the mixtures were incubated for 5 min before centrifugation.After that,the absorbance of the supernatant was measured and the absorbance collected from 400 nm to 600 nm by excited at 295 nm.The correction of emission intensity was referred to the previous study as follow equation[11]:
WhereFcorrandFmare the corrected and measured fluorescence intensities of enzyme,respectively,andAexandAemare the absorbance of inhibitors at the excitation and emission wavelengths,respectively.
Fluorescence quenching mechanisms was studied by Stern-Volmer equation.
WhereI0is fluorescence intensity of inhibitors;I1is fluorescence intensity of inhibitors with quencher;τ0(10–8s) is the average fluorescence lifetime of the fluorophore without a quencher;Kqis biomacromolecule quenching rate constant;KSVis Stern-Volmer constant;[Q] is concentration of inhibitors.
Kaandnvalues obtained from followed equation:
WhereKais the binding constant;nis binding site.
UV-vis spectroscopy measurement was carried out to identify the mechanisms of CEase and TA directly.PC with the concentration of 43.4 μmol/L and TA with the concentration of 10 μmol/L were added to CEase aqueous and mixed for 5 min at the ratio of 1:1,and the UV spectra was recorded at 190−400 nm.
The fluorescence resonance energy transfer phenomena of CEase and PC or TA were performed as the previous study with slight modifications[17].The difference was that the concentration of PC was 0.131 mmol/L,and the concentration of TA was 0.013 3 μmol/L.
2.7 Circular dichroism (CD) study
The CD spectra of enzyme aqueous and CEase-inhibitors (PC and TA) complexes was recorded according to a previous research[20].The concentration of PC and TA solution was 0.1 mg/mL.The conformation of CEase and CEase-inhibitors was calculated by CDNN 2.1 software.
2.8 Thermodynamic analysis
Thermodynamic measurements were studied by Nano ITC analyzer at 310 K[11].Whole of the samples were dissolved in buffer solution (0.1 mol/L PBS,pH 7.0) and filtered by 0.22 μm membrane and degassed at vacuum for 10 min.50 μL PC or TA aqueous and 300 μL CEase (0.25 mmol/L) were placed in sample syringe and sample cell,respectively.After attaining equilibrium,the titration began with the reaction condition of stirring rate (300 r/min) and each titration with 200 s intervals.Thermodynamic parameters were analyzed using Nanoanalyze Software.
2.9 Molecular docking study
The specific interaction mode and binding sites could be obtained from molecular docking studies by Autodock 4.2 and vina software[21,22].The 3D structure of PC,TA and CEase molecules could be downloaded from NCBI website into PDB format and energy minimized by the Cambridge Soft ChemBioOffice Ultra (Version 14.0).Before running the docking program,both the ligand and receptors were subjected to pretreatments such as water removal and hydrogenation through AutoDockTools,and then calculated the vintage binding mode.At last,the conformation of receptor-ligand complexes was visualized by the program PyMOL and ligplot+software.
2.10 Molecule dynamic simulations
To assess the system stability,a molecular dynamic simulation was used to determine the docking information in detail.The proteinligand complexes with the lowest energy values were selected as the starting conformation for molecular dynamics (MD) simulations by the AMBER force field at a stable temperature (305 K) and pH 7.0.The protein was solvated by SPC water in a cubic box.Sodium ions were added to neutralize the system charge.Systems were energy minimized using the steepest descent method and then equilibrated under NVT followed by NPT condition.The simulation results model was selected to run a MD analysis of 40 000 ps.An integration step was set to 2 fs.The MD tracks were used to analyze the root mean square deviation (RMSDs) and radius of gyration (Rg)[23].
2.11 Statistical analysis
All experiments were conducted in duplicate.One-way analysis of variance (ANOVA) and Duncan’s new multiple range tests were utilized to evaluate any significant differences among the data from different groups with the aid of SPSS Statistics 19.0 (SPSS,Chicago,USA).The differences were considered significant atP<0.05.
3. Results and discussion
3.1 Transmittance and particle size analysis
The interaction between CEase and inhibitors (PC or TA molecules) was investigated by transmittance analysis and particle size measurements.The transmittance of CEase-inhibitors mixed solution was significantly decreased when the PC and TA solutions were added into CEase solution,and the transmittance of CEase with TA solution was lower than that with PC (Fig.1A).Similarly,as shown in Fig.1B,the particle size of CEase aqueous remarkably increased with adding PC and TA solutions,and the particle size of CEase-TA complex was larger than CEase-PC.The above results indicated that both PC and TA molecules were able to bind with CEase molecules,and the binding capacity of TA molecules was more potent than that of PC,which might be ascribed to a higher molecular weight,more hydrophilic structure and more flexibility and -OH groups for non-covalent interaction with the enzyme of TA molecules as compared with PC[24].

