Metabolic profiling of four synthetic stimulants, including the novel indanyl-cathinone 5-PPDi, after human hepatocyte incubation
Dvid Fregt-Sfont , Mrie Mrdl , Jun V. Sncho , Fˊelix Hernˊndez ,Kristin Linnet , Mrí Iˊ~nez ,*
a Research Institute for Pesticides and Water, University Jaume I, Avda. Sos Baynat s/n,12071, Castellˊon, Spain
b Department of Forensic Medicine, Section of Forensic Chemistry, Faculty of Health and Medicinal Sciences, University of Copenhagen, Frederik V's vej 11,2100, København Ø, Denmark
c Department of Occupational and Environmental Medicine, University Hospital of Northern Norway, Sykehusvegen, Tromsoe, Norway
Keywords:
ABSTRACTSynthetic cathinones are new psychoactive substances that represent a health risk worldwide. For most of the 130 reported compounds,information about toxicology and/or metabolism is not available,which hampers their detection (and subsequent medical treatment) in intoxication cases. The principles of forensic analytical chemistry and the use of powerful analytical techniques are indispensable for stablishing the most appropriate biomarkers for these substances. Human metabolic fate of synthetic cathinones can be assessed by the analysis of urine and blood obtained from authentic consumers;however, this type of samples is limited and difficult to access. In this work, the metabolic behaviour of three synthetic cathinones (4-CEC, 4-CPrC and 5-PPDi) and one amphetamine (3-FEA) has been evaluated by incubation with pooled human hepatocytes and metabolite identification has been performed by high-resolution mass spectrometry. This in vitro approach has previously shown its feasibility for obtaining excretory human metabolites.4-CEC and 3-FEA were not metabolised,and for 4-CPrC only two minor metabolites were obtained. On the contrary, for the recently reported 5-PPDi, twelve phase I metabolites were elucidated. Up to our knowledge, this is the first metabolic study of an indanylcathinone. Data reported in this paper will allow the detection of these synthetic stimulants in intoxication cases, and will facilitate future research on the metabolic behaviour of other indanyl-based cathinones.
1. Introduction
Synthetic cathinones are one of the largest groups of new psychoactive substances (NPS), with around 130 compounds being monitored by the European Monitoring of Drug and Drug Addiction(EMCDDA) [1]. They constitute the second group most frequently seized in Europe(around 33%of seizures containing NPS),being α-PVP,4-CMC,3-CMC,4-CEC and 3-MMC the most commonly found compounds [1]. In 2017, 12 novel synthetic cathinones were detected for the first time in Europe [1], which suggests the continuous expansion of this non-legal market.
The fast emerging of these NPS is evidenced by a number of research papers reporting novel cathinones [2-8]. This fact remarks the need of analytical strategies for monitoring the large number of cathinones already reported but also the constant apparition of new ones. Regarding consumption products, highresolution mass spectrometry (HRMS) is one of the most powerful techniques for cathinones identification, as it allows the nontarget and suspect analysis based on full-spectrum mass-accurate data provided by this technique [4,5,9,10]. However, facing the problem that synthetic cathinones consumption cannot be limited to only the analysis of legal highs and seizures,forensic and clinical analytical chemistry plays an important role in the monitoring of NPS in biological matrices. Several UHPLC-MS/MS and UHPLCHRMS target methodologies have been applied for the determination of synthetic cathinones in hair [11], oral fluid [12], urine [13],and even wastewater [14]. Nevertheless, most current methodologies are focused on the determination of synthetic cathinones as unaltered compounds.The lack of information about the metabolic fate of these substances makes their detection difficult in intoxication cases,as they are commonly partially or totally metabolised.So, it is of utmost importance to study the pharmacology and metabolic behaviour of these compounds, trying to establish the most suitable biomarkers for their monitoring, especially for the most recently detected cathinones in the continuously moving“market” of NPS.
Ideally,human samples,mainly blood and urine,should be used but these samples are difficult to access and limited to authentic intoxication cases[15-21].As an alternative,thein vitroapproach,using pooled human liver microsomes (pHLM), pooled human S9 fraction (pS9) or pooled human hepatocytes (pHH) can be applied to the study of human excretory metabolites[22,23].Among them,pHH seem to provide the best realistic pattern of metabolites[24],with most of them being detected in authentic urine samples from intoxication cases [20,25,26].
