LIN28A inhibits DUSP family phosphatases and activates MAPK signaling pathway to maintain pluripotency in porcine induced pluripotent stem cells
Xiao-Long Wu, Zhen-Shuo Zhu, Xia Xiao, Zhe Zhou, Shuai Yu, Qiao-Yan Shen, Ju-Qing Zhang, Wei Yue,Rui Zhang, Xin He, Sha Peng, Shi-Qiang Zhang, Na Li,*, Ming-Zhi Liao, Jin-Lian Hua,*
1 College of Veterinary Medicine, Shaanxi Centre of Stem Cells Engineering and Technology, Northwest A & F University, Yangling,
Shaanxi 712100, China
2 College of Life Science, Northwest A & F University, Yangling, Shaanxi 712100, China
ABSTRACT LIN28A, an RNA-binding protein, plays an important role in porcine induced pluripotent stem cells(piPSCs). However, the molecular mechanism underlying the function of LIN28A in the maintenance of pluripotency in piPSCs remains unclear. Here, we explored the function of LIN28A in piPSCs based on its overexpression and knockdown. We performed total RNA sequencing (RNA-seq) of piPSCs and detected the expression levels of relevant genes by quantitative real-time polymerase chain reaction(qRT-PCR), western blot analysis, and immunofluorescence staining. Results indicated that piPSC proliferation ability decreased following LIN28A knockdown. Furthermore, when LIN28A expression in the shLIN28A2 group was lower (by 20%) than that in the negative control knockdown group (shNC), the pluripotency of piPSCs disappeared and they differentiated into neuroectoderm cells. Results also showed that LIN28A overexpression inhibited the expression of DUSP (dual-specificity phosphatases) family phosphatases and activated the mitogen-activated protein kinase (MAPK) signaling pathway. Thus,LIN28A appears to activate the MAPK signaling pathway to maintain the pluripotency and proliferation ability of piPSCs. Our study provides a new resource for exploring the functions of LIN28A in piPSCs.
Keywords: LIN28A; MAPK; Pluripotency; piPSCs; DUSP
INTRODUCTION
LIN28A has two nucleic acid-binding domains: i.e., cold shock domain and zinc-knuckle domain (Moss et al., 1997), which bind specific sequences and play vital roles in physiology(Balzer et al., 2010; Hafner et al., 2013; Polesskaya et al.,2007; Shyh-Chang & Daley, 2013; Xu et al., 2009; Zhu et al.,2011). In addition to RNA binding, LIN28A also functions as a transcription factor during reprogramming (Liao et al., 2008;Yu et al., 2007).LIN28Acan regulate the naïve to primed state conversion by regulating stem cell metabolism in induced pluripotent stem cells (iPSCs) (Zhang et al., 2016).Knockdown ofLIN28Apromotes the transformation of embryonic stem cells (ESCs) into the naïve state(Chandrasekaran et al., 2017; Kumar et al., 2014; Marks et al.,2012), andLIN28Aregulates pluripotent state transformation in mouse ESCs by inhibitingDPPA3expression (Sang et al.,2019). However, few studies have exploredLIN28Ain porcine iPSCs (piPSCs) and the mechanism underlyingLIN28Afunctions in piPSCs remains unclear. Previous research has indicated that inhibitingLIN28Aexpression via miR-370 may reduce piPSC proliferation ability and alkaline phosphatase(AP) activity, and up-regulate the expression of differentiationrelevant genes (Zhang et al., 2017). This finding differs from studies onLIN28Ain human and mouse PSCs (Zhang et al.,2017). Thus,LIN28Amay play a different role in piPSCs.
The mitogen-activated protein kinase (MAPK) signaling pathway plays important roles in controlling cell cycle,differentiation, proliferation, and apoptosis (Pearson et al.,2001; Shaul & Seger, 2007). Activation of the mitogenactivated protein kinase kinase (MEK)/extracellular signalregulated kinase (ERK) signaling pathway can promote the differentiation of mouse ESCs (mESCs), while its suppression can prevent mESC differentiation (Burdon et al., 1999;Deathridge et al., 2019). MESCs can be cultured in the 2i(CHIR99021 and PD0325901) system using MEK1 and glycogen synthase kinase-3 (GSK3) inhibitors (Ying et al.,2008).Invitroactivation of MAPK signaling helps maintain the primed state, whereas repression of MAPK signaling through ERK inhibition reverts PSCs to the naïve state (Chen et al.,2015; Hackett & Surani, 2014; Kalkan et al., 2019; Ying et al.,2008). The dual-specificity phosphatase (DUSP) family dephosphorylates MAPK signaling and plays an important role in regulating the duration, magnitude, and spatiotemporal profiles of MAPK activity (Caunt & Keyse, 2013; Chen et al.,2019). According to previous reports, the pluripotency of piPSCs is rapidly lost following treatment with 1.0 μmol/L MEK1 inhibitor PD0325901 (Gao et al., 2019). Thus, piPSCs may differ from mESCs in MEK/ERK signaling requirements and inhibiting MAPK may impair their pluripotency.
