Phylogeography and Cryptic Species Diversity of Paramesotriton caudopunctatus Species Group (Salamandridae:Paramesotriton) in Guizhou,China
Tao LUO,Huamei WEN ,Kai GAO,Jun ZHOU and Jiang ZHOU*
1 School of Karst Science,Guizhou Normal University,Guiyang 550001,Guizhou,China
2 School of Life Sciences,Central China Normal University,Wuhan 430000,Hubei,China
3 Key Laboratory for Biodiversity and Ecological Engineering of Ministry of Education,College of Life Sciences,Beijing Normal University,Beijing 100875,China
Abstract The Paramesotriton caudopunctatus species group is mainly distributed in the karst mountain ecosystems of Guizhou,China.Although some species have been included in previous phylogenetic studies,the evolutionary relationships and divergence-time of members of this species group as a whole remain unexplored.In this study,we report the sequencing of one protein coding mitochondrial gene fragment (ND2) and one nuclear gene (POMC),and use a combination of phylogenetic analyses and coalescent simulations to explore the cryptic diversity and evolutionary history of the P.caudopunctatus species group.Phylogenetic relationships revealed that the P.caudopunctatus species group is composed of two major groups,i.e.,East Clade and Western-South Clade.The divergence-time and ancestral area estimation suggested that the P.caudopunctatus species group likely originated in the Doupeng Mountains in Guizhou,China at 12.34 Ma (95% HPD:8.30-14.73),and intraspecific divergence began at about 2.17 Ma (95% HPD:1.39-2.97).This timing coincides with the orogenesis of the Miaoling Mountains during the Late Miocene to early Pleistocene.The delimitation of species in the P.caudopunctatus species group supports the existence of the currently identified species,and consensus was confirmed across methods for the existence of least to two cryptic species within what has been traditionally considered to be P.caudopunctatus species group.This study is of significance for understanding the species formation,dispersal,and diversity of the tailed amphibians in the karst mountains ecosystem of Guizhou and the role of the Miaoling Mountains as a geographical barrier to species dispersal.
Keywords P.caudopunctatus species group,phylogeography,cryptic diversity,biogeography,species delimitation,Guizhou
1.Introduction
TheParamesotriton
Chang,1935 of the Salamandridae are distributed in northern Vietnam and southwest-central,and southern China (Feiet al
.,2006;Frost,2020).This genus currently contains 14 species (Frost,2020),and noticeably,seven of these have been described in the past decade (Liet al
.,2008a,2008b;Wuet al.
,2010;Guet al
.,2012a;Wanget al
.,2013;Yuanet al
.,2014,2016a).Except forP.deloustali
(Bourret,1934),all other species are endemic to China,and 14 of them are assigned into two species groups (Feiet al
.,2006;Yuanet al
.,2014):(1) theP.caudopunctatus
species group,five species:P.caudopunctatus
(Liu and Hu,1973),P.wulingensis
Wang,Tian,and Gu,2013,P.longliensis
Li,Tian,Gu,and Xiong,2008,P.zhijinensis
Li,Tian,and Gu,2008 andP.maolanensis
Gu,Chen,Tian,Li,and Ran,2012;(2) and theP.chinensis
species group,nine species:P.aurantius
Yuan,Wu,Zhou,and Che,2016,P.chinensis
(Gray,1859),P.deloustali
(Bourret,1934),P.fuzhongensis
Wen,1989,P.guangxiensis
(Huang,Tang,and Tang,1983),P.hongkongensis
(Myers and Leviton,1962),P.labiatus
(Unterstein,1930),P.qixilingensis
Yan,Zhao,Jiang,Hou,He,Murphy,and Che,2014,andP.yunwuensis
Wu,Jiang,and Hanken,2010.The diversity ofParamesotriton
spp.has been surveyed in several southern provinces of China,and several new species have been described in the provinces of Guizhou (Liet al
.,2008a,2008b;Guet al
.,2012a;Wanget al
.,2013),Guangxi (Wuet al
.,2009),Guangdong (Wuet al
.,2010),Hubei (Yuanet al
.,2016a),and Jiangxi (Yuanet al
.,2014).Given the taxonomic diversity of these salamanders,additional investigations are needed to develop effective plans for their conservation and management.TheP.caudopunctatus
species group represents several taxa of mountains stream salamanders,narrowly distributed in medium to high elevation areas in the Guizhou Province,China (Feiet al
.,2016).Historically it was considered a single species (Feiet al