Fig.1 Transmittance (A) and particle size analyses (B) of the CEase-PC/TA complexes.
3.2 CEase activity inhibition and kinetics
It is noteworthy that polyphenols derived from plants exhibit considerable effects on anti-hyperlipidemia through inhibiting CEase activity and decelerating the absorption of cholesterols,which was considered to be a potential mechanism for preventing atherosclerosis and cardiovascular diseases[20].
As presented in Table 1,Figs.2A and E,both tannins exhibited remarkable inhibitory effects against the CEase activity.Results showed that the IC50of PC and TA were (45.53 ± 2.33) and(5.30 ± 0.67) μg/mL,respectively.The IC50of inhibitors was associated with the determination methods and conditions (e.g.,enzyme concentration,substrate type,reaction time,temperature,and pH)[25].Thus,under the identical determination methods and conditions,the inhibitory capacity of TA against CEase was higher than that of PC,indicating that the affinity properties were associated with the complicated spatial structure of inhibitors[24].This result was in accordance with the previous study that catechin polymers and catechin could significantly suppress the activity of CEase,and the inhibitory capacity of catechin polymers was higher than that of catechin monomers,which suggested that the inhibitory capacity enhanced with increasing polymerization degree of catechin[26].Thus,the inhibitory effect of TA on CEase was stronger than that of PC,and both PC and TA exhibited great potentials to prevent hypercholesterolemia and cardiovascular diseases.

Table 1 Enzymatic kinetic parameters of inhibitors against CEase.
The kinetic studies of the enzyme by the Lineweaver-Burk plots in the different PC concentrations were shown in Table 1 and Fig.2B.The results gave series of straight lines,intersected within the second quadrant for CEase-PC,suggesting that the inhibitory mode of PC towards CEase belongs to the mixed-competitive manner[27].PC could compete withp-NPB in terms of affinity with CEase and bind to the enzyme-substrate complex,thereby forming a PC-p-NPB-CEase tertiary complex.It was also indicated that PC could be bound to the active catalytic site of CEase or other sites[28].

Fig.2 Reaction and inhibitory kinetic curves.(A,E) Inhibitory effects of PC and TA on CEase.(B,F) Lineweaver-Burk plots of CEase in the presence of PC and TA.(C,G) Dose-effect curves of PC and TA with orlistat for CEase.(D,H) Fa-CI plots of the combination of PC and TA with orlistat for CEase.
As shown in Table 1 and Fig.2F,the graph of 1/Vvs.1/[S]suggested that the cueves of CEase-TA were intersected at the negativeX-axis,revealing that the inhibitory mode of TA towards CEase belonged to the non-competitive manner.In other words,TA might inhibit the catalytic activity of CEase by independently combing with both free CEase and the CEase-p-NPB complex instead of hindering the binding between substrate and enzymes[29].It was also revealed that TA might not bound to the active catalytic site of CEase.As suggested from all of the mentioned findings,the inhibitory manners and abilities were related to the structure of tannins,because PC (mw=576) could enter the cavity of CEase and bound with catalytical sites through hydrogen bonds or hydrophobic interaction.However,TA (mw=1 702) was too large to enter the cavity of CEase.Thus TA molecules changed the conformations and activities by interacting with the residues surrounded around the cavity of CEase.Overall,PC and TA are natural CEase inhibitors to prevent the excessive intake of cholesterol.
