基于加权关联网络分析的辣椒(Capsicum annuum L.)果实颜色转变相关基因的鉴定
金威恒 MUMTAZ Muhammad Ali 何成尧 夏禹 郝园园 李彩朝 汪志伟


Weighted Correlation Network Analysis Reveals Candidate Genes Related to Fruit Color Transition in Pepper ( L.)
JIN WeihengMUMTAZ Muhammad AliHE ChengyaoXIA YuHAO YuanyuanLI CaichaoWANG Zhiwei
1. Key Laboratory for Quality Regulation of Tropical Horticultural Crops of Hainan Province / School of Horticulture, Hainan University, Haikou, Hainan 570228, China; 2. Sanya Nanfan Research Institute of Hainan University / Hainan Yazhou Bay Seed Laboratory, Sanya, Hainan 572025, China
Pepper belongs to the annual or perennial pepper genus ( L.) in the Solanaceae. Amid all vegetables grown in China, pepper is ranked first as per its sowing area and output value. Fruit color is one of the most intuitive economic traits of pepper, which directly affects people's purchase choice. Nevertheless, the regulation of pepper fruit color is affected by many factors, especially the key regulatory factors in the process of fruit pigment formation and the regulation of related pigment biosynthesis are not clear. To understand the mechanism of pepper fruit color formation, physiological testing, transcriptome sequencing and weighted gene co-expression network analysis (WGCNA) was performed using germplasm HNCA0076 as the experimental material, presenting five colors during fruit development. During the five different color periods of the fruit, the chlorophyll content was significantly different, the carotenoids content showed an increasing trend, the flavonoid content was relatively stable, and the anthocyanin content decreased gradually after purple fruit stage, and increased after orange fruit stage. The quality of the RNA sequencing data met the requirements for further analysis. The number of differentially expressed genes (DEGs) up-regulated and down-regulated in each period was 826 (276), 390 (588), 1248 (1800) and 1528 (2485). As the fruit matures, the number of differential genes in adjacent comparison combinations gradually increased. DEGs were analyzed by gene ontology (GO) enrichment with a main focus on molecular functional such as tetrapyrrole binding, heme binding, transferase activity, oxidoreductase activity, iron ion binding and hydrolase activity. After KEGG enrichment analysis, DEGs were found mainly enriched in carotenoid biosynthesis, flavonoid biosynthesis, starch and sucrose metabolism. Among these, the expression of the three genes from the carotenoid metabolic pathway were up-regulated during the transition from purple to yellow, and the expression of 11 genes from the flavonoid metabolic pathway were down-regulated during the transition from yellow to red. The differentially expressed genes of four comparative combinations were annotated to 8877, 8736, 8689, and 8573 transcription factors (TFs), and together to 8049 TFs, including AP2F-boxMYB, and UDPGT. We performed a WGCNA using non-redundant DEGs associated with four modules including chlorophyll, carotenoid, coloring, and flavonoid content. Further analysis indicated that 29 TFs such as p450 (), Pkinase (, ), F-box (), and zf-MYND () may be involved in the regulation of pepper fruit color conversion. This study would provide an important data for further exploring the key regulatory factors in the process of pepper fruit coloring and analyzing the regulation of related pigment biosynthesis.
: ; color transition; carotenoids; flavonoids; transcription factor
: 10.3969/j.issn.1000-2561.2022.06.005
1. 海南省热带园艺作物品质调控重点实验室/海南大学园艺学院,海南海口 570228;2. 海南大学三亚南繁研究院/海南省崖州湾种子实验室,海南三亚 572025
摘 要:辣椒属于茄科(Solanaceae)辣椒属( L.)一年生或多年生植物,其播种面积和产值在中国均居蔬菜首位。果色是辣椒众多经济性状中最直观的性状之一,直接影响人们的购买选择。辣椒果色受多种因素调控,相关色素生物合成的分子调控还需进一步解析。本研究以果实发育过程中呈现5种颜色的辣椒种质HNCA0076为实验材料,进行了生理检测、转录组测序以及WGCNA分析。在果实5个不同颜色时期,叶绿素含量变化较大,类胡萝卜素含量整体呈现递增趋势,类黄酮含量无明显变化,花色苷含量在紫色果期后逐渐降低,橙色果期后又逐渐升高。RNA测序数据质量满足进一步分析要求。4个转色期的上调(下调)表达基因数分别为826(276)、390(588)、1248(1800)和1528(2485),隨着果实的逐步发育,相邻比较组合中的差异基因的数量逐渐增加。差异表达基因经GO富集分析,主要集中在分子功能中的四吡咯结合、血红素结合、转移酶活性、氧化还原酶活性、铁离子结合和水解酶活性等过程;KEGG富集分析主要富集在类胡萝卜素生物合成、类黄酮生物合成和淀粉、蔗糖代谢等信号通路,其中类胡萝卜素代谢途径的3个基因在紫色时期向黄色时期的转变过程中表达上调,类黄酮代谢途径的11个基因在黄色时期向红色时期的转变过程中表达下调。4个对比组合差异基因分别注释到8877、8736、8689、8573个转录因子,共同注释到8049个转录因子,包括AP2、F-box、MYB和UDPGT等类型。利用非冗余的差异表达基因进行了加权关联网络分析,关联到4个与叶绿素、类胡萝卜素、花色苷、类黄酮含量相关的模块,进一步分析表明p450()、Pkinase()、F-box()和zf-MYND()等29个转录因子可能参与辣椒果实转色的调控。本研究为进一步探索辣椒果实色素形成过程中的关键调控因子以及分析相关色素生物合成的调控提供了重要数据。
关键词:一年生辣椒;转色;类胡萝卜素;类黄酮;转录因子
中图分类号:S641.3 文献标识码:A
Pepper ( L.) is one of the most widely consumed vegetable crops worldwide for its color, unique taste, and nutritional benefits. In 2020, the global pepper plantation area was 199 000 hectares, and the output was 39.28 million tons. Its sown area ranks first among vegetables in China. Fruit color is one of the most intuitive economic traits of pepper, which directly affects its marketability.