Our laboratory previously reported that the doxycycline(DOX)-inducible porcine PSC line (DOX-piPSC) can be cultured with cytokines (leukemia inhibitory factor (LIF), basic fibroblast growth factor (bFGF)), signaling inhibitors(CHIR099021, SB431542), feeder cells, and serum (Ma et al.,2018; Zhu et al., 2021). PiPSCs can be maintained in the pluripotent state with the addition of DOX, but differentiate after its withdrawal (Ma et al., 2018; Zhang et al., 2017; Zhu et al., 2021). In this study, we explored the function ofLIN28Aby knockdown and overexpression. Results showed that proliferation ability and colony size decreased significantly whenLIN28Awas knocked down. Furthermore, piPSCs overexpressingLIN28Amaintained colonies after DOX withdrawal. We also performed total RNA sequencing (RNAseq) of negative control knockdown in the OEOCT4-piPSC group (OEOCT4-shNC) andLIN28Aknockdown in the OEOCT4-piPSC group (OEOCT4-shLIN28A2) after withdrawal of DOX. Based on RNA-seq analysis, a reduction inLIN28Aexpression up-regulated differentiation-relevant gene expression and promoted neuroectoderm differentiation.LIN28Aalso inhibited the expression ofDUSPfamily members and activated the MAPK signaling pathway to maintain the pluripotency of piPSCs.
MATERIALS AND METHODS
Cell culture
HEK293T cells were cultured in 6-well plates (140675,Thermo Fisher Scientific, USA) using Dulbecco’s Modified Eagles Medium (DMEM) (Hyclone, USA) with 10% fetal bovine serum (FBS) (VIS, New Zealand). Mouse embryonic fibroblasts (MEFs) were cultured in 100 mm vessels (140675,Thermo Fisher Scientific, USA) using DMEM (10% FBS) and treated with mitomycin for 2.5 h. The mitomycin-treated MEFs were then passaged at 1×105cells/well into 12-well plates(140675, Thermo Fisher Scientific, USA) as the culture matrix for piPSCs. DOX-piPSCs were cultured in the LB2i system,which included 15% FBS, 0.1 mmol/L nonessential amino acids (NEAA) (Gibco, USA), 1 mmol/L L-glutamine (Gibco, USA),10 ng/mL LIF (14890-HNAE, Sino Biological, China),10 ng/mL bFGF (10014-HNAE, Sino Biological, China),0.1 mmol/L β-mercaptoethanol (M3148, Sigma-Aldrich, USA),3 μmol/L CHIR99021 (HY-10182, MCE, USA), 2 μmol/L SB431542 (S1067, Selleck, USA), and 4 μg/mL DOX (D9891,Sigma-Aldrich, USA). The piPSCs were passaged using TrypLE™ Select (Invitrogen, USA) into single cells at 2×104cells/well in a 12-well plate every 5-6 days (Ma et al.,2018).
Cell growth curve
To obtain the cell growth curves for the negative control knockdown group (shNC),LIN28Aknockdown group(shLIN28A1/2), negative control overexpression group(OENC), andLIN28A-overexpression group (OELIN28A), cells were cultured in 24-well plates at an initial density of 1×104cells/well. The piPSCs of theshNC,shLIN28A1/2, OENC,and OELIN28A groups were cultured in the LB2i system for 5 days and cell number in each group was counted daily using a blood counting chamber.
Vector construction and cloning
All lentivirus backbone vectors were derived from pCDH-CMVMCS-EF1-GreenPuro using a Seamless Cloning and Assembly Kit (Novoprotein, China).
Construction of short hairpin RNA (shRNA) vector
Here,shRNA of porcine LIN28A was designed using BLOCKiT™ RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress/design.do) (Supplementary Table S1) and the interference fragments were synthesized (enzymatic cleavage sites were BamHI and EcoRI). The double-stranded fragment was then connected to pCDH-U6-MCS-EF1-GFP-T2A-PURO linearized by BamHI and EcoRI via the T4 DNA ligase. All interference vectors were transfected into DOX-piPSCs and their interference efficiencies were verified, i.e., 80.78%(shLIN28A1) and 85.14% (shLIN28A2).
Construction of overexpression vector
Porcine testis cDNA was used as a template, and the porcine LIN28A fragment was successfully obtained using Prime Star Max DNA Polymerase (R045B, Takara, Japan). The fragment was then connected to pCDH-EF1-MCS-T2A-PURO linearized by BamHI and EcoRI via the T4 DNA ligase. The overexpression vector of porcine LIN28A was transduced to DOX-piPSCs and overexpression efficiency was detected.