.,2006).Currently,P.caudopunctatus
is listed as near threatened by the IUCN (https://www.iucnredlist.org/species/59456/11945015) and as vulnerable on the Red List of China’s Vertebrates due to habitat degradation and overfishing,which has led to a decline in its population (Feiet al
.,2012;Jianget al
.,2016).P.longliensis
,P.zhijinensis
,andP.maolanensis
are listed as endangered due to their narrow area of distribution and limited population size (Feiet al
.,2012,2016).Previous taxonomic studies have shown that theP.caudopunctatus
species group is composed of several species,includingP.caudopunctatus
,P.wulingensis
,P.longliensis
,P.zhijinensis
,andP.maolanensis
(Yuanet al
.,2014).This suggests that the species diversity of this group may be severely underestimated.Consequently,a comprehensive assessment of the species number contextualized with their evolutionary history is necessary to identify the species conservation status.This is essential in order to develop an effective management plan and a better understanding of the biological history of theP.caudopunctatus
species group.Mountains ecosystems are characterized by high biodiversity,with evidence of species showing a wide range of evolutionary adaptations (Elsenet al
.,2015;Körneret al
.,2002;McCainet al
.,2011).They also serve as sanctuaries for many endemic and threatened species and thus play a major role in maintaining biodiversity (Favreet al
.,2016).Mountains ecosystems provide key ecological service functions and provide important natural resources that are utilized by local human populations (Grêt-Regameyet al
.,2012;Körneret al
.,2002).Thus,mountains species are currently at higher risk of extinction due to their limited distribution,unique environmental adaptability,and geographical isolation.They also are highly threatened in the context of climate change (Wanget al
.,2018;Yanet al
.,2018;Huet al
.,2019).In the case of salamanders,the mitochondrial NADH dehydrogenase subunit 2 gene (ND2) and the nuclear proopiomelanocortin protein gene (POMC) are commonly used molecular genetic markers in species identification and amphibian phylogeny (Vieiteset al
.,2007;Wanget al
.,2018).These two genes can effectively identify evolutionarily significant units and management units in tailed amphibians (Stuartet al
.,2010;Guet al
.,2012a;Wuet al
.,2010;Yuanet al
.,2014,2016a).Here,based on the most complete sampling (including two putative species) to date,we examine the phylogeny and biogeography of theP.caudopunctatus
species group using both mitochondrial and nuclear data.Specifically,we aim to:(1) explore interspecific relationship within theP.caudopunctatus
species group;(2) infer the center of origin and estimate dates for each divergence event;and (3) employ a series of speciesdelimitation methods to clarify species boundaries and to identify candidate species among taxa in theP.caudopunctatus
species group.2.Materials and Methods
2.1.Taxon sampling and molecular data collection
TheP.caudopunctatus
species group is mainly distributed in the Wuling Mountains,Miaoling Mountains and Wumeng Mountains in Guizhou.The molecular data from the vast majority of these areas have been stored in the National Center for Biotechnology Information (NCBI,USA).However,samples from the southern part of the Miaoling Mountains and the northern part of the Wumeng Mountains are still lacking.Therefore,10 samples were collected from both regions for molecular analysis,they are:Huishui (HS,six individuals) and Dafang (DF,four individuals) (Figure 1).All muscle samples were attained from euthanized specimens and then preserved in 95% ethanol and stored at -40 °C.Additional information for these samples is presented in Table 1 and Figure 1.In addition,we downloaded 44 sequences from the NCBI database for molecular analysis and determined whether the sequences were from the same individual based on the specimen voucher number.Finally,42 ND2 sequences and 35 POMC sequences were used in the molecular analyses.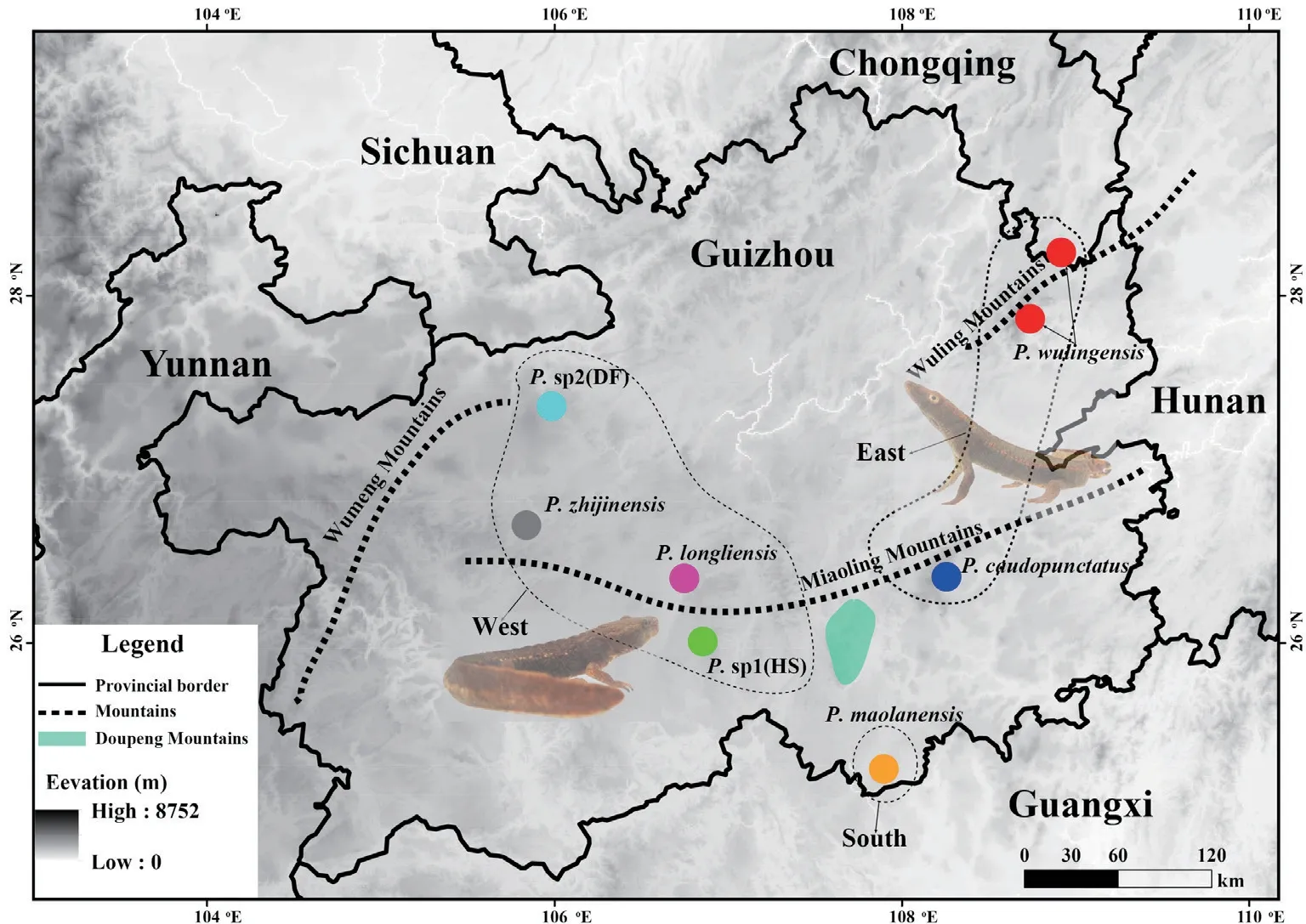
Figure 1 Map showing the distribution of the P.caudopunctatus species group in Guizhou Province,China used in this study.Colors in the pie-diagrams correspond to the time tree in Figure 2.