3.3 Combined inhibitory effect of CEase
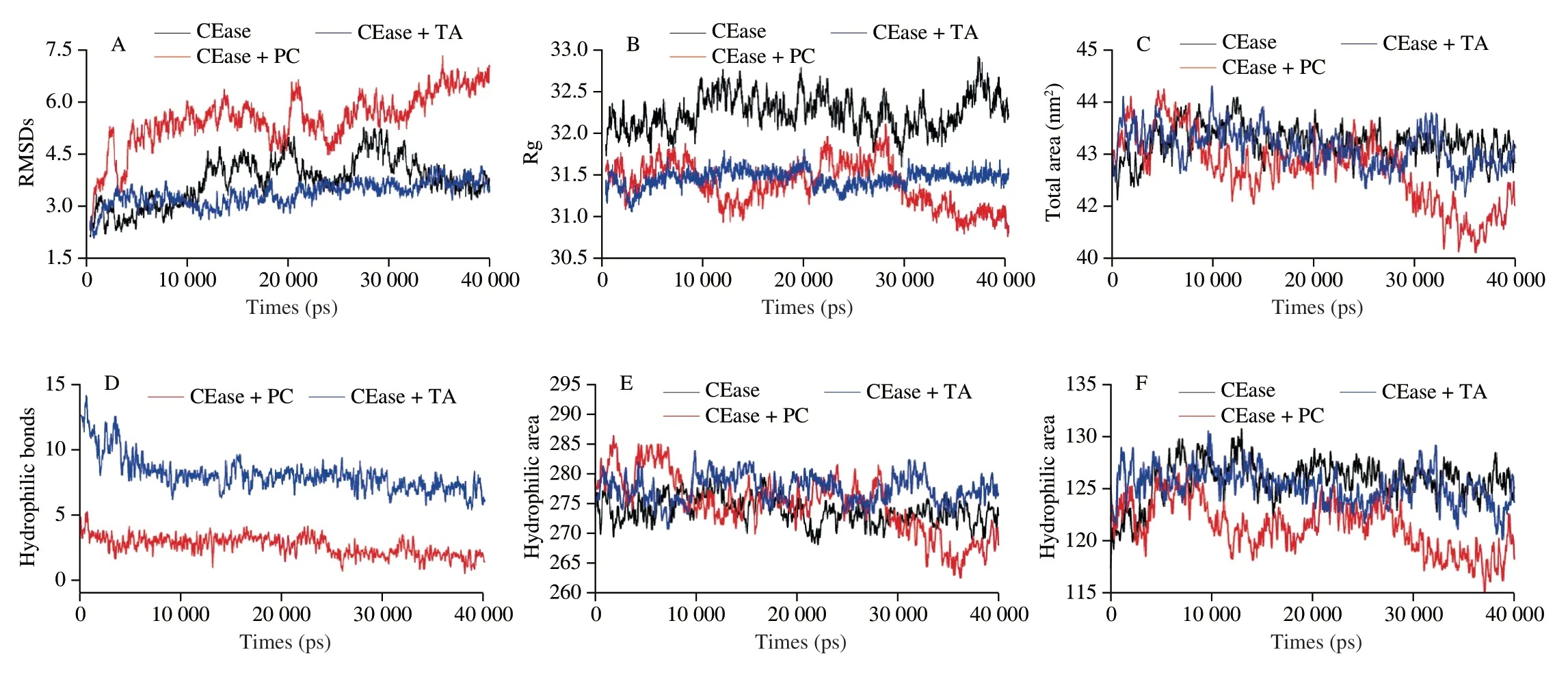
The dose-effect curve of the combinational PC/TA and orlistat on CEase was shown in Figs.2C and G,and the inhibition effect was expressed as fa (0 Fluorescence spectroscopy was employed to explore the interaction of proteins with small molecules.CEase covers several Trp that contribute to the intrinsic fluorescence of CEase.For this reason,the Trp residues located in CEase can be reflected by the alternation of fluorescence intensity,which could further expound the interaction mechanisms of molecules[31].The fluorescence spectra quenched by PC and TA were collected and presented in Fig.3. Fig.3 Interaction analysis by multi-spectroscopy methods.(A,E) Fluorescence quenching effect of PC or TA on CEase fluorescence intensity at 310 K.(B,F)UV-visible absorption spectra of CEase (0.03 mg/mL) in the absence and presence of PC and TA.Pure PC and TA (red),pure protein (black),inhibitors-protein complex (blue),and sum of absorbance of PC or TA and protein (green).(C,G) The spectral overlap of UV-vis absorption spectrum of PC or TA and fluorescence emission spectra of CEase.(D,H) Secondary structure of CEase in the absence and presence of PC or TA. Fig.4 Thermograms of titration of CEase solution by PC or TA.(A) Titration curve of CEase solution by PC.(B) Thermograms parameters of CEase-PC titration. As presented in Figs.3A and E,the fluorescence spectra of CEase was reduced remarkably by adding inhibitors solution with different concentrations,demonstrating that the microenvironment of Trp in CEase could be changed by interacting with inhibitors (PC and TA).Accordingly,the tertiary structure of enzyme was altered by these two reactions[32].Moreover,spectra data exhibited a blue-shift(approximately 2 nm) with adding inhibitors,demonstrating a decrease in micro-polarity and an elevation of hydrophobicity[33].The mentioned results also suggested that theβ-sheet of CEase increased with the addition of the inhibitors (PC and TA),complying with the results listed in Table 2. Table 2 Conformations (%) of CEase and CEase-inhibitors calculated by CDNN. The relative quenching parameters were showed in Table 3.The Stern-Volmer plots were presented in Fig.S1,displaying a good linear relationship with quenchers concentrations.The mentioned results indicated that the inhibition type belonged to a single type of quenching (static quenching or dynamic quenching)[34].The values ofKqcalculated from the Stern-Volmer plots were 7.10 × 1010and 3.63 × 1012L/(mol·s) for CEase-PC and CEase-TA,respectively,more than the constant in dynamic quenching of the maximal value(2.0 × 1010L/(mol·s))[35].Thus,the quenching of CEase by PC and TA belonged to the static quenching by forming complexes with non-covalent bonds.There are large numbers of -OH groups and benzene rings in PC and TA molecules,so they are easy to form hydrogen bonds and hydrophobic interaction with the enzyme.The binding constant (Ka) of CEase with TA was higher than that with PC,demonstrating that the binding affinity of CEase with TA was higher,which could be attributed to the more complex structure and higher molecular weight of TA. Table 3 Fluorescence quenching parameters for the interaction of CEase and inhibitors. As presented in Figs.3B and F,the CEase-inhibitors (PC and TA) complex exhibited a lower UV absorption compared with the combination of inhibitors (PC and TA) and CEase,which further demonstrated that novel complexes formed,complying with the result obtained from fluorescence quenching (Figs.3A and E). Referring to Foster’s non-radiative energy transfer theory,as presented in Figs.3C and G,the energy was efficiently transferred from CEase to inhibitors due to donor-acceptor distance <8 nm,proving that the reactions belonged to static quenching.Likewise,the mentioned conclusion could be demonstrated by the observations of fluorescence spectroscopy (Figs.3A and E). To further study the interaction of inhibitors (PC and TA)and CEase,CD spectra was utilized to analyze the structural conformations of CEase in the solution.The CD spectra was