Lentiviral packaging
HEK293T cells were cultured in a 6-well plate (140 675,Thermo Fisher Scientific, USA) at a density of 80%-90%.Lentiviral plasmids and packaged viral vectors (pVSV-G and psPAX2) were transfected into the HEK293T cells using polyethyleneimine (PEI, Sigma-Aldrich, USA). In total, 1 μg of pVSV-G, 1 μg of psPAX2, and 2 μg of the lentiviral vectors were mixed in 12 μL of PEI (1 mg/mL). The plasmid mixture was then rested for 15 min at room temperature, after which 200 μL of optiMEM was added. After 12 h, the culture medium was replaced with DMEM. The HEK293T cells were then cultured for 48-72 h to produce lentiviral particles. The lentiviral particles (culture supernatant) were gathered and filtered through a 0.45 μm filter to remove cell debris.
Lentiviral particle transduction
The piPSCs were cultured in a 12-well plate covered with MEF at 37 °C for 12 h. The lentiviral particles (supernatant) and piPSC medium were mixed at a 1:1 ratio and 4 μg/mL polybrene was added to the mixture. The piPSCs were cultured with mixed medium at 37 °C for 8-12 h, which was then replaced by new iPSC medium. Fluorescence was observed after 2-3 days and puromycin was used to select puro-positive cells after the piPSCs were cultured for 1 week.
Total RNA extraction, reverse-transcription polymerase chain reaction (PCR), and quantitative real-time PCR(qRT-PCR)
Total RNA was extracted using the RNAiso Plus reagent (9108,Takara, Japan) via the guanidine isothiocyanate phenolchloroform method (Chomczynski & Sacchi, 2006). Extracted RNA quality was detected using a NanoDropTMspectrophotometer (Thermo Fisher Scientific, USA) and agarose gel electrophoresis. Then, 2 μg of RNA was reverse transcribed to obtain cDNA using a FastKing RT Kit (with gDNase) (KR116, day root). Quantitative RT-PCR was performed using a SuperReal PreMix Plus (SYBR Green)(FP215, Tiangen, China) via a three-step process. The primers used for qRT-PCR are shown in Supplementary Table S2.
Western blot analysis
The piPSCs cultured for 5 days were digested by TrypLE™Select (Invitrogen, USA), and the same volume of DMEM+was added to neutralize the reaction. The mixture was then transferred to a 1.5 mL tube and centrifuged at 5 000gat 4 °C for 3 min. The supernatant of the culture medium was discarded and RIPA lysate (P0013B, Beyotime, China) with 10 mmol/L protease inhibitor PMSF (Sigma-Aldrich, USA) and phosphatase inhibitor was used to lyse the piPSCs for 30 min.Then, 5×SDS-PAGE loading buffer (JC-PE007, GENSHARE G, China) was added, followed by heating at 100 °C for 5 min.The protein samples were added to 8%-12% SDS-PAGE gel and run at 100 V for 1.5 h, then transferred onto polyvinylidenefluoride (PVDF) membranes at 15 V for 45 min using a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell(BioRad, USA). The membranes were then blocked using 8%skim milk (5% bovine serum albumin (BSA) in phosphop44/42 MAPK and p44/42 MAPK) at room temperature for 2 h.Primary antibodies, including LIN28A (1∶400; Santa Cruz Biotechnology, USA), proliferating cell nuclear antigen (PCNA)(1∶500; Boster, China), FLAG (1∶1 000; Sigma-Aldrich,USA), β-actin (1∶4 000; Sungene Biotech, China), phosphop44/42 MAPK (1∶1 000; Cell Signaling Technology, USA),and p44/42 MAPK (1∶1 000; Cell Signaling Technology,USA), were diluted in TBS-T buffer (20 mmol/L Tris HCl/pH 8.0,150 mmol/L NaCl, 0.05% Tween 20) according to the instructions and then incubated at 4 °C for 12 h.
The PVDF membranes were washed using TBS-T buffer at room temperature for 15 min. AffiniPure Goat Anti-Mouse/Rabbit IgG (H+L) was then used to combine the antibody at 37 °C for 1 h (Hu et al., 2020). The membranes were washed using TBS-T buffer at room temperature for 15 min and a Tanon-5200 automatic chemiluminescence image analysis system (Tanon, China) was used to detect the horseradish peroxidase (HRP) signal. The relative grays of the western blots were analyzed by ImageJ.
Immunofluorescent staining
After twice washing with PBS, the piPSCs (cultured for 5 days)were fixed in 4% paraformaldehyde (pH 7.4) at room temperature for 15 min. We used 0.1% Triton-100 to perforate the membranes at room temperature for 10 min. Then, 10%FBS was used to block the membranes at room temperature for 1 h. The membranes were incubated with primary antibodies, including LIN28A (1∶200; Santa Cruz Biotechnology, USA) and FLAG (1∶1 000; Sigma-Aldrich,USA), for 12 h at 4 °C and then washed three times with PBS.The membranes were then incubated with goat anti-mouse IgG (H+L) secondary antibody Alexa Fluor 488 conjugate(1∶500; ZSGB-BIO, China) at room temperature for 1 h and nuclei were stained with Hoechst33342 (1∶1 000) at room temperature for 5 min (Ma et al., 2019; Wei et al., 2021).