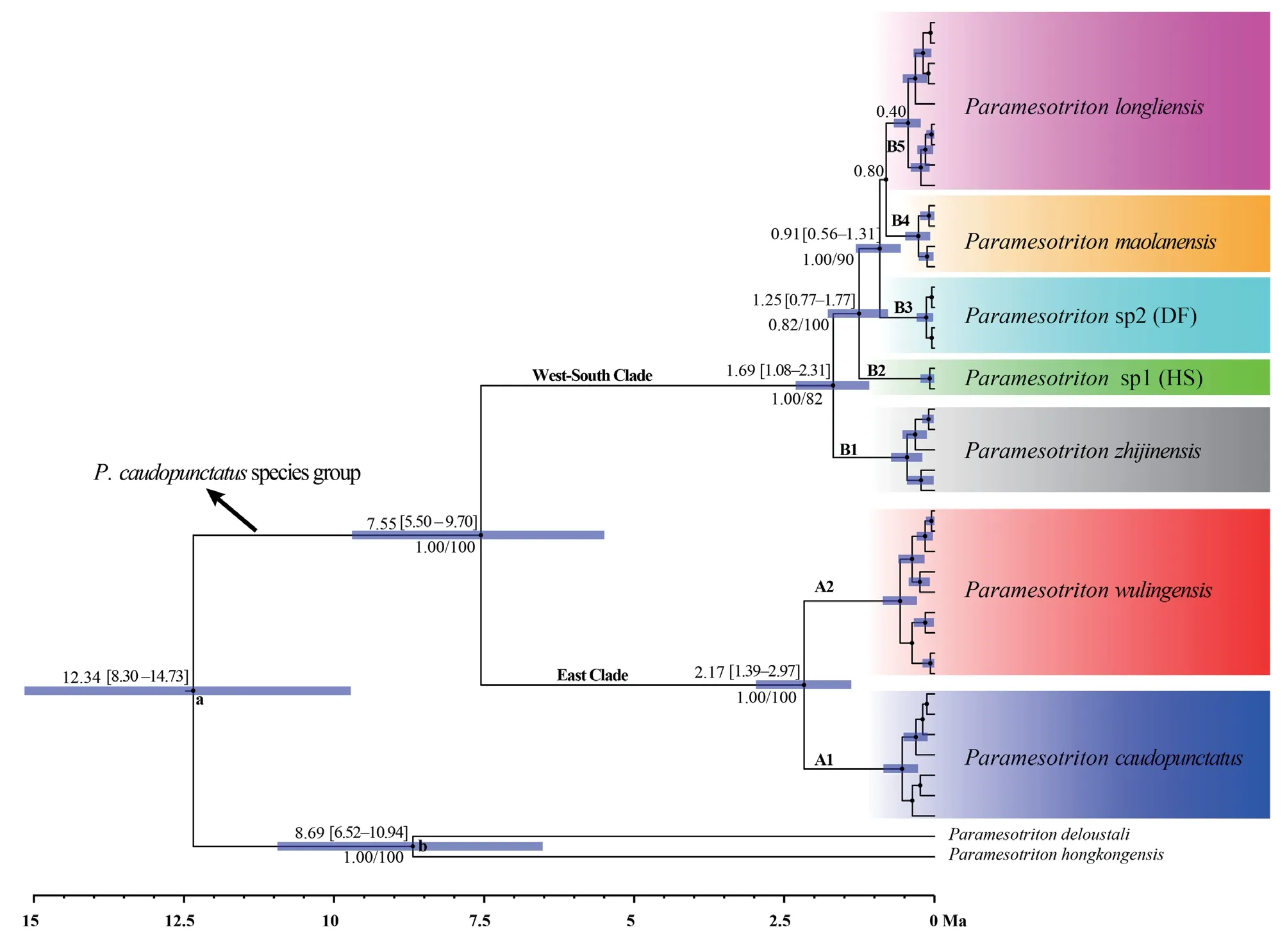
Figure 2 Mitochondrial and nuclear gene sequences phylogeny of the P.caudopunctatus species group.The values below the node indicate Bayesian posterior probabilities and ML ultrafast bootstrap support (shown as a percentage).Letters (a and b) indicate the calibration points.The blue line at each node correspond to the 95% highest posterior density of the age of that node.The bottom axis is in millions of years.