presented in Figs.3D and H,the conformations of enzyme and enzyme-inhibitors complexes were displayed in Table 2.Results showed that the spectra of enzyme changed and red-shifted with the inhibitors (PC and TA) added,suggesting that the binding presented in enzyme-inhibitors systems,and the inhibitors changed the structure of enzyme significantly.The catalytic action of CEase was related to its native structure,so the alteration of CEase conformations due to the addition of inhibitors might lead to a decrease of CEase activity[36].Moreover,the CD spectra of the enzyme presented a negative peak in the region around 206 and 222 nm,demonstrating that theα-helix existed in enzyme structural.The negative peak at around 214 nm was characteristics of theβ-sheet element[37].The conformations of CEase were analyzed by CDNN software in detail. As presented in Table 2,CEase alone contained 38% ofα-helix,14% ofβ-sheet,17% ofβ-turn,and 32% random coils.With PC solution added,the content ofα-helix decreased to 23%.Besides,the proportion ofβ-sheet was up-regulated to 27%,and the proportion ofβ-turn as well as random coil remained unchanged.With the addition of TA solution,the content ofα-helix was reduced to 30%.Besides,the content ofβ-sheet and random coil was up-regulated to 17% and 37%,respectively,while the content ofβ-turn was constant.The mentioned results suggested that the addition of inhibitors (PC and TA) caused the variation of the secondary structure of CEase,which further led to the alteration of its catalytic activity[19]. Isothermal titration calorimetry (ITC),a vital analysis technique to determine the interaction between two molecules in solution could measure the major thermodynamic parameters directly.In the present study,the ΔHof the affinities between CEase and TA was determined by an ITC analyzer at 37 °C.The generated raw thermograms and parameters were presented in Figs.4,S2 and Table 4.The stoichiometry value (1.48,n≈ 1) of CEase with PC was higher than that of CEase with TA (1.03,n≈ 1),suggesting that per mole of CEase was able to bind more molecules of PC than TA[37].As shown in Table 4,theKDvalue of CEase-TA system(5.55 × 10−5mol/L) was lower than that of CEase-PC system(5.90 × 10−5mol/L),demonstrating that the combined capacity of CEase with TA was stronger than that with PC.TheKDvalue of the whole bindings was less than 1 × 10−3mol/L,proving that a stronger interaction occurred between CEase and inhibitors.The ΔGvalues for CEase-PC and CEase-TA systems were negative,suggesting that both reactions were spontaneous. Table 4 Binding and thermodynamic parameters for the interaction between CEase and inhibitors. For CEase-TA and CEase-PC systems,the ΔHwas negative,suggesting that the above experiments belonged to exothermic process.The enthalpies of the above reactions were far less than 200 kJ/mol,which is a value that should be achieved to form covalent bonds,demonstrating that enzymes combined with TA or PC through non-covalent forces[38].The negative ΔHand the positive entropy change (ΔS) demonstrated that the main thermodynamic driving force was both entropy and enthalpy,and that the binding occurred via hydrogen bonds and hydrophobic interaction[39].Considering notably that the ΔHvalue of CEase-PC and CEase-TA (−8.153 and −11.49 kJ/mol,respectively) was relatively low,suggesting that the interactions between CEase and PC or TA also included electrostatic interactions,which played an essential role in the binding[40]. Molecular docking,a common technology to predict the binding sites and main interaction between protein and ligand,was employed to investigate whether non-covalent interactions account for CEase-inhibitors complexes[25,30].In this study,the three-dimensional molecular structures of PC,TA and CEase were built from PubChem-Open Chemistry Database.The energy of three-dimensional molecular structures of PC and TA was minimized by the Cambridge Soft ChemBio Office Ultra with a MM2job. The docking conformations of CEase-PC complex were displayed in Figs.5A-D.By Autodock Vina simulating (Figs.5A and B),PC was detected in the active pockets of CEase