Color change as peppers grow is an indication of the maturity level. It also heralds a change in flavor of the pepper fruit. Pepper fruit can show different colors during development, such as green, yellow, red, purple, etc. Pepper fruit color is mainly determined by the relative content of chlorophyll, anthocyanins, carotenoids and other pigment substances in the fruit. Anthocyanin is one of the main pigments determining the color of fruit. It is synthesized through flavonoid pathway under the control of multiple structural and regulatory genes, and mainly accumulated in the outer epidermis of pepper fruit. The large accumulation of anthocyanin in immature fruit leads to purple pepper fruit. Previous studies suggested that structural genes and regulatory genes were related to anthocyanin synthesis. Structural genes encode key enzymes required for anthocyanin synthesis, and regulatory genes indirectly regulate anthocyanin accumulation by regulating the expression of key genes in anthocyanin synthesis. The key structural genes in anthocyanin biosynthesis pathway include chalcone synthase (CHS), chalcone isomerase (CHI), etc. Some transcription factors, such as MYB, bHLH and WD40, indirectly regulate the anthocyanin content of pepper fruit by regulating the transcription of structural genes for anthocyanin synthesis. Maize gene is the first MYB transcription factor isolated and identified. It promotes the synthesis of anthocyanins in maize stems and leaves by activating the expression of four flavonoid structural genes. In , 13 MYB transcription factors controlling flavonoid synthesis have been identified, of which nine are positively correlated with flavonoid synthesis, while the other four MYB transcription factors inhibit flavonoid synthesis by suppressing the expression of key genes from phenylpropane pathway and the downstream flavonoid synthesis pathway. And in seed coat, transcription factor TT1 controls the synthesis of procyanidins by activating the expression of the gene. To determine the expression pattern of flavonoid synthesis structural genes in pepper with different fruit colors, STOMMEL detected the expression abundance of flavonoid structural genes (CHS, DFR, ANS) and transcription factors (Myc / bHLH, MYB, WD) by qRT-PCR. It was found that the expression of structural genes and regulatory genes bHLH and MYB in black pepper were significantly higher than that in green pepper, it is speculated that the accumulation of anthocyanins in pepper fruits is mainly regulated by CHS, DFR, ANS, bHLH, and MYB genes. With the development of pepper fruit, flavonoids and chlorophyll are gradually degraded in plastids, and carotenoids are synthesized and accumulated. Three loci and were proposed to explain the variation of carotenoid composition, which led to different colors of pepper fruits. The locus corresponds to theGene, and the locus is identified as the gene involved in capsanthin synthesis. Although some genes related to fruit color have been cloned in pepper, how these genes regulate the synthesis of carotenoids and anthocyanins is still a mystery due to the lack of effective mutants.
High throughput sequencing technology is widely used in functional genomics of non model species. Through transcriptome data, a large number of gene expression information can be obtained at the RNA level, and candidate genes would be identified to be associated with the target traits. In this study, the fruits of pepper germplasm HNCA0076 showed five color changes during development. We carried out comparative transcriptome analysis on the fruits at different color stages and identified some key genes involved in carotenoid and anthocyanin synthesis by weighted gene coexpression network analysis (WGCNA). These results provide useful information for further study of the regulation of the biosynthesis of various pigments in the process of pepper fruit coloring.
