Possible genetic reproductive isolation between two tilapiine genera and species: Oreochromis niloticus and Sarotherodonmelanotheron
LI Si-Fa, ZHAO Yan FAN Wu-Jiang CAI Wan-Qi XU Ying-Fang
(1. Key Laboratory of Aquatic Genetic Resources and Utilization Certificated by the Ministry of Agriculture, Shanghai Ocean University, Shanghai 201306, China; 2. Chinese Academy of Fishery Science, Yangtze River Fisheries Research Institute, Jingzho 434000, China)
Possible genetic reproductive isolation between two tilapiine genera and species:Oreochromis niloticusandSarotherodonmelanotheron
LI Si-Fa1,*, ZHAO Yan1, FAN Wu-Jiang1, CAI Wan-Qi1, XU Ying-Fang2
(1. Key Laboratory of Aquatic Genetic Resources and Utilization Certificated by the Ministry of Agriculture, Shanghai Ocean University, Shanghai 201306, China; 2. Chinese Academy of Fishery Science, Yangtze River Fisheries Research Institute, Jingzho 434000, China)
Successful crossbreeding betweenOreochromis niloticusandSarotherodon melanotheronto produce a commercial hybrid has been difficult. The karyotypes and isoenzyme of these two species and their reciprocal hybrids (O. niloticus♀ ×S.melanotheron♂, S.melanotheron♀ ×O. niloticus ♂,the last not included in the isoenzyme study) were investigated via metaphase chromosomes obtained from head kidney cells and electropherogram of lactate dehydrogenase (LDH) isoenzymes from the liver, kidney, white muscle, heart, and eye balls. The diploid chromosome number (2n=44) and the fundamental number (NF=50) of the four tilapia genotypes were the same. However, the karyotype ofO. niloticushad three pairs of sub-metacentric (sm), twelve pairs of sub-telocentric (st), and seven pairs of telocentric (t) chromosomes, whileS.melanotheronhad one pair of metacentric (m), two pairs of sm, 12 pairs of st, and seven pairs of t chromosomes. The reciprocal hybrids both showed a mixed karyotype range between their parents: 0.5 pair of m, 2.5 pairs of sm, 12 pairs of st, and seven pairs of t chromosomes. In view of the electropherogram of isozymes, only the LDH of the kidney showed significant clear bands, with five bands inO.niloticus, three bands inS.melanotheron,and duplicated six bands in the hybrids. The bands varied depending on their activities and mobilities. We considered that the differences in karyotype and isoenzyme were related to the genetic mechanism for post-mating isolation, and provided some additional basic genetic background of their taxonomy.
Tilapiine;Oreochromis niloticus;Sarotherodon melanotheron;Karyotype; Isoenzyme; Reproductive isolation; Taxonomy
Belonging to the order Perciformes, family Cichlidae, Tilapiini is a highly diverse tribe with more than 70 species (Trewavas, 1983), commonly called tilapia. Tilapia is a generic term used to designate a group of commercially important Cichlid fish, which consists of three genera:Oreochromis(maternal mouthbreeder), Sarotherodon(paternal mouthbreeder) andTilapia(substrate breeder).Tilapia classification relies on differences in morphology and breeding behavior. In aquaculture, interspecific hybridization within the genus (such asO. niloticus×O. aureusfor the purpose of producing all male fish andO. niloticus×O. mossambicusfor the purpose of producing red tilapia) is commonly performed.
Culture of tilapia in brackish and salt waters remains poorly developed because few species possess both rapid growth and high adaptability to salinity. Since 1999, we have used fast growingO. niloticusand high salt toleratingS. melanotheronfor a crossbreeding program to produce a new variety of tilapia possessing both high salt tolerance and good growth rate (Li et al, 2008a, b; Liu et al, 2009). However, natural hybridization betweenO. niloticusandS. melanotheronproved impossible and the artificial reciprocal hybridization was unpredictable and unsuccessful, with only several hundred F1individuals produced from over 100 batches of artificial hybridization each year. Despite this, the F1fish were fertile and produced hundreds of F2off-spring, which were similar to those of F1and had ideal aquaculture characteristics such as: (1) better salt tolerance――can stand 15‰−25‰ salinity with good survival and growth, (2) reasonable grow rate―over 500 g in 5−6 months in south China, and (3) better testy than tilapia cultured in freshwater. However, the major obstacle for commercialization of this hybrid was the bottleneck of F1 production. In addition to ecological habits, we attempted to understand their mechanisms of reproductive isolation, including karyotype and isoenzyme point of view.