AP staining
The piPSCs (cultured for 5 days) were fixed in 4%paraformaldehyde (pH 7.4) at room temperature for 15 min,with AST Fast Red TR and α-naphthol AS MX phosphate(Sigma-Aldrich, USA) then used to stain the cells according to the manufacturer’s instructions. The piPSCs were incubated in 1.0 mg/mL Fast Red TR, 0.4 mg/mL α-naphthol AS-MX, and 0.1 mmol/L Tris-HCL 8.8 buffer at room temperature for 20 min. The AP-positive piPSCs clones showed red (Zhang et al., 2017). Images were obtained using a Nikon phase difference microscope.
5-Ethynyl-2’-deoxyuridine (EdU) staining
EdU detection was performed on 3 d cultured piPSCs according to the Cell-Light EdU Apollo567In-VitroKit instructions (RiboBio, China). The PiPSCs were exposed to 50 μmol/L EdU medium at 37 °C for 20 min, and then fixed in 4% paraformaldehyde (pH 7.4) at 37 °C for 15 min. After this,2 mg/mL glycine was added to neutralize excess aldehyde.The piPSCs were then exposed to 1×Apollo staining solution at 37 °C for 30 min and washed three times with PBS. We then used 0.1% Triton-100 to perforate the membranes for 10 min at 37 °C and the nuclei were stained with Hoechst33342 (1∶1 000).
RNA-seq
To explore the molecular mechanism of LIN28A in piPSCs,theshNC andshLIN28A2 groups with two biological replicates underwent RNA-seq. Total RNA was extracted using RNAiso Plus reagent (9 108, Takara, Japan) with the guanidine isothiocyanate phenol-chloroform method (Chomczynski &Sacchi, 2006). Extracted RNA quality was detected using a NanoDropTMspectrophotometer (Thermo Fisher Scientific,USA) and agarose gel electrophoresis. Total RNA was treated using Oligo dT-enriched mRNA and purification. Fragmented RNA was then reverse-transcribed using random N6 primers to form double-stranded DNA (second-strand cDNA synthesis with dUTP instead of dTTP). Next, 3'adenylated and adaptor ligation was added to the end of the synthesized doublestranded DNA, which was amplified using PCR with specific primers. The PCR products were thermally denatured into a single strand and a bridging primer was used to form a loop to obtain a single-stranded circular DNA library. Quality control(QC) was conducted on the DNA library, and sequencing was then performed on the DNBSEQ platform. Raw reads were filtered through QC using SOAPnuke. Finally, Bowtie2 was used to align the clean reads. Heatmaps were plotted through pheatmap (v1.0.12) to represent specific gene expression levels. Both Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted using clusterProfiler (v3.12.0). An adjustedP-value of <0.05 and Q-value of <0.05 were used to define the working threshold for statistical significance.
Statistical analysis
Two-tailedt-tests were used to determine significant differences between two groups and one-way or two-way analysis of variance (ANOVA) was used to determine significant differences between three groups. All data are shown as mean±standard error of the mean (SEM).Differences were considered significant whenP<0.05.
RESULTS
Effects of LIN28A on piPSC proliferation ability
LIN28AmRNA expression fluctuated under different concentrations of DOX (Supplementary Figure S1A), reaching a maximum level with the addition of 2 μg/mL DOX and then gradually decreasing with increasing concentrations of DOX(4 μg/mL to 16 μg/mL). Interestingly, colony size decreased withLIN28Aexpression after the administration of increasing concentrations of DOX (4 μg/mL to 16 μg/mL) (Supplementary Figure S1B).
Therefore, we next explored the relationship between the expression level ofLIN28Aand colony size. We designed two pairs ofshRNA and constructed aLIN28Ainterference vector.The interference efficiency of the vector was detected by qRTPCR. Results showed that the mRNA and protein expression levels ofLIN28Adecreased significantly in theshLIN28A1/2 groups (Figure 1A, B). Compared with theshNC group, colony size and AP activity decreased followingLIN28Aknockdown(Figure 1C). The cell growth curve showed that proliferation ability and protein expression of proliferating cell nuclear antigen (PCNA) decreased significantly whenLIN28Awas knocked down (Figure 1B, D). These results indicate that the proliferation ability of piPSCs decreased afterLIN28Aknockdown.
To further explore its function, porcineLIN28Awas overexpressed in piPSCs (OELIN28A group) and its expression level was detected by qRT-PCR, western blotting,and immunofluorescent staining (Supplementary Figure S2A,B). Compared to the negative control overexpression group with DOX (OENC+DOX+), colony size and AP activity in the OELIN28A+DOX+ group did not change significantly(Supplementary Figure S2C). Furthermore, compared to the OENC+DOX+ group, the percentage of EdU-positive cells in the OELIN28A+DOX+ group did not change significantly(Supplementary Figure S2D, E). The cell growth curve also showed that there was no significant change in proliferation ability after overexpression ofLIN28A(Figure 1D).