The pepper germplasm HNCA0076 ( L.) was used as a plant material. The pulp of pepper fruit was taken after removing the placenta and seeds at the purple fruit stage (QP, 25 days after flowering), white fruit stage (QI, 30 days after flowering), yellow fruit stage (QY, 35 days after flowering), orange fruit stage (QO, 45 days after flowering) and red fruit stage (QR, 50 days after flowering). Six representative fruits for each stage were collected in three biological replications and frozen immediately in liquid nitrogen and stored at –80℃ for further analysis.
Ultraviolet spectrophotometry was used to determine chlorophyll and carotenoids, and a commercially available kit (Beijing Solarbio Science & Technology Co., Ltd.) was used to determine the content of anthocyanins and flavonoids as per the manufacturer’s protocol.
RNA was extracted from the frozen pulp with a total RNA extraction kit (NEBNext Ultra? RNA Library Prep Kit for Illumina), Purified RNA was obtained by mRNA enrichment method. cDNA was synthesized by reverse transcription with random N6 primers to form a DNA double-strand. Single-stranded circular DNA library was obtained by PCR amplification using specific primers.
After the construction of the library, Qubit 2.0 fluorometer was used for preliminary quantification, the library was diluted to 1.5 ng/μL, and then Agilent 2100 Bioanalyzer was used to detect the insert size of the library. qRT-PCR was used to accurately quantify the effective concentration of the library (the effective concentration of the library was higher than 2 nm) to ensure the quality of the library.
Since the original sequencing data contains low-quality sequences, adapter sequences, etc., to ensure the reliability of the information analysis results, a series of data processing is required to filter these impurities (raw reads), including removing reads with adapters and removing reads containing adapters. N (N means that the base information cannot be determined) reads, and low-quality reads (reads with a Qphred≤20 base number accounting for more than 50% of the entire read length) were removed. At the same time, Q30 and GC content calculations were performed on clean data. Only high-quality clean data were used for subsequent analyses.
Clean reads were mapped to the pepper genome (Zunla-1 version) as the reference genome using HISAT2 v2.0.5 software to align clean reads to the reference genome.
DESeq2 software (1.20.0) was used to analyze the differential expression between samples. Genes with logFold Change>1 and False Discovery Rate (FDR)<0.05 were defined as differentially expressed genes (DEGs).
GO enrichment of differential expressed genes was analyzed by GO seq. Kyoto Encyclopaedia of Genes and Genomes (KEGG) was used for the pathway enrichment analysis.
gene coexpression network TFs were searched by iTALK (version 1.2) software (http:// plntfdb.bio.uni- potsdam.de/v3.0/) .
WGCNA was used to identify genes for network construction, gene cluster, and visualization. The gene correlation and soft thresholding power analysis were based on the Pearson correlation matrix and network topology. The networks were constructed by software Cytoscape (V3.8.0).
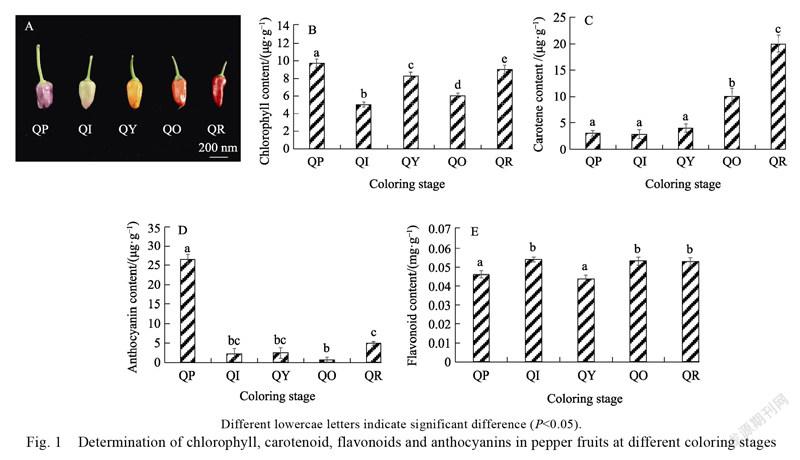
The ripening process of pepper fruit is accompanied by the synthesis and degradation of various pigments. In this experiment, pepper fruits in different coloring stages (Fig. 1A) were used to determine the chlorophyll content, carotenoid content, anthocyanin content, and flavonoid content (Fig. 1B, Fig. 1C, Fig. 1D, Fig. 1E). The chlorophyll content was significantly different in all five stages (Fig. 1B); the carotenoid content showed an increasing trend as a whole (Fig. 1C); the flavonoid content was relatively stable in all five stages, with no significant differences (Fig. 1E); whereas the anthocyanin content gradually decreased from purple to white stage and again increased till the red stage (Fig. 1D).