In this work,incubation with pHH was applied for investigating the metabolic behaviour of three synthetic cathinones and one amphetamine: the recently reported 3,4-trimethylene-α-pyrrolidinobutiophenone (5-PPDi) [27], 4-chloro-α-pyrrolidinopropiophenone (4-CPrC or 4-chloro-PPP [3]), 4-chloroethcathinone (4-CEC [28]), and 3-fluoroethamphetamine (3-FEA [29]) (Fig. 1).Metabolite identification was performed by UHPLC-HRMS. The results showed that 3-FEA and 4-CEC were not metabolised by pHH, while only 2 metabolites were detected for 4-CPrC. On the contrary, up to 12 phase I metabolites were elucidated for 5-PPDi.To our knowledge, this is the first study reporting the metabolic profile of an indanyl-cathinone. Data reported in this paper will allow to focus future analysis of urine from intoxication cases and hospital emergences associated with the consumption of cathinones and amphetamines.
2. Materials and methods
2.1. Reagents and chemicals
Research chemical samples containing 5-PPDi, 4-CPrC, 4-CEC and 3-FEA were kindly provided by Energy Control (Asociaciˊon Bienestar y Desarrollo,Barcelona,Spain),and compound purity was tested by nuclear magnetic resonance.MDMA-d5(used as internal standard) was purchased from Cerilliant (Round Rock, TX, USA).Diclofenac was purchased from Sigma-Aldrich(St.Louis,MO,USA).Reagents used in the incubation (0.4% trypan blue solution, fetal bovine serum, Leibovitz's L-15 medium, Gibco cryopreserved hepatocyte recovery medium), solvents (methanol, acetonitrile, ultrapure water), formic acid (LC-MS grade) as well as mass calibration solutions(Pierce®LTQ Velos ESI positive Ion calibration solution, Pierce™ESI negative ion calibration solution) were obtained from Fisher Scientific (Leicestershire, UK). The pHH were purchased from Lonza (Basel, Switzerland), and stored in liquid nitrogen until use.

Fig. 1. Structure of the three cathinones (5-PPDi, 4-CPrC and 4-CEC) and the amphetamine (3-FEA).
2.2. pHH incubation
Cryopreserved pHH were used in this work. Information about the incubation procedure applied can be found in literature [23].
The four tested compounds and diclofenac (used as positive control to test the feasibility of incubations) were added at a final concentration of 10 μM. Controls without the addition of hepatocytes were run simultaneously to identify hydrolysis products and artefacts.Aliquots of 20 μL were collected after 0,60,and 180 min of incubation and mixed with 80 μL of an ice-cold acetonitrile solution containing 100 ng/mL of internal standard. The extracts were frozen until analysis.
2.3. Instrumentation
Extracts were analysed using a Dionex Ultimate 3000 UHPLC system from Thermo Scientific(Germering,Germany)coupled to a Q Exactive high resolution mass spectrometer from Thermo Scientific (Bremen, Germany) equipped with a hybrid quadrupole-Orbitrap mass analyzer.
Chromatographic separation was performed using an Acquity HSS C18(2.1 mm×150 mm,1.8 μm)analytical column from Waters(Wexford,Ireland).The mobile phase consisted of water with 5 mM ammonium formiate and 0.1% (v/v) formic acid (solvent A), and acetonitrile with 0.1%(v/v)formic acid(solvent B).Flow rate was set to 0.25 mL/min,maintaining the column at 40°C.A 14 min gradient was used, changing the composition as follows: 5% B (0-0.5 min),70%B(0.5-9.95 min),and 99%B(9.9-10.0 min),where it remained isocratic for 2 min and followed by re-equilibration for 2 min.Samples were kept at 5°C, and the injection volume was 3 μL.