2.2.DNA extraction and sequencing
Total DNA was extracted using the standard phenol-chloroform extraction protocol (Sambrooket al
.,1989).All samples were sequenced for one mitochondrial gene and one nuclear gene,i.e.the partial NADH dehydrogenase subunit 2 gene (ND2) and the proopiomelanocortin gene (POMC).Primers used for ND2 were Ile3700L (5'-AGGRRYYACTTTGATARAGT-3') and COI5350H (5'-AGGGTGCCRATRTCYTTRTGRTT-3') following Zhanget al
.(2008),and for POMC were POMC-F (5'-ACTGCAGGAAATAAGAGAGAAG-3') and POMC-R (5'-GAGTCATTAGAGGTTTTGACT-3') from this study.Amplification was performed in 25-μL reaction mixes using the following procedure:initial denaturation at 95°C for 5 min;followed by 35 cycles of denaturation at 95°C for 45 s,annealing at 50°C (for ND2) /48°C (for POMC) for 1 min,and extension at 72°C for 70 s;and a final extension at 72°C for 10 min.The amplified PCR products were purified and sequenced in both directions using an ABI 3730 automated genetic analyzer.All sequences were deposited to GenBank.Some homologous DNA sequences of voucher specimens of related species were downloaded from GenBank and included in phylogenetic analyses (Table 1).For the molecular analysis,a total of 75 nucleotide sequences of theP.caudopunctatus
species group were used,including 40 ND2 sequences and 35 POMC sequences.In addition,two sequences were downloaded from GenBank as outgroups (i.e.P.deloustali
andP.hongkongensis
).Detailed information of these materials is shown in Table 1.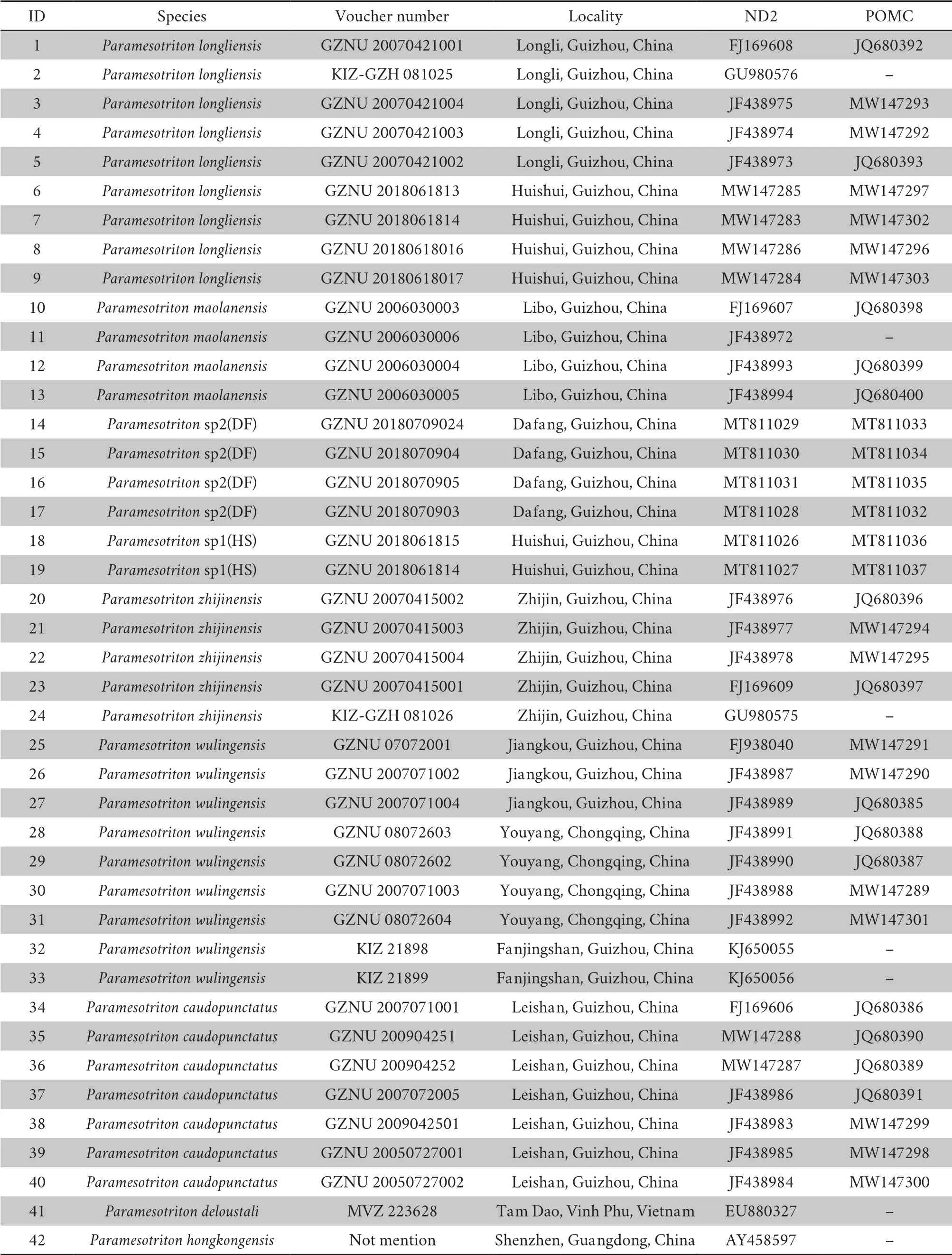
Table 1 Specimens included in the molecular phylogenetic analysis.An asterisk indicates newly obtained nucleotide sequence data from this study.GZNU:Guizhou Normal University,China;KIZ:Kunming Institute of Zoology,Chinese Academy of Sciences,China.