while the interaction between PC and CEase occurred under the lowest energy.It could be observed that 15 amino acids surrounded the PC molecular.Among the mentioned amino acids,hydrophobic interaction residues consisted of Ala-117,Gly-116,Tyr-125,Gly-112,Gly-106,Leu-110,Gly-107,Leu-323,Ser-194,Leu-274,Ala-108,His-435,and Phe-324.Meantime,hydrogen bonding interaction was identified between PC and Ala-113 or Met-111 of CEase.Moreover,PC could interact with Ser-194 and His-435 (the catalytic sites) in CEase by hydrophobic interaction (Figs.5C and D)[41].The mentioned results suggested that the inhibiting mode of PC towards CEase belonged to the mixedcompetitive mode,which was similar with the result of reaction kinetic analysis.And PC located in the vicinity of hydrophobic pocket and catalytic triad,which also indicated that PC discouraged the catalytic activity of CEase in reaction substrate.Thus,the catalytic effect of CEase was inhibited by PC significantly. Likewise,with the help of Autodock Vina simulating,TA was identified in the active pockets of CEase while the interaction formed between TA and CEase under the lowest energy (Figs.5E-H).Furthermore,it could be identified that 15 amino acids surrounded the TA molecular.Among the mentioned amnio acids,hydrophobic interaction residues contained Ala-117,Met-111,Leu-439,Ala-436,Leu-69,Phe-119,and sodium taurocholate (Tch-601)(Fig.5H).Moreover,the hydrogen bonds were identified between TA and amino acid residues (Try-125 and Try-75) of CEase (Figs.5G and H).According to the mentioned result,TA could interact with the residues of CEase,which did not include the active catalytic sites(Fig.5G);the inhibiting mode of TA towards CEase belonged to the non-completive mode,complying with the result from the result of CEase activity inhibition kinetics.TA was located in the vicinity of hydrophobic pocket and catalytic triad,revealing that TA discouraged the catalytic activity of CEase in reaction substrate.Overall,the catalytic effect of CEase was inhibited by TA noticeably. MD simulations were carried out to obtain detailed molecularlevel pictures of the CEase-inhibitors aggregates at different timescales[12,42].The RMSDs was used to determine the statistical deviation of the MD results from a target conformation.The RMSDs in the dynamic simulations were calculated to estimate the structural drift from the initial coordinates and the atomic fluctuation[43].The RMSD values of the heavy atoms in the free CEase,CEase-PC and CEase-TA complex systems were shown in Fig.6A.The RMSD values showed some apparent disturbance in the free CEase system,but increased apparent disturbance in the CEase-PC complex system and decreased apparent disturbance in the CEase-TA complex system.The sequence of structure stability was: the CEase-TA complex system > the free CEase system > the CEase-PC complex system,indicating that an increase and decrease of the degrees of freedom for CEase motions upon the binding of PC and TA to the enzyme,respectively[13].Thus,based on the slight structural drift present in the MD simulations,the CEase-TA complex showed relatively low atomic fluctuations and increased stability compared with CEase-PC complex system. Fig.6 Molecular dynamic simulation of CEase-PC/TA complex.(A) The plots of RMSDs as a function of simulation time (ps) in different regions.(B) The plots of Rg as a function of simulation time (ps) in different regions.(C) The solvent-accessible surface area (SASA) of CEase as a function of simulation time (ps) in different regions.(D) The amounts of hydrogen bonds as a function of simulation time (ps) in different regions.(E) The plots of hydrophobic area as a function of simulation time (ps) in different regions.