Karyotypes are chromosome complements of related individuals (species) and the number of chromosomes is often used as an indicator of relationship among species and to determine hybridization potential between species. The application of electrophoretic methods to the taxonomic study of various fish groups has revealed that electropherograms of muscle, serum and hemoglobin tissue proteins are species specific (Thompson, 1960; Nyman, 1965; Li, 1998), and therefore are a valuable tool for the analysis of phylogenetic relationships between different groups within a family (Tsuyuki & Roberts, 1965; Li & Cai, 1995). Additionally, some isoenzymes have strong specificity in activity of a substrate, and genetic information can be easily obtained by using the electropherogram of isoenzymes such as lactate dehydrogenase (LDH), malate dehydrogenase (MDH), and esterase (EST).
We conducted a karyotype and isoenzyme study of these two tilapia genera and their hybrid to: (1) understand the genetic mechanism of the hybridization barrier between the two species, and (2) provide some genetic basis for their taxonomy.
1 Materials and Methods
1.1 Karyotype study
1.1.1 Sample collection
We collectedO. niloticus, S. melanotheronand their reciprocal hybrids (about 100 g of body weight) from the State Zhongji Tilapia Farm, Hebei Province. Ten fish of each genotype were used for the study. Fish were maintained for one week in aerated aquaria (1m×1.5 m) with constant temperature of 30 ℃. Each specimen was injected intra-peritoneally with freshly prepared phytohemagglutinin (PHA) at 10 μg/g body weight, then returned to the aquarium for 16−18 h, and later injected with 0.01% of colchicine solution of pure colchicine at a dosage with 2 μg/g body weight.
1.1.2 Cell harvest
Three hrs after injection with colchicine, fish were euthanized by cutting the anterior cardinal aorta and vein. The anterior head kidney was taken out and pulverized with isotonic solution of NaCl. Cells drifting away from the tissue were moved to 0.075 mol/KCl solution at 37 ℃for 30 min for low osmotic pressure treatment and then centrifuged (8 min, 1 000 r/min). After removing the supernatant, the samples were fixed in fresh Carnoy’s solution (methanol:acetic acid=3:1) for 15 min, and then centrifuged (8 min, 1 000 r/min).
1.1.3 Spreading of cells and staining
The centrifuged cell suspension was spread by Pasteur pipette on slides washed by 95% ethanol and then swirled in distilled water. The slides were air dried on flame and 24 h later stained with 3% Giemsa in Sorensen’s buffer with pH of 6-8 for 40 min. The slides were rinsed in distilled water, air dried and mounted.
1.1.4 Examination of the karyotype
Spread chromosomes in metaphase were observed using an Olympus compound microscope and 50 fields from each specimen under x400 and x1 000 (oil immersion) magnifications, respectively, were photographed by an automatic camera fixed on the microscope. The number and total length of chromosomes were evaluated, and the chromosomes were arranged in pairs from the longest to the shortest. The centromeric position was determined according to the classification of Levan et al (1964).
1.2 Isoenzyme study
1.2.1 Specimens collection and preservation
LiveO. niloticus, S. melanotheronand their hybrids (about 300-600 g of body weight) were collected from the ponds at the State Zhongji Tilapia Farm, Huanghua city, Hebei province. Ten fish of each genotype were used for the study. The liver, kidney, white muscle, heart, and eye ball were taken after releasing blood by cutting the anterior cardinal aorta and vein. Tissues were put into small marked bags and preserved in liquid nitrogen, then transferred to a –80 ℃ refrigerator upon arrival to the laboratory.
1.2.2 Electrophoresis
Following National Standard GB/T 18654.13 (2008), electrophoresis was run at the DYY-12 electrophoresis set (Beijing Liuyi Company) with 7.0% polyacrylamide vertical continuous swimming, 300 V and 30 s for prepare-swimming and pre-swimming before and after adding the samples. Formal swimming lasted 5 h at 200 V. After staining, gel banding patterns were densitometrically analyzed and interpreted to quantify genetic variability among the tilapia genotypes. Lactate dehydrogenase (LDH), malate dehydrogenase (MDH), and esterase (EST) were examined.