HRMS was equipped with a heated electrospray ionization source (HESI-II) working in positive (ESI+) and negative (ESI-)ionization modes. The capillary temperature was 350°C, and the spray voltage was 4.0 kV in ESI+and -4.0 kV in ESI-. Data were acquired using data-dependent acquisition (dd-MS2, DDA) and parallel reaction monitoring (PRM, MS/MS). Full-scan data (FTMS)were collected in a scan-range ofm/z200-900 using a resolution of 70,000 FWHM.DDA MS/MS was acquired at a resolution of 35,000 FWHM, while PRM were acquired at 17,500 FWHM. Nitrogen was used as the collision gas at normalized collision energy(NCE)of 10,30 or 50 eV. Data processing was performed using Compound Discoverer 2.0 software (Thermo Scientific) for a preliminary compound identification, and FreeStyle 1.3 (Thermo Scientific) for working with raw data. Further details about HRMS instrumentation can be found in literature [23].
3. Results and discussion
3.1. Analytical strategy for metabolite identification
Compound Discoverer 2.0 software was selected for UHPLCHRMS DDA data processing. Based on the structure of the selected compounds, a list of expected biotransformations (and their combinations)(Table S1)were selected for obtaining potential metabolites. Extracted ion chromatograms (EIC) with a mass window of ±5 ppm were automatically performed for searching the(de)protonated molecule of the expected metabolites in the fullscan function of the DDA acquisition. If a chromatographic peak was detected, it was compared with the blank and stability incubations,in order to discard false positives.Additionally,a binary comparison between blank incubations and compound incubations was performed for unexpected metabolites detection. Once the potential metabolites were selected, MS/MS spectra acquired during DDA acquisition were manually processed using FreeStyle 1.3 software. When a potential metabolite presented a fragmentation which fits with its expected biotransformation, it was re-analysed using PRM acquisition at three different collision energies (10, 30 and 50 eV). The PRM fragmentation of the metabolite was compared with the fragmentation pathway of parent compound.Based on this information, the position, where the biotransformation occurred,was determined,and the structure of the metabolite was suggested.In order to enhance the confidence in the structure elucidation, fragmentation pathways were proposed for all metabolites. The present work includes an (extremely) detailed explanation of the identification of metabolites based on the observed PRM fragmentation.
For the studied compounds, no additional metabolites in ESIwere observed.
3.2. Fragmentation pathway of 5-PPDi
The first step in the metabolite elucidation strategy applied was the in-depth study of the mass fragmentation behaviour of the parent compound (C17H24NO+,m/z258.1851, -0.9 ppm). Table 1 shows the information obtained on the 5-PPDi fragmentation,including accurate mass, elemental composition and mass error.Fig.2B shows the MS/MS fragmentation spectrum at 50 eV(MS/MS spectra at 10 and 30 eV can be found in Fig. S1).
Fig. 2A shows the proposed fragmentation pathway for 5-PPDi.The fragmentation starts with the disconnection of the different bonds of the α-carbon atom. On the one hand, the minor product ion atm/z229 corresponded to the homolytic dealkylation from parent compound.On the other hand,a product ion atm/z187 was obtained after the loss of the pyrrolidine moiety. A subsequent CO loss and structure rearrangement led to the ionm/z159. Product ions atm/z117 and 131 were obtained after an alkylic disconnection from m/z 159, depending on the C-C bond disconnected. Product ionm/z145 corresponded to the disconnection of the bond between the carbonyl and the α-carbon atom, releasing the alkylic chain and pyrrolidine moiety. Alternatively, product ionm/z117 could be also obtained after a CO loss from product ionm/z145.The pyrrolidine moiety ion(product ionm/z72)was also observed as a product ion,similarly to the ion corresponding to the alkylic chain and pyrrolidine moiety (product ionm/z112).In this case,m/z112 produced also the product ions atm/z84 and 70,corresponding the first one to the alkylic chain C-C bond disconnection and the second one to theN-dealkylation.
3.3. 5-PPDi metabolites elucidation
12 phase I metabolites were found after incubation.Six of these metabolites corresponded to hydroxylated compounds (M1-M6).Similarly, two metabolites corresponding to hydroxylation plus oxidation (M10 and M11), and two with ring opening and carboxylation (M8 and M9) were detected. The correct position of the biotransformation was determined after an accurate study of the observed PRM fragmentation. Additionally, metabolites corresponding to oxidation (M7)and double oxidation(M12)were also found. Table 1 shows the accurate mass, elemental composition,observed fragmentation, and mass error for the 12 identified metabolites.