2.3.Phylogenetic analysis
The resulting nucleotide sequences were first assembled and edited using DNASTAR LASERGENE 7.1.Sequences from ND2 and POMC genes were aligned using MUSCLE (Edgar,2004) in MEGA 7.0 (Kumaret al
.,2016).In cases for which only mitochondrial data or nuclear loci were available for a species,we treated nuclear or mitochondrial partitions for these individuals as missing data.Based on the dataset using ND2+POMC,phylogenetic analyses were conducted using the maximum likelihood (ML) and Bayesian inference (BI) methods,as implemented in IQTREE 1.6.5 (Nguyenet al
.,2014) and MRBAYES 3.2 (Ronquistet al
.,2012),respectively.Prior to analyses,the best-fitting substitution models and partitioning schemes were selected in PARTITIONFINDER 2.1.1 using the Bayesian information criterion (Lanfearet al
.,2012).BI analyses were conducted with 2×10generations and sampled every 1,000 generations.Convergence was assessed in TRACER 1.7.1 (Rambaut and Drummond,2007) based on the average standard deviation of split frequencies (< 0.01) and ESS values (> 200).We used the remaining samples to generate the consensus tree after omitting the first 25% trees as burn-in.ML analyses were conducted in IQ-TREE using ultrafast bootstrapping (Hoanget al
.,2018) and run until these converged at a correlation coefficient of ≥ 0.99.Nodes in the trees were considered well supported when Bayesian posterior probabilities (BPP) were ≥ 0.95 and ML ultrafast bootstrap values (UFB) was ≥ 95%.Pairwise distance (p-
distances,1 000 replicates) between lineages was calculated in MEGA 7.0 (Kumaret al
.,2016).2.4.Divergence time estimation and ancestral area reconstruction
In this study,our divergence time estimates were based solely on mitochondrial and nuclear genetic data.To understand the divergence times of theP.caudopunctatus
species group,we used a relaxed molecular clock assumption in BEAST 1.8.2 (Drummondet al
.,2012).No reliable ranid fossil record is presently available to provide proper calibration within theP.caudopunctatus
species group.Therefore,two divergence time calibration points were included in this study following the recommendations of Zhanget al
.(2008):(1) the calculated age of 9.9-16.5 Ma for the split betweenP.caudopunctatus
andP.deloustali
+P.hongkongensis
(normally distributed calibration density;mean= 13.2 Ma);(2) the calculated age of 4.4-11.8 Ma for the split betweenP.deloustali
andP.hongkongensis
(normally distributed calibration density;mean= 7.5 Ma).Divergence times were estimated using the optimal partitioning strategy as determined by PARTITIONFINDER and normal prior distributions.BEAST analyses were run for 1×10generations,with samples taken every 5,000 generations under a relaxedmolecular clock and a Yule tree prior.Stationarity was assessed using TRACER 1.7.1,with ESS values >200 taken as evidence of convergence.Maximum clade credibility (MCC) trees were obtained using TREEANNOTATOR 2.4.1 by applying a burnin of 25%.
The ancestral range was reconstructed using Dispersal-Vicariance Analysis (S-DIVA) implemented in the program RASP 3.2 (Yuet al
.,2014).We reconstructed an mtDNA and nDNA genes tree using one representative of each taxon based on the same sets as BI analyses,and a total of 1000 post-burnin trees were used.Each sample was coded to represent the distribution of the species,and five distributional areas were used:Leigong Mountains (LM),Wuling Mountains (WL),Southern Guizhou (SG),and west of Miaoling Mountains (WM).The maximum number of areas was kept as two.2.5.Species delimitation
We used SPLITSTREE 4.16.1 (Huson and Bryant,2006) to construct a phylogenetic network based on uncorrectedp
-distances with heterozygous ambiguities averaged and normalized,using the neighbor-net ordinary least squares variance and equal angle algorithm and 1000 bootstrap replicates to assess branch support.We used the Bayes factor (BF) approach (Grummeret al
.,2014) to estimate the best fitting model to our dataset between alternative models (M1:7 species;M2:6 species;M3:5 species;M4:4 species;M5 3 species;M6:2 species;M7:1 species),defined by the estimates of population structure identified using the above phylogenetic tree.Marginal model likelihood used in Bayesian model comparisons (Bergstenet al
.,2013) was calculated via both path-sampling (PS;Baeleet al
.,2012) and stepping-stone (SS;Xieet al
.,2011) methods in BEAST 1.8.2 (Drummondet al
.,2012).Substitution models for mtDNA and nDNA were determined using PARTITIONFINDER 2.1.1.The Marginal likelihood estimate (MLE) of each model was estimated and the BF between pairs of modes was calculated as BF= 2 × (best MLE model - worst MLE model),with values for BF between 0 and 1 indicating very weak support for model 1 over 2,values between 1 and 3 indicating some support,albeit little,for model 1,values between 3 and 5 indicating strong support for model 1,and values >5 indicating decisive support for model 1 (Kass and Raftery,1995).Two independent runs for each model were performed in BEAST 1.8.2 to assess convergence of the MCMC runs.*BEAST was run each time for 1×10generations of the MCMC algorithm sampling every 1 000 generations and discarding the first 25% of the iterations as “burn-in”.The clock model parameter settings were a strict clock normal model of lineage variation,a Yule Process prior for the branching rates,and an HKY model of sequence evolution.For MLE analysis,the applied parameters were as follows:1×10generations,sampling every 1 000 generations and default settings for the other parameters.The results for different runs were combined using LogCombiner.Based on the MLE results,the species tree ofP.caudopunctatus
species group was determined.Convergence of all model parameters was assessed by examining the trace plots and histograms in TRACER 1.7.1.In addition to the Bayesian methods tested,we also applied three tree-based species-delimitation methods.They are the single-threshold General Mixed Yule Coalescent model (sGMYC;Fujisawa and Barraclough,2013),the multiple threshold GMYC model (mGMYC;Monaghanet al
.,2009),and the Bayesian implementation of the Poisson Tree Processes model (bPTP;Zhanget al
.,2013).All three analyses were calculated using an online server (http://species.h-its.org/).BEAST’s ultrametric tree with two outgroups (P.deloustali
,P.hongkongensis
) was used for the sGMYC,mGMYC and bPTP models with default parameter settings in the server.The parameters of these three analyses were set as follows:500,000 generations,a thinning of 500 and burn-in of 10%.Convergence of models were assessed by visualizing plots of the MCMC iteration vs.the Log likelihood results.Lastly,we used the computationally efficient distance-based species-delimitation method ABGD (Puillandreet al
.,2012),which can quantify the barcode gap location that separates intra-from interspecific distances.During the calculation,default settings were used for the prior range of maximum intraspecific divergence (0.001,0.1) and minimum slope increase (X) of 1.5 (default) and 1.0.K80 corrected distances were used to compare species delimitation results.3.Results
3.1.Sequence information
The aligned ND2 and POMC genes from theP.caudopunctatus
species group and outgroups consisted of 1 864 bp nucleotide positions after trimming.This included 1 404 bp from ND2 and 460 bp from POMC.The trimmed data were used for genealogical reconstructions,including 1 570 constant and 294 variable sites.The partitions and best-fit evolutionary models for the data were as follows:HKY+I for the first and the second positions of ND2,HKY+G for the third position of the ND2,F81+I for POMC.All novel sequences generated have been deposited in GenBank (Table 1).3.2.Phylogenetic analysis
The ML and BI analyses resulted in essentially identical topologies and were integrated in Figure 2.In the phylogenetic tree,except for the outer-groups,the monophyly of theP.caudopunctatus
species group was strongly supported (BPP=1.00,UFB=100%).The inner-group contained the East Clade,which consisted ofP.caudopunctatus
andP.wulingensis
regions in the east of Guizhou,and West-South Clade,which was composed of theP.longliensis
,P.