2 Results
2.1 Karyotype
The arm ratio and type of chromosome sets ofO. niloticus, S. melanotheronand the hybrids are shown in Tab. 1. The metaphase spread of the chromosomes of the four investigated tilapia genotypes is shown in Fig.1. Chromosomes size ranged from 2.1 to 8.0 μm. We found 78%, 83%, and 79% of metaphases studied contained 44 chromosomes for each of the three groups, respectively, therefore the diploid chromosome number for all was 2n=44. The karyotype ofO. niloticushad three pairs of sub-metacentric (sm), 12 pairs of sub-telocentric (st), and seven pairs of telocentric (t) chromosomes, and a fundamental number (NF) of 50. The karyotype ofS. melanotheronhad one pair of metacentric (m), two pairs of sm, 12 pairs of st, and seven pairs of t chromosomes, andNFof 50. However, the karyotype of the reciprocal hybrids was the same: 0.5 pairs of m, 2.5 pairs of sm, 12 pairs of st, seven pairs of t, andNFof 50 (Fig. 2)
2.2 Isoenzymes
Only the LDH from the kidney showed clear bands (Fig. 3). There were five bands (LDH1, LDH2, LDH3, LDH4and LDH5) inO. niloticus,three bands (LDH1, LDH2and LDH3) inS. melanotheron, and six bands (LDH1, LDH2, LDH3, LDH4, LDH5and LDH6) in the hybrid. All LDH isoenzymes were at the anodal side of the gel, and were numbered from the anodal side, with the most anodal being LDH1. Tab.2 lists the relative activity and relative mobility of the LDH bands in the kidneys ofO. niloticus, S. melanotheronand their hybrids. The densitometric patterns are shown in Fig. 3.
3 Discussion
Taxonomy is the theory and practice of naming, describing and classifying organisms. However, the taxonomic and evolutionary relationship among the Tilapiini species remains controversial. Trewavas (1983) reclassified the tilapiini into three genera:Oreochromis, SarotherodonandDanakiliaand suggested that the first two mouth brooding genera could have arisen as one or possibly two splits from the ancestral line, withSarotherodonbeing conservative andOreochromismore progressive. Unfortunately, there is a lack of genetic information to support this classification. Sodsuk and McAndrew (1991) analyzed 44 enzyme loci of 15 species from the three genera and concluded that despite the minor rearrangements within the major classes, Trewavas’s (1983) classification not only clearly described the biological characteristics of the species within each genus, but also reflected the evolution of this group. Previous research has clarified that the LDH isoenzyme of fish is coded by two genes on different loci, which is common among the various groups of vertebrates, and its electrophoretic pattern shows marked differences among fish species (Markert & Faulhaber,1965). This isoenzyme pattern has also used as evidence for the hypothesis of the evolution of vertebras from fish to mammals and is realized mainly by gene duplication (Ohnoet al, 1968).