In this study, a total of 101.36 Gb clean data was obtained. Among the 15 samples, the clean reads in the QI6 sample were the least with 39 640 268 reads, and the clean reads in the QR8 sample were the most with 49 433 112 reads. The overall sequencing error rate of the data was within 0.02%, the percentage of bases with a Phred value greater than 30 to the total bases was greater than or equal to 94.18%, and the GC content was above 40%. These data indicate high sequencing quality for subsequent analysis.
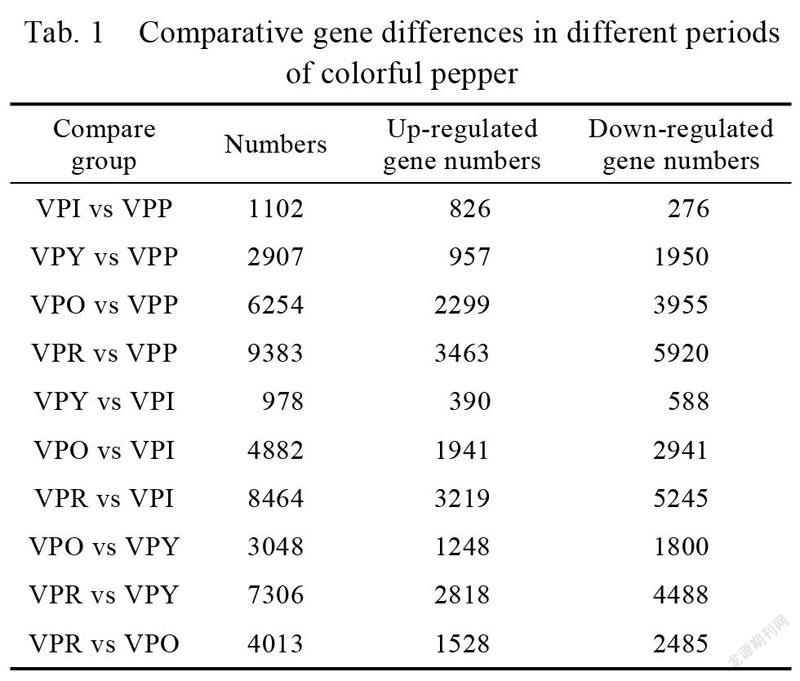
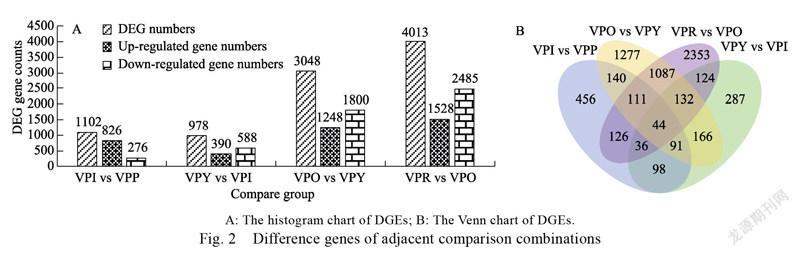
The apparent characteristics of pepper fruit at different maturity stages were significantly different, so differential genes at different developmental stages were analyzed in pairs (Tab. 1). In the ten sets of data, the most differential genes were identified in comparison (VPR vs VPP) in the red and purple fruit periods, a total of 9383 genes were differentially expressed (3463 upregulated and 5920 downregulated). The least differential genes were identified in the yellow and white fruit stage comparisons (VPY vs VPI), a total of 978 genes were differentially expressed (390 upregulated and 588 downregulated). We can see that, except for the combination of VPI vs VPP, the number of up-regulated genes was less than down-regulated genes in the comparison of different stages of coloration; as the fruit matures, the number of differential genes in adjacent comparison combinations gradually increased (Fig. 2).