3.3.1. Metabolite M1
The molecular formula obtained for metabolite M1 (m/z274.1799,C17H24NO2+,-0.9 ppm)corresponded to a hydroxylation.Product ion atm/z161 suggested the hydroxyl group to be in the aromatic moiety, based on the 16 Da shift respect the corresponding product ion of 5-PPDi (m/z145). Moreover, the product ion atm/z185 (C13H13O+) revealed that this product ion corresponded to the 187 observed for the parent compound(C13H15O+) but with a -2 Da shift, this is, with an additional insaturation. This would point out where the water loss had occurred.Based on both premises, the hydroxyl group was located on the trimethylene moiety (if it was in the aromatic ring, a water loss would not be possible). The other observed product ions could be satisfactorily justified based on the proposed structure. Fig. S2 shows the PRM spectra for M1, and Fig. S3 the proposed fragmentation pathway.
3.3.2. Metabolite M2
M2 was detected atm/z274.1800 (C17H24NO2+, -0.7 ppm). In this case,the observed fragmentation(Table 1,Fig.S4)indicated the hydroxyl group to be on the aromatic ring.Thus,the product ion atm/z158 (C11H10O+) could only be justified if the hydroxyl group was in the aromatic ring (Fig. S5). This allowed the loss of a water molecule from the carbonyl group (m/z256), as the subsequent insaturation could be stabilised by the aromatic ring. After that,product ions atm/z227 and 143 were produced, depending on which bond was disconnected. With this assumption, the fragmentation pathway was proposed, fitting the observed fragmentation with the proposed structure. In order to facilitate the visualisation of the electronic rearrangement during fragmentation pathway,the hydroxylation has been placed in a certain atom of the aromatic ring.
3.3.3. Metabolites M3 and M4
M3 and M4 were observed atm/z274.1798 and 274.1799 corresponding to C17H24NO2+(-1.1 and -0.9 ppm, respectively). Both metabolites presented exactly the same PRM fragmentation(available in Fig. S6 and S8). The productions atm/z72 (C4H10N+)and 70(C4H8N+)indicated that the hydroxylation did not occur in the pyrrolidine moiety.The product ionm/z203 was the equivalent to the 187 of 5-PPDi,but with a chemical shift of 16 Da.This would locate the hydroxyl group to be on the aromatic moiety or the alkylic chain. Product ion atm/z131 excluded the possibility of hydroxylation in the aromatic moiety whereas product ion atm/z112 suggested that the alkylic chain remained unaltered. Nevertheless,if the hydroxylation point was located in the alkylic chain,it was possible to obtain the product ionm/z112 after an intramolecular nucleophilic substitution (type SN2). Thus, M3 would correspond to the hydroxylation in the methyl group of the alkylic chain and M4 in the methylene (or vice-versa). Finally, fragmentation pathways of M3 (Fig. S7) and M4 (Fig. S9) were proposed(similarly to M2,the hydroxylation is placed in a certain atom of the alkylic chain in order to facilitate the visualisation of the electronic rearrangement).
3.3.4. Metabolite M5
The molecular formula obtained for metabolite 5 corresponded,again, to a hydroxylated metabolite (m/z274.1799, C17H24NO2+,-0.7 ppm). In this case, the PRM at 50 eV showed a complex fragmentation spectra, with up to 12 product ions (Table 1). As some of the product ions were also observed for 5-PPDi (m/z159,145,131,117),the hydroxylation in the aromatic ring was discarded.Product ionsm/z245,128 and 100 (all of them with a 16 Da shift from 5-PPDi product ions) were crucial for locating the correct position of the biotransformation. Thus, the hydroxylation was situated in the α-carbon atom.The product ion atm/z85(C4H5O2+),corresponding to the alkylic chain and the carbonyl, enhanced the confidence on the metabolite structure.The proposed structure for M5 could justify all the observed PRM product ions (Fig. S10), and its fragmentation pathway(Fig. S11) was proposed.