sp1 (HS),P.zhijinensis
,P.
sp2 (DF),andP.maolanensis
regions in the west and south of Guizhou (BPP=1.00,UFB=100%).East Clade consisted of two lineages,i.e.lineages A1 and A2.Lineage A1 was composed of seven samples from the Leigong Mountains,and lineage A2 was composed of nine samples from two populations (from Jiangkou and Youyang) in the Wuling Mountains area.The West-South Clade consisted of five lineages.Lineages B1 and B3 were composed of five samples from Zhijin County in the eastern part of the Wumeng Mountains and four samples from Dafang County;lineages B2 and B4 were composed of two samples from Huishui County,located south of the Miaoling Mountains and four samples from Libo County.Lineage B5 was composed of nine samples from the southern part of the Miaoling Mountains (from Longli and Huishui).Overall,this dataset yielded well-supported phylogenetic trees (BI and ML;Figure 2),reflecting the same topological structure previously identified for theP.caudopunctatus
species group (Guet al
.,2012b;Yuanet al
.,2014).3.3.Divergence dating and ancestral-area estimation
The two BEAST runs converged after 1×10generations,yielding ESS values > 200 for all parameters,and producing highly congruent trees and dates.Figure 2 reveals the divergence times between different species of theP.caudopunctatus
species group.Results indicate with weakly support (Probability=0.50) that theP.caudopunctatus
species group likely originated in the middle range of the Miaoling Mountains (current location of the Doupeng Mountains,including Majiang and Guiding) during the late Miocene (12.34 Ma;95% HPD:8.30-14.73).The divergence time of West-South Clade and Eastern Clade of theP.caudopunctatus
species group was estimated to 7.55 Ma (95% HPD=5.50-9.70).The divergence time ofP.caudopunctatus
andP.wulingensis
was estimated to be 2.17 Ma (95% HPD=1.39-2.97),and that ofP.zhijinensis
and (HS,DF (P.maolanensis
,P.longliensis
)) was 1.69 Ma (95% HPD=1.08-2.31).3.4.Species delimitation and species tree
The phylogenetic network of theP.caudopunctatus
species group contained the same groupings observed use phylogenetic methods (Figure 3).For the Bayes-factor species delimitation,we compared the starting delimitation of seven species derived from mitochondrial and nuclear gene sequences reciprocal monophyly using seven alternative models (Table 2) that assumed fewer species.PS and SS ranked the seven delimitation models identically and with highly similar marginal likelihood estimates.The starting delimitation received the highest likelihood value,and the five-species model yielded the lowest value.Between the starting and alternative delimitations,BF ranged from 12.84 to 189.13 by PS,and from 12.70 to 190.06 by SS,decisively supporting the recognition of seven species.Because the seven-species delimitation constituted the optimal model,we used the corresponding *BEAST species-tree for further analyses (Figure 4).Noteworthy is the fact that nodal support was low for basal relationships.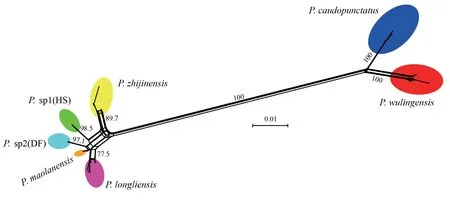
Figure 3 Network constructed from the ND2 sequence of the P.caudopunctatus species group samples based on uncorrected p-distances using SPLITSTREE.The value at each node indicates bootstrap support (only values above 70% are shown).

Figure 4 Species tree inferred from the seven-species delimited by BEAST.The values on nodes are Bayesian posterior probabilities.
Four out of five of the species-delimitation methods consistently identified seven species,while the bPTP identified more than six (~ 6.55).The areas of Leishan,Jiangkou,Youyang,Zhijin,Dafang,Libo,and Longli consistently presented one species per area.Finally,in terms of genetic differences,average pairwise sequence divergence varied markedly among these species,from 0.61% (P.maolanensis
vsP.longliensis
) to 9.74% (P.caudopunctatus
vsP.zhijinensis
) (Table 3).
Table 2 Species delimitation results of the P.caudopunctatus species group based on the BF method.“MLE” represents “Marginal likelihood estimate”;“BF ” represents “Bayes factor”;“PS” represents path-sampling method;“SS” represents the stepping-stone method.