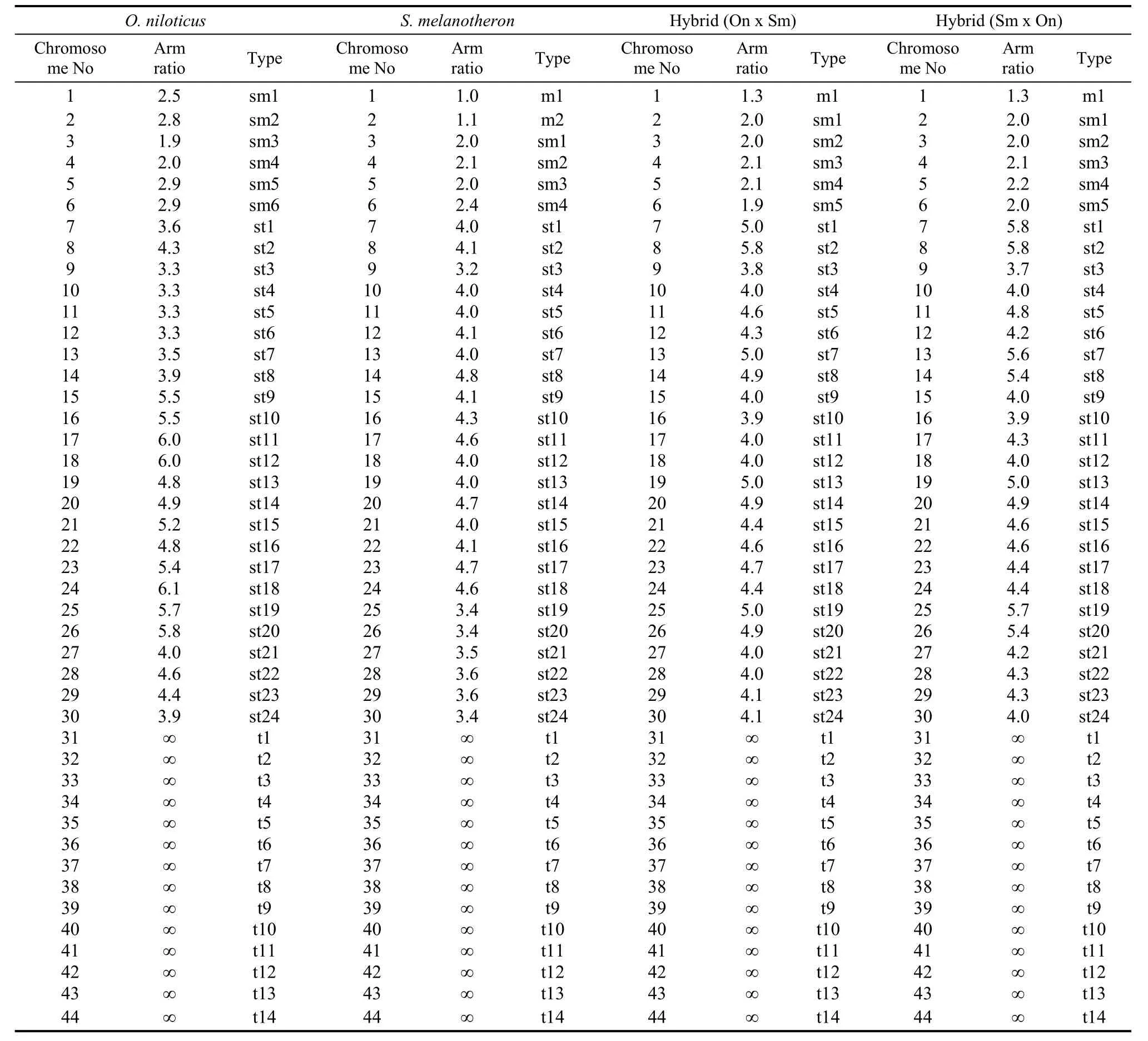
Tab. 1 Arm ratio and type of the chromosomes set of Oreochromis niloticus, Sarotherodon melanotheron and their reciprocal hybrids
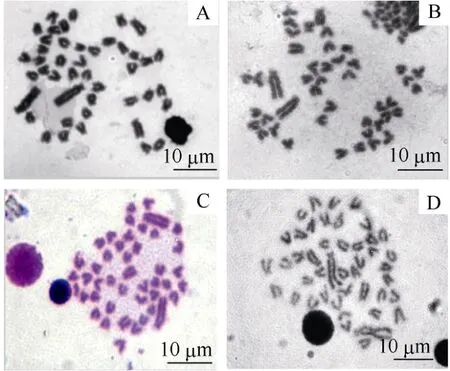
Fig. 1 Metaphase chromosome of the Cichlid fish
BothO. niloticus and S. melanotheronare the most representative species in the generaOreochromisandSarotherodon. They occupy different ecosystems in Africa, withS. melanotheroncovering a natural geographical area of the brackish estuaries and lagoons along the cost of Zaire to Senegal and O.niloticusliving in fresh waters from Senegal to Gambia, Volta to Chad, Nile, Ethiopia, and Baringo (Trewavas, 1983). Due to geographical isolation, no natural hybrid has been reported between these two species. In addition, well known differences in breeding habits exist between the two species, e.g.,O. niloticusis a maternal mouthbreeder andS. melanotheronis paternal mouthbreeder, which provides a reasonable ecological and physiological explanation for their reproductive isolation.