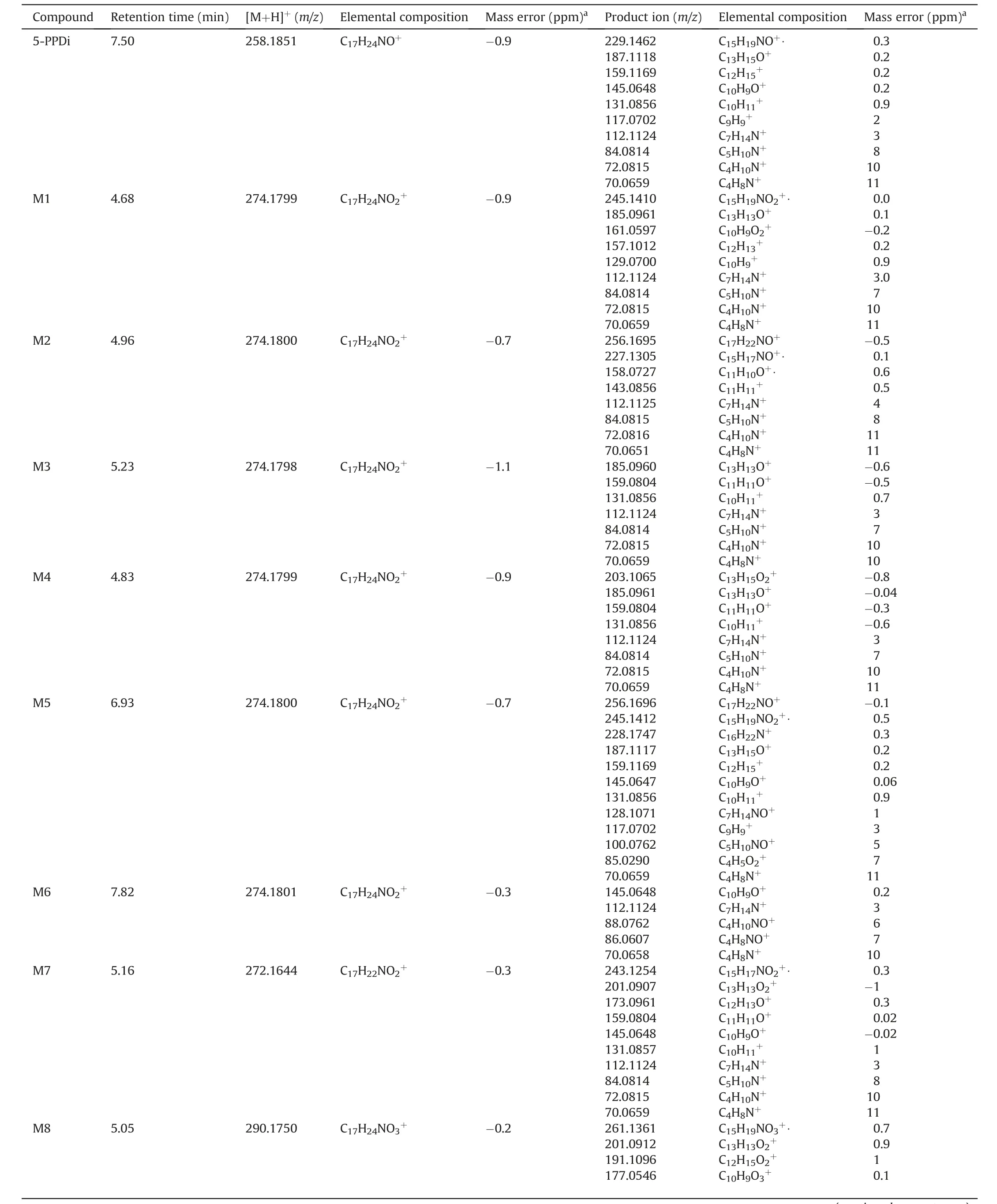
Table 1Mass spectrometric detection of 5-PPDi and its metabolites. Data obtained from PRM spectra.