(F) The plots of hydrophilic area as a function of simulation time (ps) in different regions. In addition,the Rg values were calculated to assess the stability of the backbone atoms of the system[44].Rg is the distance between every atom and the center of the molecule.Smaller Rg values indicate that the structure is more compact.If the enzyme is folded stably,Rg can keep a relatively steady state,and if the enzyme is unfolded,Rg changes with time[45].As shown in Fig.6B,the Rg values of the CEase-PC complex system and the CEase-TA complex system were less than that of the free CEase system,which indicated that the structure of enzyme became compact after the addition of PC and TA molecules due to their interaction.Fig.6B also showed that only the Rg value of CEase-TA complex system maintained constant,suggesting that CEase was folded stably by binding with TA. As shown in Fig.6C,the total area of CEase decreased with adding PC and TA solution from 439 to 422 nm2and 435 nm2,demonstrating that the degree of exposure of the protein surface to the solvent was reduced,which resulted in the formation of hydrophobic interactions within CEase.As shown in Fig.6D,the number of hydrogen bonds formed in CEase-TA system was more than that in CEase-PC system,which contributed to a higher affinity of CEase-TA system as compared to CEase-PC system.Further,as shown in Figs.6E and F,the hydrophilic area of CEase changed insignificantly with adding PC and TA molecules in general,and the hydrophobic area of CEase present fluctuation with interacting with PC molecules.The above results suggested that the TA changed the microenvironment of the CEase,leading to conformational changes in the CEase during the MD simulation. In the present study,the interaction mechanisms of CEase and PC or TA were analyzed by kinetics analysis,fluorescence spectra measurements,far-UV CD method,ITC,molecular docking,and dynamic simulation.As indicated from the reaction kinetics and combined measurements with orlistat,PC and TA inhibited the catalytic action of CEase by binding with them,and the combination of PC/TA and orlistat showed the synergistic inhibition on CEase at relatively low concentrations.As suggested from the analysis of ITC,the combination occured via hydrogen bonds,hydrophobic interaction,and electrostatic interactions,and it was considered to be spontaneous and exothermic.Moreover,the mentioned results were demonstrated by molecular docking and dynamic simulation.As revealed from the fluorescence data,the quenching of PC and TA for CEase pertained to static quenching by forming novel complexes that exhibited non-convent interaction.As suggested from the far-UV CD data,PC and TA indeed affected the secondary structure of CEase.As confirmed by the mentioned results,PC and TA inhibited the catalytic activity of CEase by forming new complexes and altering CEase conformations.It was also revealed that TA might not bound to the active catalytic site of CEase.As suggested from all of the mentioned findings,the different inhibitory manners and abilities of PC and TA were related to the structure of tannins,because PC could enter the cavity of CEase with low molecular weight and bind with catalytical sites,but TA was too large to enter the cavity of CEase and could change the conformations and activities by interacting with the residues surrounded around the cavity of enzyme.The findings of this study could provide additional health-promoting benefits of polyphenol by inhibiting the activity of CEase. Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Acknowledgements This work was supported by the National Basic Research Program of China (‘973’program,2013CB127106). Appendix A.Supplementary data Supplementary data associated with this article can be found,in the online version,at http://doi.org/10.26599/FSHW.2022.9250030.3.4 Fluorescence quenching of CEase by PC and TA




3.5 UV-vis absorption spectra
3.6 CD spectra
3.7 ITC analysis

3.8 Molecular docking
3.9 Molecular dynamic simulation
4. Conclusions
- 食品科学与人类健康(英文)的其它文章
- GUIDE FOR AUTHORS
- Targeting gut microbiota in osteoporosis: impact of the microbial based functional food ingredients
- Weizmannia coagulans: an ideal probiotic for gut health
- Natural sources,refined extraction,biosynthesis,metabolism,and bioactivities of dietary polymethoxyflavones (PMFs)
- A review of salivary composition changes induced by fasting and its impact on health
- Minerals in edible insects: a review of content and potential for sustainable sourcing