The DOX-piPSCs were maintained in the pluripotent state with DOX but differentiated after its withdrawal (Figure 1E, first column), consistent with previous studies (Ma et al., 2018;Zhang et al., 2017). The colonies in the negative control overexpression group without DOX (OENC+DOX-) gradually disappeared (Figure 1E, first column). However, colonies in the OELIN28A group without DOX (OELIN28A+DOX-) were still observed, although their size and number decreased significantly compared with the OENC+DOX+ and OELIN28A+DOX+ groups (Figure 1E, F). These findings indicate that LIN28A can maintain typical colonies after withdrawal of DOX. Pluripotent-relevant genes were detected by qRT-PCR. Results showed that the expression levels ofSALL4andNANOGincreased and decreased, respectively, in the OELIN28A groups (Figure 1G). Thus, the knockdown and overexpression experiments showed thatLIN28Aplays a vital role in maintaining the proliferation ability of piPSCs.
Effects of LIN28A on piPSC pluripotency
LIN28AmRNA expression levels in the piPSCs fluctuated with DOX concentrations (Supplementary Figure S1A). Therefore,we explored the function ofLIN28Aafter excluding the influence of DOX. Results showed that OEOCT4-piPSCs maintained colonies and proliferation ability in the absence of DOX (Supplementary Figure S3A). Therefore, experiments were performed on piPSCs overexpressingOCT4(OEOCT4-piPSCs).
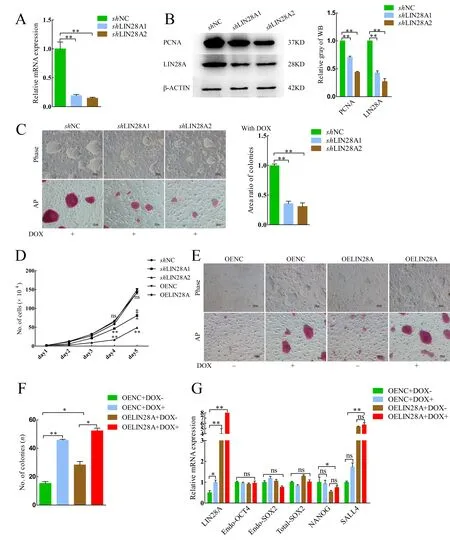
Figure 1 Effects of LIN28A on piPSC proliferation ability

Figure 2 Effects of LIN28A on piPSC pluripotency
The interference efficiency of shLIN28A1 and shLIN28A2 was similar, but the proliferation rate of shLIN28A2 decreased more obviously according to the cell growth curve and expression of PCNA, so subsequent experiments were performed on the shLIN28A2 groups. Compared with the OEOCT4-shNC group without DOX, colony size decreased in the OEOCT4-shLIN28A2 group without DOX (Figure 2A, B),consistent with the shLIN28A2 group results (Figure 1C). TheLIN28AmRNA expression level was detected again and was significantly decreased (Figure 2C). The percentage of EdUpositive cells also decreased significantly in the OEOCT4-shLIN28A2 group (Figure 2D, E). The AP staining assays showed that AP activity and colony size decreased significantly in the OEOCT4-shLIN28A2 group without DOX(Figure 2F). Based on AP staining assays (Sang et al., 2019),colonies can be classified into three shapes: i.e., typical primed diffuse-shape, compact dome-shape, and mixedshape. Compact dome-shaped colonies in the OEOCT4-shNC group accounted for 60% of total colonies in this group, while compact dome-shaped colonies in the OEOCT4-shLIN28A2 group accounted for ~50%; however, the total number of colonies in the OEOCT4-shLIN28A2 group decreased significantly and colonies in the OEOCT4-shLIN28A2 group basically disappeared (Figure 2G).
The results obtained for the OEOCT4-shLIN28A2 group with DOX were similar to that of the OEOCT4-shLIN28A2 group without DOX. Compared to the OEOCT4-shNC group with DOX, colony size (Supplementary Figure S3B, E) and AP activity in the OEOCT4-shLIN28A2 group with DOX decreased significantly (Supplementary Figure S3E). The percentage of EdU-positive cells also decreased significantly in the OEOCT4-shLIN28A2 group with DOX (Supplementary Figure S3C, D). The number of compact dome-shaped colonies in the OEOCT4-shLIN28A2 group with DOX decreased significantly compared with that in the OEOCT4-shNC group (Supplementary Figure S3F). These results indicate that pluripotency decreased after LIN28A knockdown and LIN28A plays a vital role in maintaining the pluripotency of piPSCs.