Table 1 (continued)
3.3.5. Metabolite M6
M6 was the last hydroxylated metabolite, observed atm/z274.1801 (C17H24NO2+, -0.3 ppm). The position of the hydroxylation was easily assigned to be in the pyrrolidine moiety based on the product ions observed atm/z88 and 86(16 Da shifted from 5-PPDi product ions at m/z 72 and 70,respectively).In this case,only 5 product ions were observed in PRM spectra(Fig.S12),being all of them justified based on the proposed structure and fragmentation pathway(Fig. S13).
3.3.6. Metabolite M7
M7 corresponded to an oxidized metabolite, based on its elemental composition(m/z272.1644,C17H22NO2+,-0.3 ppm).This metabolite presented a similar fragmentation to M1(hydroxylation in the trimethylene moiety of the indanyl),but with some product ions shifted 16 Da(m/z201 vs 185,173 vs 157,and 145 vs 129).It is not surprising that some hydroxylated metabolites become oxidized,generating a ketone group which cannot be lost as water molecule. With this premise, the ketone group was placed on the indanyl ring, trying to justify all the observed PRM product ions(Table 1, Fig. S14). Product ions atm/z173 and 159 guarantee the biotransformation location. The proposed fragmentation pathway(Fig. S15) fitted perfectly with this biotransformation.
3.3.7. Metabolite M8
M8 exhibited a molecular formula containing 2 oxygen atoms more than parent compound (m/z290.1750, C17H24NO3+,-0.2 ppm), which could fit with a double hydroxylation or with a ring opening followed by a carboxylation.Product ionm/z177,with a 32 Da chemical shift respect them/z145 of 5-PPDi, revealed a possible double hydroxylation on the indanyl moiety.Nevertheless,it is hardly difficult to observe double hydroxylations in cathinones after pHH incubation [24], and the most common biotransformation described in literature for these compounds is the pyrrolidine ring opening followed by carboxylation [19,21,24]. So, it was proposed a ring-opening and carboxylation in the indanyl moiety,which perfectly fit with the observed PRM fragmentation (Table 1,Fig.S16).The fragmentation pathway(Fig.3A)was finally proposed,taking into account the fragmentation pathway described in literature for MDPV[19].Product ion atm/z119 was determined,as it is more feasible to produce this fragmentation with a ring opening carboxylation, instead of double hydroxylation. Nevertheless, it is not possible to completely rule out the double hydroxylation based only on the presence of product ionm/z119. For an unequivocal identification, analytical standard of both compounds should be needed for retention time and fragmentation comparison, or the use of additional analytical techniques such as nuclear magnetic resonance.
3.3.8. Metabolite M9
M9 presented the same elemental composition than M8 (m/z290.1749, C17H24NO3+, -0.3 ppm). In this case, the rich PRM fragmentation spectra obtained (Fig. S17), and more concretely the product ions atm/z272 (C17H22NO2+), 187 (C13H15O+) and 87(C4H7O2+), suggested the pyrrolidine ring opening and carboxylation of the alkylic chain obtained [19,21,24]. The 16 product ions observed in PRM spectra were satisfactorily justified using the fragmentation pathway reported for MDPV-COOH metabolite[19],and the fragmentation pathway of M9 was established (Fig. S18).

Fig. 2. Mass spectrometric behaviour of 5-PPDi. (A) Proposed fragmentation pathway. (B) PRM spectrum at 50 eV.
3.3.9. Metabolites M10 and M11
M10 and M11 presented the same proposed elemental composition (m/z288.1593 and 288.1594, C17H22NO3+, -0.30 and 0.01 ppm, respectively), which could match with a hydroxylation plus oxidation (carbonyl formation). In both cases, the observed PRM fragmentation (Table 1, Figs. S19 and S21) revealed that both indanyl and pyrrolidine moieties presented biotransformations.So,it was assumed that M10 and M11 should have a hydroxyl in the indanyl group and a ketone in the pyrrolidine, and vice-versa.