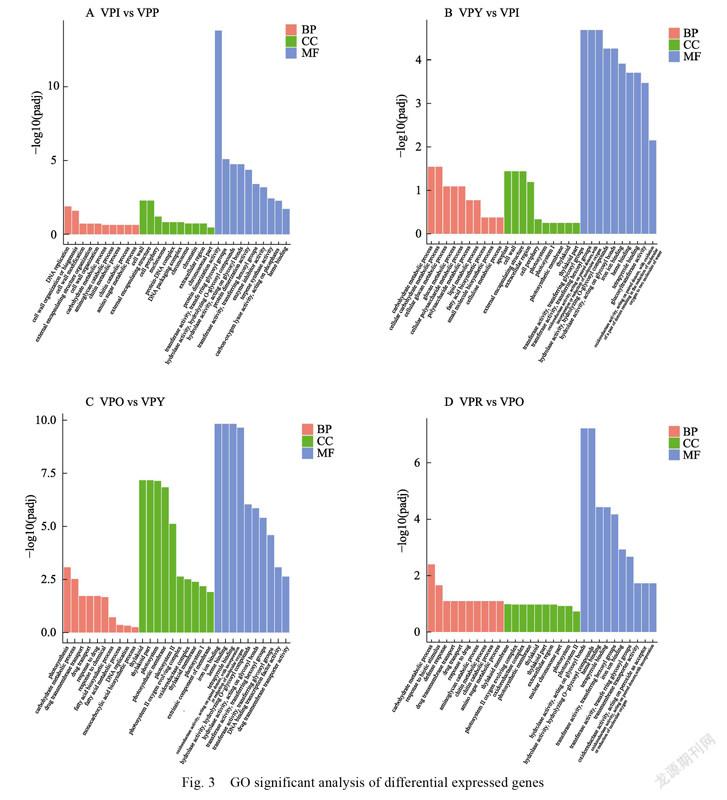
According to the statistical results of go classification of differentially expressed genes, We can know that DEGs were annotated into 3 categories and 30 subclasses of go classification in the comparison of VPI vs VPP, in which the biological process was mainly concentrated in DNA replication and cell wall tissue; The cell components were mainly concentrated in the cell wall; The molecular functions are mainly concentrated in protein heterodimerization activity, transferase activity and transferase hydrolase activity (Fig. 3A). DEGs were annotated into 3 categories and 30 subclasses of go classification in the comparison of VPY vs VPI, and the number of annotated genes was the least, in which the biological process was mainly concentrated in the process of carbohydrate metabolism; The cell components were mainly concentrated in the cell wall; The molecular functions are mainly concentrated in transferase activity, hexoses transferase activity and oxidoreductase activity (Fig. 3B). DEGs were annotated into 3 categories and 30 subclasses of go classification in the comparison of VPO vs VPY, in which the biological
processes were mainly concentrated in photosynthesis, carbohydrate metabolism and drug transmembrane transport; The cell components were mainly concentrated in thylakoid; The molecular functions are mainly concentrated in iron ion binding, heme binding and tetrapyrrole binding (Fig. 3C). DEGs were annotated into 3 categories and 10 subclasses of go classification in the comparison of VPR vs VPO, and the number of annotated genes was the largest, in which the biological process was mainly concentrated in the process of carbohydrate metabolism and response to biological stimulation; There are 10 subclasses of molecular functions, which are mainly concentrated in hydrolase activity and heme bindin (Fig. 3D). Meanwhile, in five periods, we found that tetrapyrrole binding, heme binding, hydrolase activity, transferase activity, oxidoreductase activity, iron ion binding and hydrolase activity were enriched most significantly in the MF category.
Using the method of hypergeometric test, the metabolic pathway with
-value≤0.05 is used as the pathway with signifi
cant enrichment of differentially expressed genes,
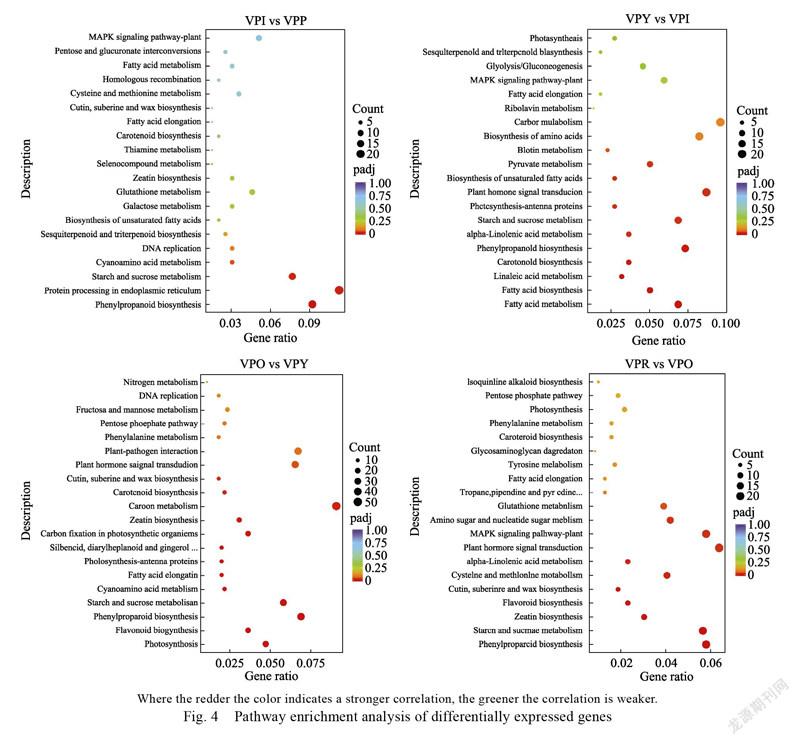