Fig. 2 Karyograms of the Cichlid fish
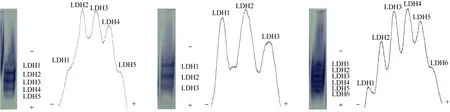
Fig. 3 Electropherograms and densitometric patterns of lactate dehydrogenase (LDH) in kidney of the Cichlid fish
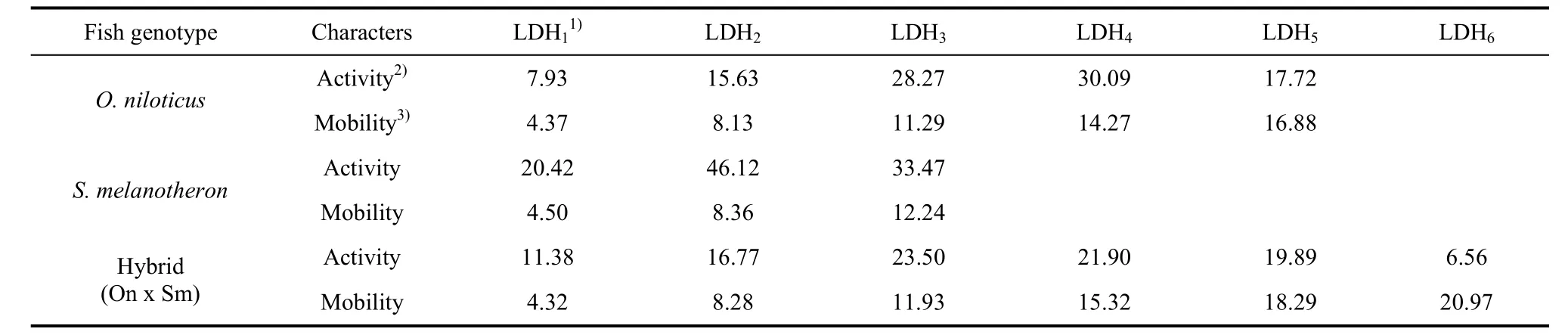
Tab. 2 Relative activity and mobility of bands of lactate dehydrogenase (LDH) in kidney of Oreochromis niloticus, Sarotherodon melanotheron and their hybrid (O. niloticus × S. melanotheron)
Some tilapia species are appropriate for both intensive and extensive pisciculture due to positive characteristics such as fast growth, tolerance for poor water quality, and wide range of feed habits. However, their uncontrolled and prolific breeding at a small size in mixed sex culture is a constraint on their efficient production. The interspecific hybrids ofO. niloticus♀×O. Aureus ♂produced a high male ratio, which resolves the over reproduction problem in tilapia culture.
The culture of tilapia in brackish and salt waters remains poorly developed because few species have both rapid growth and ability to withstand marked variations in salinity. For example,O. niloticusis characterized by a high growth rate but low salt tolerance, whileS. melanotheronis characterized by high salt tolerance but poor growth rate. We expected that the hybridization between these two species could produce a hybrid sharing both good salt tolerance and growth. In our study, hybridization experiments betweenO. niloticusandS. melanotheronwere conducted in two national tilapia seed farms located in south and north China, respectively. Firstly, no natural matings occurred in the ponds (under semi-natural conditions). Secondly, it was difficult to produce hybrids through artificial fertilization, with only a few offspring (F1) produced by reciprocal practice (Li et al, 2008b). Thirdly, successful fertilization during artificial fertilization, even if eggs and sperm met, was rare or absent, possibly due to a lack of effective interaction between the sperm and eggs. Finally, even if fertilization occurred, most embryos failed to develop properly. The above issues are in contrast to the well known phenomenon that interspecific and intraspecific crossbreeding is rather common in Tilapiini, both in the wild and in hatcheries. Many closely related species produce fully viable and fertile F1and backcross progeny in semi-natural conditions and are commercially utilized. For example,O. niloticus×O. mossambicusandO. niloticus×O. aureus, which belong to the same genus, have of same chromosome number (2n=44). Harvey et al (2002a) reported that differences in chromosome number does not prevent the production of inter-specific hybrids betweenO. niloticus(2n=44) andO. karongae(2n=38),although the experiment was only at the laboratory scale with very small number and makes no mention of whether the hybrids were sterile or not.