The product ions atm/z86(C4H8NO+,common to M6 and M9),98(C5H8NO+),126(C7H12NO+,also shared with M9)confirmed the ketone group to be in the pyrrolidine ring; the one atm/z157(shared with M1)confirmed the structure of M10.In a similar way,M11 was elucidated based on product ions atm/z173 (C12H13O+)and 159 (C11H11O+, shared with M4 and M7) and 128 (C7H14NO+,common to M5), which indicated that this metabolite presented a ketone group in the pyrrolidine moiety, and a hydroxyl in the indanyl, respectively. The structures of M10 and M11 were confirmed after proposing the fragmentation pathway for both compounds (Figs. S20 and S22, respectively).

Fig. 3. Mass spectrometric behaviour of M8. (A) Proposed fragmentation pathway. (B) PRM spectrum at 50 eV.
3.3.10. Metabolite M12
M12 was detected atm/z286.1434 (C17H22NO3+, -1.4 ppm),being the compound that presented the lowest response. In this case, only two product ions were observed (Fig. S23) atm/z159(shared with M7 and M11) andm/z126 (shared with M10). M12 was then proposed as a compound with two ketone groups,one in the indanyl moiety and the other in the pyrrolidine ring. The proposed fragmentation pathway (Fig. S24) fitted with the structure and fragmentation observed.
3.4. 5-PPDi metabolic fate
Once elucidated the 12 5-PPDi metabolites formed after pHH incubation,the metabolic pathway of this compound was proposed(Fig. 4). This indanyl-cathinone is hepatically metabolised mainly through hydroxylation and oxidation processes.
In the case of the pyrrolidine moiety, hydroxylation (M6),oxidation(ketone formation,M10 and M12)and oxidative opening(M9) occurred. These biotransformations have been reported for pyrrolidine-cathinones, such as MDPV[19],α-PHP [21],α-PBP [17]or 4-methoxy-α-PVP [25]. M3 and M4 corresponded to hydroxylations in the methyl or methylene groups of the alkylic chain,biotransformations were also observed for MDPV[19],α-PVT [26],PV8 [20] and α-PHP [21]. Nevertheless, no information about hydroxylations in the α-carbon of cathinones(M5)has been found in literature.Regarding the indanyl moiety,hydroxylation(M1,M10),ketone formation(M7,M11,M12)and oxidative ring-opening(M8)were observed.These biotransformations are quite similar to those observed for pyrrolidine moieties in literature.In some cases,more than one biotransformation was found,such as hydroxylation plus ketone formation (M10, M11) or formation of two ketones(M12).
At the end of the pHH incubation, the presence of 5-PPDi in media was around 67% respect to its initial response, indicating a relative high hepatic stability. In the case of the metabolites, M9 presented the higher response after 180 min (around 3.3% respect to 5-PPDi initial response),together with M8(3.2%),M4(3.1%),M2(1.6%) and M3 (1.4%). The remaining metabolites were found at responses lower than 0.4%, the lowest one being M12 with a relative response of 0.001%.These results show that the metabolic fate of 5-PPDi goes to the generation of carboxylic acid that involves ring-opening reactions(in this case,indanyl and pyrrolidine).Based on these results, the potential biomarkers proposed for detecting the consumption of 5-PPDi in intoxication cases by the analysis of urine and blood samples are 5-PPDi itself and the metabolites M8 and M9. Both compounds are the major metabolites after pHH incubation,and may be expected to be major metabolites of 5-PPDi in urine as well.Nevertheless,for analysis of urine samples,a previous enzymatic hydrolysis with β-glucuronidase should be performed,in order to de-conjugate M8/M9 glucuronide and detect the phase I metabolites.

Fig. 4. Proposed metabolic pathway for 5-PPDi.
3.5. Metabolic behaviour of 3-FEA, 4-CEC and 4-CPrC
Data obtained in this work revealed that amphetamine 3-FEA and the cathinone 3-CEC were not metabolised by the hepatocytes, probably due to the high polarity of both compounds.Differentin vitroexperiments performed with similar compounds also obtained few metabolites, and at low concentrations. As an example, 3-FEA was compared with the amphetamines 5-APDB
[30] and 2-AEPB [31] using pHLM, which usually present higher metabolic activity than pHH, identifying three metabolites per compound.In the case of 4-CEC,analogous compounds,such as 4-MEC[32],3-BMC and 3-FMC[33],have been studied,showing they are highly metabolised using pHLM (between 4-7 metabolites).Nevertheless, these incubations were performed at high concentration levels:50 μM for 4-MEC[32]and 250 μM for 3-BMC and 3-FMC [33]. In our work, the concentration tested (10 μM) was selected after considering the typical dose used by authentic consumers.Moreover,pHH were used in order to simulate the human hepatic metabolic fate for the studied compounds. The absence of metabolites for 3-FEA and 4-CEC suggest they are excreted as unaltered compounds in human urine, and so, the consumption biomarker proposed is the parent compound.