and the metabolic pathway with significant enrichment of differentially expressed genes is found through significance analysis. VPI vs VPP, VPY vs VPI, VPO vs VPY, and VPR vs VPO had 196, 218, 552, and 689 differentially expressed genes enriched in 81, 92, 108, and 114 pathways, respectively. The DEGs of VPI vs VPP are mainly enriched in metabolic pathways such as phenylpropane biosynthesis (cann00940), protein processing in the endoplasmic reticulum (cann04141), starch and sucrose metabolism (cann00500), and cyanoamino acid metabolism (cann00460); the DEGs of VPY vs VPI is mainly enriched in fatty acid metabolism (cann01212), carotenoid biosynthesis (cann00906), photosynthetic antenna protein (cann00196), plant hormone signal transduction (cann04075) and other metabolic pathways; the DEGs of VPO vs VPY are mainly enriched in photosynthesis (cann00195), flavonoid biosynthesis (cann00941), carotenoid biosynthesis (cann00906) and other metabolic pathways; the DEGs of VPR vs VPO are mainly enriched in phenylpropane biosynthesis (cann00940), starch and sucrose metabolism (cann00500), zeatin biosynthesis metabolic pathways such as synthesis (cann00908) and flavonoid biosynthesis (cann00941), indicating that the differentially expressed genes at different transition stages are enriched in carotenoid biosynthesis (cann00906) and flavonoid biosynthesis (cann00941) (Fig. 4). Notably, we can know that the DEGs enriched in the
two pathways of carotenoid biosynthesis (cann 00906) and flavonoid biosynthesis (cann00941) are in different color transition stages. Gene was up-regulated in the flavonoid biosynthesis pathway in the combination of VPO vs VPY and VPR vs VPO; and all exhibited down-regulation; genes and were up-regulated in carotenoid biosynthesis pathway in VPY vs VPI and VPO vs VPY combinations; gene was also down- regulated. These results strongly suggest the contribution of these genes to fruit color regulation.
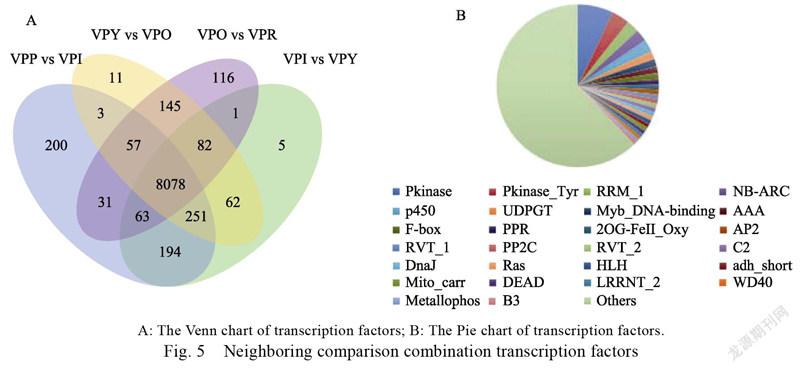
3.4.3 Transcription factor analysis of differentially expressed gene annotation Through the transcriptome analysis of different coloring stages of pepper fruit, we identified the transcription factors that regulate the coloring of pepper fruit, and understand their mechanisms of action, and provide a theoretical basis for the regulation and genetic improvement of pepper fruit color quality. The differentially expressed genes of VPI vs VPP, VPY vs VPI, VPO vs VPY, and VPR vs VPO were jointly annotated to 8078 transcription factors, belonging to 618 TF families, including Pkinase (572), NB-ARC (197), P450 (197), UDPGT (132), Myb (121), F-box (99), AP2 (78), HLH (67), WD40 (53), B3 (50) and other transcription factors (Fig. 5)
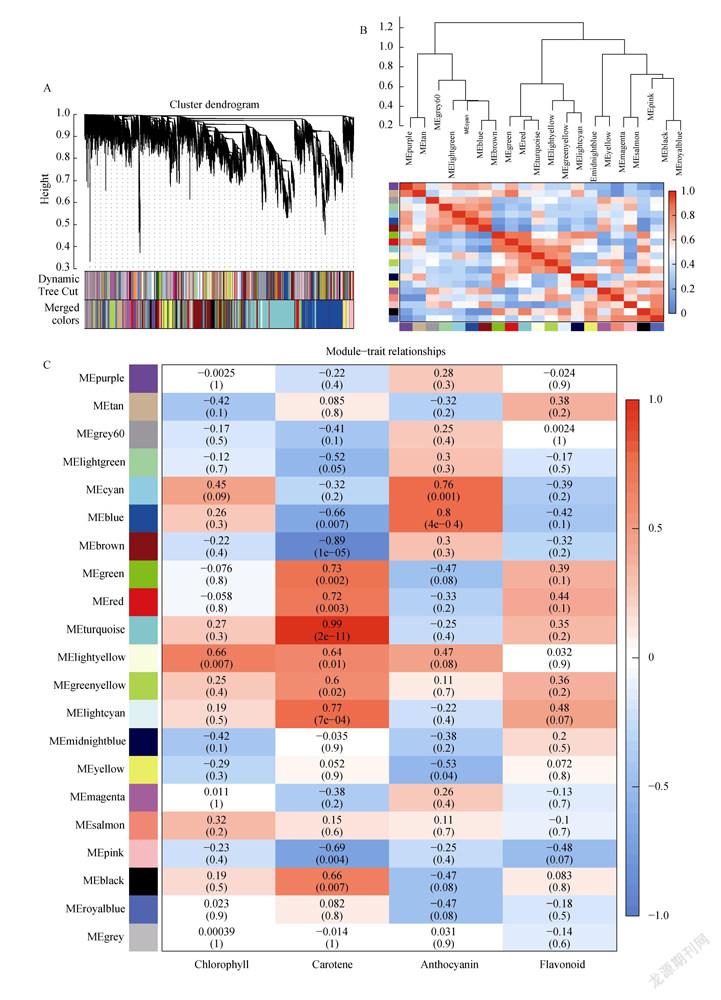