A great deal has been written about reproductive isolation mechanisms, which can be categorized into spatial and ecological (geographical) isolation and genetic isolation approaches. The geographical barrier plays a significant role in pre-mating (spatial, seasonal, habitat, and behavioral), while the genetic barrier plays an inherent role in post-mating (structure, gametic incompatibility, hybrid non-viability or weakness, and hybrid sterility, Merrell, 1981). Previous research has shown strong evidence related to geographical barriers, but little regarding genetic barriers.
It was difficult to determine generic level differences in karyotype and isoenzyme betweenO. niloticusandS. melanotheronin the present study. We recognized that these differences might not be enough to conclude that they were responsible for the obstacles of hybridization, but we believe that in addition to the differences in natural distribution and breeding behavior that influence pre-mating isolation, the differences in karyotype and isoenzymes may relate to the genetic mechanism for post-mating isolation.
Our results suggest thatO. niloticus and S. melanotheronhave a conservative but different karyotypic structure. Their diploid numbers were both 2n=44, which is in agreement with previous research (Li, 1998; Harvey et al, 2002b). The most interesting finding was the significant difference in the composition of m and sm chromosomes between the two species, specificallyO. niloticushad three pairs of sm andS. melanotheronhad one pair of m and two pairs of sm. However, the number of st and t in the two species were the same, and the hybrids were of mixed m and sm composition. Harvey et al (2002a) reported thatS. melanotheronhas 2 m + 15 sm/st + 5t and Sofy et al (2008) reported thatO. niloticushas 1sm + 13st + 7t, which differ from our present findings. These differences may reflect the difficulties in obtaining high quality metaphase chromosome preparations and in accurately measuring chromosome arm lengths.
Acknowledgements:We wish to express our gratitude to Professor Zhang K.J., Shanghai Ocean University, and Mr. Jing W. K, Tianjin Fish Farm, for their help with chromosome sample preparation, and to Professor Louis Bernatchez, Laval University, Canada, and Lin Junda, Florida Institute of Technology, USA, for assistance with English.
Harvey SC, Campos-Ramos R, Kennedy DD, Ezaz MT, Bromage NR, Griffin DK, Penman DJ. 2002a. Karyotype evolution in Tilapia: mitotic and meiotic chromosome analysis of Oreochromis karongae and O. niloticus×O. karongae hybrids[J].Genetica,115: 169-177.
Harvey SC, Powell SF, Kennedy DD, McAndrew BJ, Penman, DJ. 2002b. Karyotype analysis of Oreochromis mortimeri (Trewavas) and Sarotherodon melanotheron (Rüppell) [J].Aquac Res,33: 339-342.
Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes[J].Hereditas,52: 201-220.
Li SF, Cai WQ. 1995. Introgression of farming populations between Nile tilapia and blue tilapia[J].J Fish, 19(2): 105-111.
Li SF. 1998. Genetical Characterization of Major Freshwater Culture Fishes in China[M]. Shanghai: Shanghai Scientific & Technical Publishers. (in Chinese)
Li SF, Yan B, Cai WQ, Li TY, Jie JH, Zhang YH. 2008a. Heterosis and related genetic analysis by SSR for the salt tolerance of reciprocal hybrids between Nile tilapia (Oreochromis niloticus) and blackchin tilapia (Sarotherodon melanotheron) [J].J Fish Sci Chn,15: 189-197. (in Chinese)
Li SF, Yan B, Cai WQ, Li TY, Jie JH, Zhang YH. 2008b. Evaluation of growth, salt tolerance and parents’ heterosis contribution in reciprocal hybrids F2 between Oreochromis niloticus and Sarotherodon melanotheron [J].J Fish Sci Chn,15: 335-341. ( in Chinese)
Liu YX, Li SF, Cai WQ, Li TY, Jia JH, Zhang YH. 2009. Evaluation on the effects of back clossing for salt tolerance selection of tilapia[J].J Fish Sci Chn,16(3): 332-339. (in Chinese)
Markert CL, Faulhaber I. 1965. Lactate dehydrogenase isozyme patterns of fish[J].J Exp Zool,159: 319-332.