For the last compound studied, 4-CPrC, only two minor metabolites were found, each corresponding to less than 0.5% of the initial parent response at 180 min. The metabolite M1 corresponded to the pyrrolidine moiety-hydroxylated metabolite, and M2 to the pyrrolidine-ring opening followed by the formation of carboxylic acid. Both biotransformations have been described in literature for pyrrolidinophenone-derived cathinones, such as αpyrrolidinohexiophenone (α-PHP) [21], or 3,4-methylenedioxypyrovalerone (MDPV) [19]. Supplementary files contain the complete information about the 4-CPrC metabolism study, including the proposed metabolic pathway (Fig. S25), PRM fragmentation for parent compound and metabolites (Table S2,Figs. S26-28), and the proposed fragmentation pathways(Figs. S29-31).
4. Conclusion
This work reports the metabolic behaviour of four new psychoactive substances-3 synthetic cathinones and 1 amphetamineafter incubation with pooled human hepatocytes.The stimulants 3-FEA and 4-CEC were not metabolised, and therefore the parent compounds are proposed as biomarkers for monitoring the consumption of these compounds.4-CPrC also presented high hepatic stability,and only 2 phase I metabolites were detected.The parent compound and the oxidative ring-opening metabolite M2 seem to be the most suitable consumption biomarkers. In relation to the recently reported cathinone 5-PPDi, up to 12 phase I metabolites were identified, all of them resulting from hydroxylation and oxidation processes. The oxidative ring-opening metabolites M8(indanyl ring-opening)and M9(pyrrolidine ring-opening)were the major metabolites detected in terms of response. Nevertheless, at the end of the incubation, 5-PPDi gave around 67% of the initial response, suggesting again a high resistance of synthetic cathinones to hepatic metabolism.For monitoring the use of 5-PPDi,the parent compound and the metabolites M8 and M9 are proposed as consumption biomarkers. It is worth to noticing that 5-PPDi presents an indanyl moiety, and to our knowledge this is the first metabolic profile reported for an indanyl-based cathinone. The information provided in this work will be useful in forensic and clinical chemistry to focus the analytical determination of cathinones in biofluids, and also in other areas related to metabolic pathways of new psychoactive substances, such as toxicology and pharmacology.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
D. Fabregat-Safont, J.V. Sancho, F. Hernˊandez and M. Ibˊa~nez acknowledge financial support from the Ministerio de Economía y Competitividad Spain (Project CTQ2015-65603-P) and from Universitat Jaume I (UJI-B2018-19). D. Fabregat-Safont acknowledges Ministerio de Educaciˊon, Cultura y Deporte in Spain for his predoctoral grant(FPU15/02033)and for the financial support received for his research stay at the University of Copenhagen (EST17/00024).
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2019.12.006.
 Journal of Pharmaceutical Analysis2020年2期
Journal of Pharmaceutical Analysis2020年2期
- Journal of Pharmaceutical Analysis的其它文章
- Recent advances and perspectives of nucleic acid detection for coronavirus
- Molecular immune pathogenesis and diagnosis of COVID-19
- Hollow fiber-based liquid phase microextraction followed by analytical instrumental techniques for quantitative analysis of heavy metal ions and pharmaceuticals
- Quantitative computed tomography analysis for stratifying the severity of Coronavirus Disease 2019
- Erucic acid from Isatis indigotica Fort. suppresses influenza A virus replication and inflammation in vitro and in vivo through modulation of NF-κB and p38 MAPK pathway
- Analysis of TRPA1 antagonist, A-967079, in plasma using highperformance liquid chromatography tandem mass-spectrometry