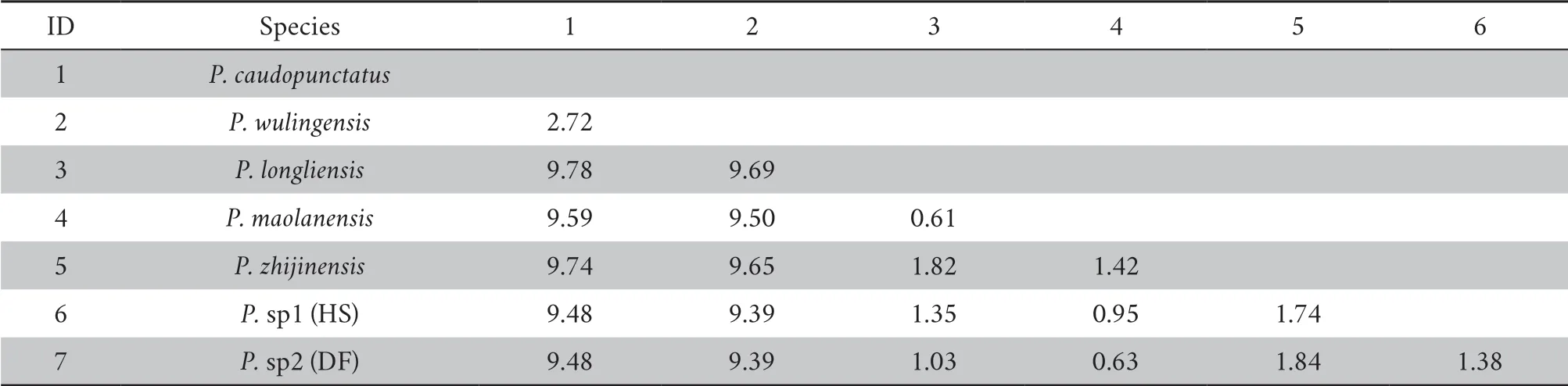
Table 3 Mean p-distance ND2 gene among the P.caudopunctatus species group used in this study.
4.Discussion
4.1.Phylogenetic relationships of the species group
In this study,we conducted a large-sample comprehensive phylogenetic analysis of theP.caudopunctatus
species group of the genusParamesotriton
,including 10 newly acquired taxonomic samples.Based on the mitochondrial gene sequence ND2 and the nuclear gene POMC,two major clades (the East and West-South Clades) were consistently identified for theP.caudopunctatus
species group in Guizhou Province,China.This topology is similar to that of previous studies (Guet al
.,2012b;Yuanet al
.,2014).The genusParamesotriton
is divided into three subgenera by Feiet al
.(2016),i.e.Paramesotriton
,Allomesotriton
,andKarstotriton
. The subgenusAllomesotriton
includesP.caudopunctatus
andP.wulingensis
;and the subgenusKarstotriton
includesP.longliensis
,P.zhijinensis
,andP.maolanensis
.Among the two subgenera previously proposed based on morphological characters,the subgenusAllomesotriton
in the East Clade and subgenusKarstotriton
in the West-south Clade were all identified as monophyletic.At present,the phylogenetic relationships between species within the subgenusKarstotriton
have not been well resolved.4.2.Biogeography of the species group
Although biogeographic studies on Asian amphibians have greatly increased in the last decade (Cheet al
.,2010;Yanet al
.,2013;Chenet al
.,2017;Wanget al
.,2018;Yuanet al
.,2018;Jianget al
.,2019;Wuet al
.,2020),little attention has been paid to salamanders.TheP.caudopunctatus
species group appears to have had a relatively short history in Southwest China.Dating analyses indicate thatP.caudopunctatus
species group originated about 12.34 Ma,and that the sequential speciation of each species took place during a period of 0.80-7.60 Ma (Figure 2).This timing coincides well with the orogenesis of the Miaoling Mountains (7-15 Ma) during the Late Miocene to early Pleistocene (Zhouet al
.,1993).Mountain building may have triggered changes in local climate,the development of complex terrain (including the uplifting of mountains and the formation of rivers) and the formation of heterogeneous environments (Shiet al
.,2006).The origin of theP.caudopunctatus
species group is spatially and temporally consistent with the time of divergence estimated for the common distribution of other biological taxa (e.g.,Quasipaa boulengeri
,Yanet al
.,2013;Scaptonyx fusicaudus
,Heet al
.,2019) in the region,and the dates of speciation are concordant with some species of frogs (Wuet al
.,2020;Chenet al
.,2017;Zhouet al
.,2017),and snakes (Guoet al
.,2020).All interspecific divergence events date to less than 3.0 Ma,which corresponds with the rapid uplifting and westward movement of the Miaoling Mountains at approximately 3.40 Ma (Zhouet al
.,1993).This is related to the rapid uplifting of the Tibetan plateau during this period (Sun,1997).The Western part of Guizhou has attracted widespread attention from biologists as a center of origin and a corridor for their dispersal for several taxa (Yanet al
.,2013;Yuanet al
.,2016b;Yaoet al
.,2018).For theP.caudopunctatus
species group,the current geographic distribution and species tree distribution are consistent with the “dispersal-vicariance-dispersal” model.After the initial split,the common ancestor of the species group expanded its distribution into the central part of the ancient Miaoling Mountains and West of Guizhou.This resulted in geographical isolation,which in turn led to the formation of East and West Clades.Continuous uplifting of the Qinghai-Tibetan Plateau led to habitat heterogeneity and ecological divergence,thus resulting in vicariance,driving speciation.Subsequent vicariance also best explains the narrow distribution of these species.Within the Eastern Clade,isolation betweenP.caudopunctatus
andP.wulingensis
species occurred early in the Pleistocene with the breakup of the Miao Mountains and the Wuling Mountains (Zhouet al
.,1993).Within West-South Clade,the phylogenetic branching pattern also suggests a general West-to-South and West-to-North dispersal pattern,because species endemic to the western most part of Miaoling Mountains represent basal clades (Figure 1).This unique pattern of dispersion may be related to the numerous rivers in the area.4.3.Species validity and cryptic species diversity
Since the description ofP.longliensis
andP.zhijinensis
in Guizhou,China,it has been suggested that the limited genetic divergence betweenP.longliensis
andP.zhijinensis
indicates that the two names may describe the same (single) species (Wuet al
.,2010).In this study,we amplified and sequenced the mitochondrial and nuclear gene sequences of both species from specimens deposited in our laboratory from the type locality.In addition,sequence information was collected from relevant literature.Two lines of evidence provide support for the validity ofP.longliensis
andP.zhijinensis
as separate species.First,the BFD,GMYC single,GMYC multiple,bPTP,and ABGD analyses support the recognition ofP.longliensis
andP.zhijinensis
.Second,In the species-tree (Figure 4),P.longliensis
andP.zhijinensis
were clearly distinguished as monophyletic,although the posterior probability between them was relatively low (BPP=0.83).Phenotypic similarity is not a clear distinguishing characteristic,especially in species with significant phenotypic divergence and small genetic differences.Most importantly,there is no evidence yet as to whether gene flow exists betweenP.longliensis
andP.zhijinensis
.Morphological analyses and karyotype data also support this conclusion.P.longliensis
morphologically differs fromP.zhijinensi.