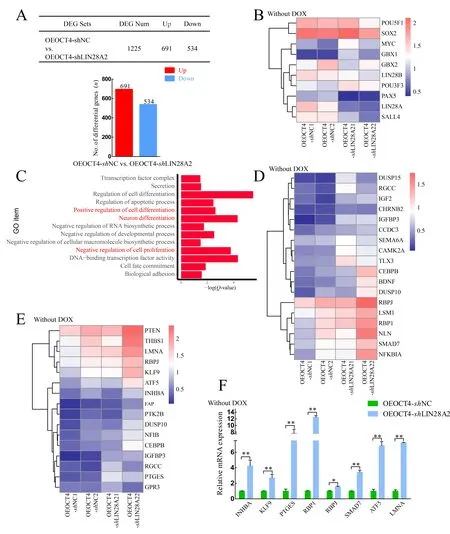
Figure 3 LIN28A inhibited expression of differentiation-related genes
LIN28A inhibited expression of differentiation-related genes
We performed total RNA-seq on two samples in the OEOCT4-shNC and OEOCT4-shLIN28A2 groups after withdrawal of DOX. The table in Figure 3A shows the number of up- and down-regulated differentially expressed genes (DEGs) in the OEOCT4-shNC and OEOCT4-shLIN28A2 groups after DOX withdrawal. Among them, the expression levels ofOCT4/SOX2/LIN28Bshowed no significant changes, but the expression level ofLIN28Adecreased in the OEOCT4-shLIN28A2 group (Figure 3B). After analyzing the RNA-seq results, GO analysis showed enrichment in positive regulation of cell differentiation, neuronal differentiation, negative regulation of cell proliferation, and DNA-binding transcription factor activity (Figure 3C). The expression of genes involved in the negative regulation of cell proliferation increased significantly in the OEOCT4-shLIN28A2 group based on heat map and qRT-PCR (Figure 3D, F), which may explain the decline in piPSC proliferation ability followingLIN28Aknockdown. In addition, the heat map and qRT-PCR results showed that the expression levels of genes involved in neuronal differentiation and positive regulation of cell differentiation also increased significantly in the OEOCT4-shLIN28A2 group (Figure 3D, F), which may explain the significant decrease in AP activity afterLIN28Aknockdown.These data indicate thatLIN28Acan inhibit the expression of differentiation-related genes in piPSCs and maintain the proliferation ability and pluripotency of piPSCs.
LIN28A inhibited expression of DUSP family and activated MAPK signaling pathway
Based on KEGG analysis, the primary enriched pathways included Axon guidance, Notch signaling pathway, and MAPK signaling pathway (Figure 4A). The MAPK signaling pathway plays an important role in pigs, and its inhibition can result in loss of pluripotency in porcine PSCs (Gao et al., 2019). Both heat map and qRT-PCR analyses showed that the mRNA expression levels ofDUSP-family members increased in the OEOCT4-shLIN28A2 group (Figure 4B, C), and the protein expression levels of ERK and phospho-ERK (p-ERK) in the OEOCT4-shLIN28A2 group significantly increased and decreased, respectively (Figure 4D). These findings indicate that the MAPK signaling pathway is inactivated following LIN28A knockdown.
TheDUSPmRNA expression levels were significantly decreased (Figure 4E) and the ERK and p-ERK protein expression levels were significantly decreased and increased,respectively, whenLIN28Awas overexpressed (Figure 4F).These results indicate that the MAPK signaling pathway was activated whenLIN28Awas overexpressed andLIN28Aactivated the MAPK signaling pathway by inhibiting theDUSPfamily phosphatases. Results showed that cell proliferation ability and AP activity decreased when the MEK1 inhibitor(PD0325901) was used, consistent with the phenomena in the OEOCT4-shLIN28A2 cells (Figure 4G). Cell proliferation ability and AP activity decreased with the addition of 1 μmol/L PD0325901, but this decrease was rescued by the overexpression ofLIN28A(Figure 4H). These results suggest thatLIN28Acan maintain the pluripotency and proliferation ability of piPSCs by activating the MAPK signaling pathway.
DISCUSSION
PiPSCs can be generated using human OCT4, SOX2, KLF4,and c-MYC lentiviruses in porcine fetal fibroblasts (Esteban et al., 2009; Ezashi et al., 2009). All piPSCs can exhibit terminal differentiation and generation of teratomainvivo, but none can produce chimeric offspring and germline transmission,suggesting that their pluripotency may be defective (Cheng et al., 2012; Esteban et al., 2009; Ezashi et al., 2009; Xue et al.,2016; Zhang et al., 2015, 2017). In the past several years,various laboratories have obtained piPSCs and modified the culture system (Cheng et al., 2012; Haraguchi et al., 2012;Hou et al., 2016; Xu et al., 2020; Xue et al., 2016; Zhang et al., 2015, 2017). Here, we attempted to obtain naïve piPSCs by modifying the patterns of gene expression.