In order to study the gene regulation network of fruit color-related regulatory network in pepper fruit, we used non-redundant DEGs to perform a weighted gene co-expression network analysis (WGCNA). These non-redundant DEGs are grouped into 20 main branches, each branch representing a module (marked with a different color) (Fig. 6A), and we also conducted a correlation analysis among the modules (Fig. 6B), and among them, purple and tan modules, blue and brown modules, red and turquoise modules, greenyellow and lightcyan modules, midnightblue and yellow modules, magenta and salmon modules, black and royalblue modules The correlation is stronger, and the gene similarity between its modules is stronger.
Subsequently, data of four horticultural physiological indicators related to the module characteristics were analyzed in the five coloring stages (Fig. 6C). The results show that the lightyellow module is highly positively correlated with the chlorophyll content (=0.66, =0.007); the turquoise module is highly positively correlated with the carotenoid content (=0.99, =2e-11); the blue module is highly positively correlated with the total anthocyanin content (=0.80, =4e-0.4); the lightcyan module is positively correlated with the flavonoid content (=0.48, =0.07). To determine the expression patterns of the genes in the lightyellow, turquoise, blue and lightcyan modules, the FPKM values of the genes in the modules were used to perform a heat map (Fig. 6D). The heat map results show that the genes co-expressed in the lightyellow module are expressed in the purple fruit and red fruit stages, and the genes co-expressed in the turquoise and lightcyan modules are only highly expressed in the red fruit stage, showing significant specificity, while the genes in the blue module only
expressed during the purple fruit stage.
Screening transcription factors that regulate pigment synthesis through WGCNA among the 20 major genes in the lightyellow module, we found eight transcription factors. These include p450 (), HMA (), Pkinase (, ), Peptidase_C1 (), F-box (), Aa_trans () and Myb (). Among the 20 major genes of the turquoise module, five transcription factors including, MFS_1 (), Peptidase_S10 (), Homeobox (), Usp (), DUF1671 () were identified. Interestingly, ten transcription factors were identified in the 20 major genes of the blue module, including 2OG-FeII_Oxy (), Glycos_transf_1 ), zf-MYND (). Moreover six transcription factors in the lightcyan module, including LRRNT_2 (), ADH_N(), TB2_DP1_HVA22 (LOC10 7875806) were identified. These transcription factors, as highly connected central genes, may have a regulating effect on color formation in pepper fruit (Tab. 2).