Merrell DJ. 1981. Ecological Genetics[M]. London: Longman.
National Quality Inspection Bureu of Peoples’ Republic China. 2008. GB/T 18654.13 National standard of PRC: Inspection of gemplasm for cultured fishes. Part 13: Isozyme electrophoresis[S]. Nyman L. 1965. Species specific proteins in fresh water fishes and their suitability for a ‘protein taxonomy’ [J].Hereditas,53: 117-126.
Ohno S, Wolf U, Atkin NB. 1968. Evolution from fish to mammals by gene duplication[J].Hereditas,59: 169-187.
Sodsuk P, McAndrew BJ. 1991. Molecular systematics of three tilapiine generaTilapia,SarotherodonandOreochromisusing allozyme data[J].J Fish Biol,39: 301-308.
Sofy HL, Layla AM, Iman MKA. 2008. Karyotypic diversity of some tilapia species[J].Nat Sci,6: 19-27.
Thompson RR. 1960. Species identification by starch gel zone electrophoresis of protein extracts. I. Fish[J].J Ass Off Agric Chem,43: 763-764.
Trewavas R. 1983. Tilapiine Fishes of the Genera Sarotherodon, Oreochromis and Danakilia[M]. London: Comstock Publishing Associates.
Tsuyuki H, Roberts E. 1965. The species specifity and constancy of muscle myogen and hemoglobin electropherograms ofOncorhynchus[J].J Fish Res Board Can,22: 215-216.
尼罗罗非鱼和萨罗罗非鱼遗传生殖隔离的初步证据
李思发1,*, 赵 岩1, 范武江1, 蔡完其1, 许映芳2
(1.上海海洋大学 农业部水产种质资源与利用重点开放实验室,上海201306; 2.中国水产科学研究院长江水产研究所,湖北 荆州434000)
罗非鱼类(Tilapiini)含3个属70余种, 种间和属间颇易人工杂交, 但尼罗罗非鱼(Oreochromis niloticus)和萨罗罗非鱼(Sarotherodon melanotheron)人工杂交难度大, 产苗概率甚低, 要获得数量足够的可用于生产的杂交子代相当困难。该文对这两种鱼及其正交(O. niloticus♀ ×S. melanotheron ♂)和反交(S. melanotheron♀ ×O. niloticus ♂)子代的头肾细胞的核型进行了比较。此外, 采用同工酶电泳方法检测肾、肝、眼、肌肉、心中乳酸脱氢酶等4种同工酶的表型差异。4种遗传型罗非鱼具有相同的染色体二倍数(2n=44)和总臂数(NF=50), 但各具不同的染色体类型, 尼罗罗非鱼为3对近中着丝点染色体(sm)、12对近端着丝点染色体(st)和7对端着丝点染色体(t); 萨罗罗非鱼为1对中间着丝点染色体(m)、2对 sm、12对 st和7对 t;正反杂交子代表现为介于双亲之间的混合类型, 为0.5对m、2.5对sm、12对st和7对t。在同工酶中, 仅见肾脏乳酸脱氢酶电泳结果有清晰差异, 尼罗罗非鱼出现5条谱带, 萨罗罗非鱼3条, 而杂交子代6条, 且所有谱带的迁移率和活性均表现出多态性。据此初步认为, 核型和同工酶方面的差异可能是导致这两种不同属罗非鱼生殖隔离的遗传原因, 这些差异也可能为这两种(属)鱼的分类学提供新的遗传背景资料。
罗非鱼类;尼罗罗非鱼;萨罗罗非鱼;核型;同工酶;生殖隔离;分类
2011-03-17;接受日期:2011-07-13
罗非鱼行业技术体系(nycytx-48-3);公益性行业(农业)科研专项:罗非鱼大规格鱼种规模化培育与生态养殖技术研究(nyhyzx07-044-01);国家科技支撑计划专题:耐盐罗非鱼新品种选育(2006BAD01A1203)
Q959.483; Q951.3 ; Q959.483.09
A
0254-5853-(2011)05-0521-07
10.3724/SP.J.1141.2011.05521
date: 2011-03-17; Accepted date: 2011-07-13
*Corresponding author (通信作者), E-mail: sfli@shou.edu.cn
book=522,ebook=492