Lin28ais the on-off switch between naïve and primed states. PSCs convert to the naïve state whenLin28adecreases, but to the primed state whenLin28aincreases(Marks et al., 2012). Interestingly, in our study, piPSC proliferation, colony size, and AP activity all decreased followingLIN28Aknockdown (Figure 1B-D). We also found that the LIN28A mRNA expression level in piPSCs fluctuated with DOX concentration as LIN28A is regulated by OCT4/SOX2 (Supplementary Figure S1A) (Buganim et al.,2012). Therefore, we further explored the function of LIN28A excluding the influence of DOX. Results showed that the DOX-piPSCs started to differentiate after the withdrawal of DOX (Figure 1E, first column). By exploring single pluripotency gene function using a lentiviral overexpression system, we found that piPSCs maintained typical colonies after withdrawal of DOX when OCT4 was overexpressed(Supplementary Figure S3A) (Zhu et al., 2021). Compared with piPSCs, the cell proliferation and AP activities of the OEOCT4-piPSCs after DOX withdrawal showed no obvious differences. Therefore, experiments were performed on OEOCT4-piPSCs, with similar results as for piPSCs. Based on RNA-seq, we demonstrated that the pluripotency of the piPSCs disappeared and piPSCs differentiated into neuroectoderm cells whenLIN28Awas knocked down.Lin28ais also highly expressed in ESCs and is downregulated in response to differentiation (Balzer et al., 2010; Richards et al.,2004; Yang & Moss, 2003). Moreover, the expression levels of LIN28B showed no significant changes, indicating that knockdown of LIN28A did not affect LIN28B expression.LIN28A also plays an important role in nervous system development (Faunes, 2020; Romer-Seibert et al., 2019;Yermalovich et al., 2020). The expression of genes involved in the negative regulation of cell proliferation, neuronal differentiation, and positive regulation of cell differentiation also increased in the OEOCT4-shLIN28A2 group (Figure 3D-F). Thus, after LIN28A knockdown, the pluripotency of piPSCs disappeared, the proliferation ability of piPSCs decreased, and the piPSCs differentiated into neuroectoderm cells. These results are consistent with previous study, which found that miR-370 can inhibit the expression of LIN28A(Zhang et al., 2017). To further explore its function, porcine LIN28A was overexpressed in the piPSCs. However, colony size, proliferation ability, and AP activity demonstrated no significant change after LIN28A overexpression. This was not obvious with the addition of DOX but was observed after the withdrawal of DOX. The OELIN28A+DOX- group maintained typical colonies, whereas the OENC+DOX- group did not(Figure 1E), suggesting that LIN28A plays an important role in maintaining the proliferation ability of piPSCs.
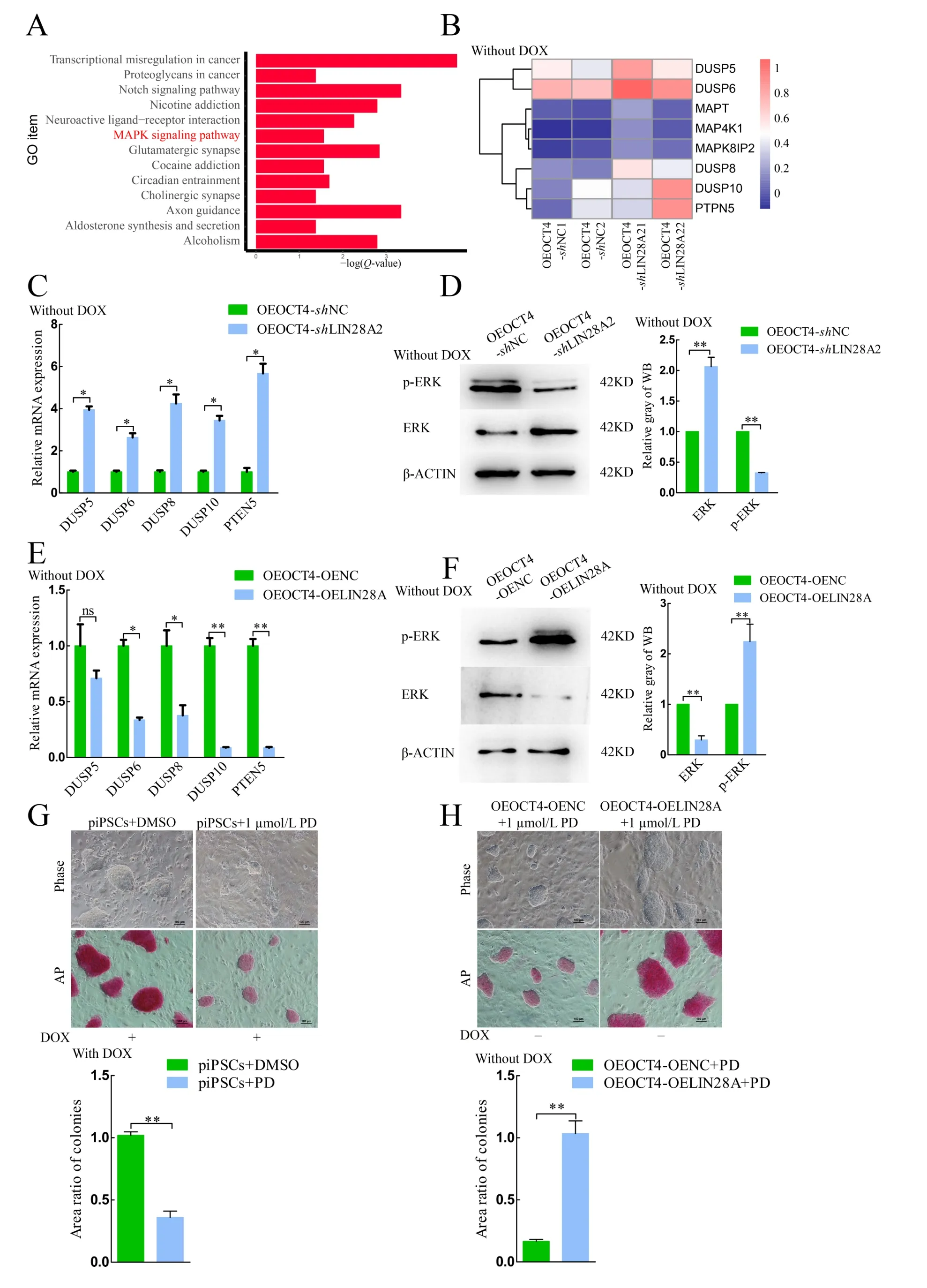
Figure 4 LIN28A inhibited DUSP-family expression and activated MAPK signaling pathway