The posterior part of the upper branchial bone points upwards (vs.had outer gills) inP.longliensis
,the fold of the tail fin behind the anus is red-orange and fades away at about 1/2 end of tail (vs.the fold of tail fin behind the anus being yellowy orange,and fades away at about 3/4 end of tail),there is a continuous earthen-yellow longitudinal stripe on each side of the abdomen (vs.lacking or intermittent),andP.longliensis
has four pairs of submetacentric chromosomes (vs.two pairs inP.zhijinensi
) (Figure 5:Liet al
.,2008a,2008b;Sunet al
.,2015).Thus,both molecular and morphological analyses supportP.longliensis
andP.zhijinensis
as valid species.In this study,our data and analyses clearly demonstrate that the species diversity of theP.caudopunctatus
species group has been underestimated.In addition to the five described species of theP.caudopunctatus
species group,our results identified two distinct additional lineages that we consider as two candidates undescribed species (Figure 2 and Table 4).We found that two areas (DF and HS) consistently showed support for the existence of two putative species across the various speciesdelimitation methods used (Table 4).In our analysis,most species-delimitation methods supported the presence of two candidate species.The genetic distance values among the five lineages were variable,ranging from 0.61% (P.longliensis
vsP.maolanensis
) to 9.78% (P.caudopunctatus
vsP.longliensis
) (Table 3).Overall,the genetic distances between the two species in question and the other five genealogies are greater than the genetic distances between the accepted species within theP.caudopunctatus
species group,and also are greater than the genetic distances between the other tailed amphibian species (1.1%,Hynobius formosanus
vsHynobius arisanensis
;Panet al
.,2019).Therefore,in this study,the species delimitation based on mitochondrial and nuclear gene sequences data revealed that there are multiple species inP.caudopunctatus
species group.Of five species-delimitation methods used,four methods strongly supported the existence of seven species,P.caudopunctatus
,P.wulingensis
,P.longliensis
,P.maolanensis
,P.zhijinensis
,P
.sp1 (HS),andP
.sp2 (DF).Underestimated species diversity indicates that theP.caudopunctatus
species group evolved in response to complex species-forming mechanisms and a history of biological dispersal.
Table 4 Number of lineages in the P.caudopunctatus species group inferred by multiple species delimitation methods.

Figure 5 Dorsal view and ventral view of living specimens of the P.caudopunctatus species group.(A) P.caudopunctatus,GZNU 20190825001,adult male;(B) P.wulingensis,GZNU 20110719,adult male;(C) P.zhijinensis,GZNU 20190815002,adult male;(D) P.longlisis,GZNU 20190922002,adult male;(E) P.sp2 (DF),GZNU 20180711001,adult male;(F) P.sp1 (HS),GZNU 20180612001,adult male.
Acknowledgments
This work was supported by the programs of the Strategic Priority Research Program B of the Chinese Academy of Sciences (CAS) (No.XDB31000000),the National Natural Science Foundation of China (Grant No.31460091),the National Animal Collection Resource Center,China (Grant No.2005DKA21402),the Application of Amphibian Natural Antioxidant Peptides as Cosmetic Raw Material Antioxidants (QKZYD [2020]4002),the National Top Discipline Construction Project of Guizhou Province,Geography in Guizhou Normal University (No.85 2017 Qianjiao Keyan Fa).We thank Ms.Huan He for his help with sample collection.We thank Professor Paul A.Garber for editing assistance during the preparation of this manuscript.We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript. Asian Herpetological Research2021年2期
Asian Herpetological Research2021年2期
- Asian Herpetological Research的其它文章
- A New Leaf Litter Toad of Leptobrachella Smith,1925 (Anura,Megophryidae) from Sichuan Province,China with Supplementary Description of L.oshanensis
- A New Species of Trimeresurus Lacépède,1804 (Squamata:Viperidae) from Southwestern China,Vietnam,Thailand and Myanmar
- A New Species of the Genus Achalinus from Huangshan,Anhui,China (Squamata:Xenodermidae)
- Phylogeny and Biogeography of Common Toad (Bufo bufo) in Xinjiang,China
- Evolution of Phenotype and Mitochondrial Genome Reveals Limbless and Body-elongated Squamates may Change Their Energy Basis for Locomotion
- Male Advertisement Call of the Endangered Leptobrachella tengchongensis (Anura:Megophryidae) from Mount Gaoligongshan,Yunnan Province,China