Previous immunohistochemical analysis reported that ERK phosphorylation is up-regulated inLin28atransgenic mice(Kobayashi & Kozlova, 2018), which, in turn, promotes the phosphorylation and protein stability of LIN28A (Tsanov &Daley, 2017; Tsanov et al., 2017). We found thatLIN28Ainhibited the expression ofDUSP-family phosphatases, which activated ERK signaling (Figure 4C-F). The mRNA expression levels ofDUSP6/8/10were markedly up-regulated afterLIN28Aknockdown. As a member of theDUSPfamily,DUSP6/8/10inhibits ERK activity to regulate MAPK signaling in ovarian epithelial cancer (Gao et al., 2020), pancreatic cancer (Liu et al., 2021), and human epidermal stem cells(Hiratsuka et al., 2020). The increase inDUSP6/8/10expression indicates inactivation of MAPK signaling(Cornacchia et al., 2019). The weakly positive AP activity in theshLIN28A1/2 and OEOCT-shLIN28A2 groups was consistent with the results obtained when DOX-piPSCs were supplemented 1.0 μmol/L MEK1 inhibitor PD0325901(Figures 1C, 2F, 4G).Invitro, MAPK is the key for maintaining the primed state, whereas PSCs transform into the naïve state with MAPK signaling repression (Chen et al., 2015; Hackett &Surani, 2014; Ying et al., 2008). However, the pluripotency of piPSCs is rapidly lost with 1.0 μmol/L MEK1 inhibitor PD0325901 (Gao et al., 2019). This indicated that inactivation of MAPK signaling impaired the pluripotency of piPSCs. We demonstrated thatLIN28Amaintained the pluripotency piPSCs by activating the MAPK signaling pathway. Therefore,further investigations on the molecular mechanisms underlying howLIN28Aregulates its downstream genes and interacts with other transcription factors in piPSCs are warranted.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
X.L.W., Z.S.Z., and J.L.H. designed the research. X.L.W.,Z.S.Z., Z.Z., and S.Y. performed the research. X.L.W., X.X,and J.L.H. wrote the paper. X.X, Q.Y.S., and M.Z.L analyzed the data. X.L.W., Z.S.Z., J.Q.Z, W.Y., R.Z., X.H., S.P., S.Q.Z.,N.L., M.Z.L, and J.L.H. modified the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
The authors thank Dr. Ying Zhang and Fang-Lin Ma for helpful comments on this paper.
- Zoological Research的其它文章
- Northern pig-tailed macaques (Macaca leonina)infected with SARS-CoV-2 show rapid viral clearance and persistent immune response
- A review of the Cypriniform tribe Yunnanilini Prokofiev,2010 from China, with an emphasis on five genera based on morphologies and complete mitochondrial genomes of some species
- Particulate matter exposure exacerbates susceptibility to SARS-CoV-2 infection in humanized ACE2 mice
- Potential aquatic environmental risks of trifloxystrobin:Enhancement of virus susceptibility in zebrafish through initiation of autophagy
- Cataract-causing allele in CRYAA (Y118D) proceeds through endoplasmic reticulum stress in mouse model
- Species bias and spillover effects in scientific research on Carnivora in China