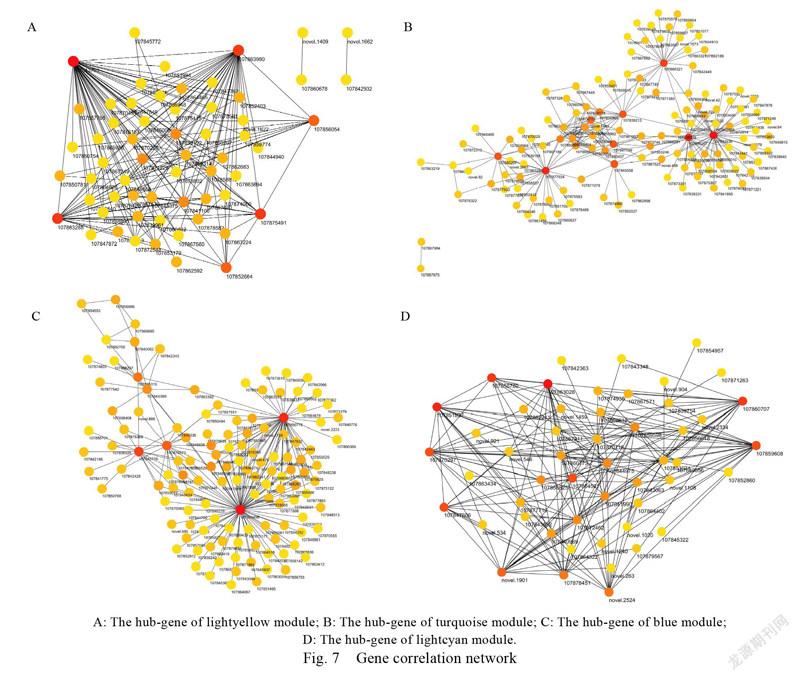
To further analyze the transcription factors involved in pigment regulation, relevant networks were constructed in four modules for analysis (Fig. 7). In the lightyellow module, 60 genes are highlycorrelated with chlorophyll synthesis (edge weight≥0.11), including hub transcription factor genes. In addition, CBS (), Abhydrolase_3 (), SIR2 (), and other non-hub transcription factor genes are also related to the regulation of chlorophyll synthesis; in the turquoise module, 127 genes are highly correlated with carotenoid synthesis (edge weight≥0.42), of which K-box () 41 non-core transcription factors such as UDPGT (), AP2 (), GST_C) have a certain relationship with carotenoid synthesis regulation; in the blue module,there are 126 genes and anthocyanins. The synthesishas a high correlation (edge weight≥0.42). Amongthem, 36 non-core transcription factors such as FAD_ binding_4 () and ADH_zinc_N () are related to the regulation of anthocyanin synthesis; in the lightcyan module, there are 203 genes related to flavonoid synthesis (edge weight≥0.35). In addition to the core gene UDPGT (), there are six non-core transcriptionfactors such as FKBP_C () and IPPT () that have a certain degree of regulation of flavonoid synthesis relation.
The formation of pepper fruit color is closely related to the content and type of anthocyanins, carotenoids, chlorophyll and other pigments. The type and content of color pigments change dramatically in different maturity stages of pepper fruit. In this study, inbred lline “HNCA0076” was used to study the color formation and regulatory network underlying fruit coloring at five different maturity stages. The anthocyanin content in the purple fruit stage was significantly higher as compared to other maturity stages, which is consistent with the previous reports as anthocyanin account for the purple color. With the fruit ripening, the carotenoid content gradually increased in the fruit development stage and reached the highest in the red fruit stage. The total carotenoid content in the mature red pepper fruit was 30 times higher as compared to the immature fruit. Pepper accumulated a higher amount of carotenoids, and the diversity of fruit color was largely related to the gene expression of carotenoid pathway. In this study, most genes in flavonoid biosynthesis pathway showed downregulation in VPO vs VPY and VPR vs VPO comparisons. Whereas genes in carotenoid biosynthesis pathway showed upregulation in VPY vs VPI and VPO vs VPY comparisons; The majority of ornamental peppers are orange at the color transition stage and red at maturity. However, some varieties remain orange after ripening, which is related to the non-expression or gene mutation of (capsaicin and capsaicin synthase). In previous studies, RFLP and specific polymerase chain reaction amplification were used to analyze the polymorphism of gene in Fgeneration of red and yellow pepper fruits. The results showed that the gene was completely separated from the red mature fruit, which proved that the yellow pepper fruit may be formed due to the deletion of gene.
In the four comparisons of VPI vs VPP, VPY vs VPI, VPO vs VPY, and VPR vs VPO, some genes were enriched in plant photosynthesis and plant- pathogen interaction. This corresponds to that flavonoids can induce or inhibit the gene expression of photosynthesis and plant pathogen pathways, and the abundant gene expression of these pathways can be used as signal feedback to plants to inhibit or promote the synthesis of flavonoids. At the same time, some genes are significantly enriched in the plant hormone signaling pathway. Plant hormones bind to receptors, through signal transmission, activate trans-acting factors, and act on the cis-acting regions of hormone-regulating genes, thereby regulating gene transcription and translation. Flavonoids is considered to be highly reactive and potentially toxic in the cytoplasm. To avoid toxicity, cytoplasmic synthetic flavonoids are transported to vacuoles for storage or isolation through transporters. ATP binding cassette (ABC) transporter is an important transporter in plants. The ABC-type transporter MRP3 is involved in the transport of Zea’s flavonoids. The enrichment of flavonoid transport genes was consistent with previous studies, and these transport genes may be involved in the transmembrane transport of flavonoids.
Furthermore, the lightyellow module gene is positively correlated with the chlorophyll content in the pepper fruit; the turquoise module gene is positively correlated with the carotenoid content in the pepper fruit; the blue module gene is positively correlated with the anthocyanin content in the pepper fruit; the gene in the lightcyan module is positively correlated with the flavonoid content in the fruit positive correlation. By analyzing the core genes of the module, 29 transcription factors such as Pkinase, F-box, and zf-MYND were found, which may be involved in the synthesis and regulation of various pigments in pepper fruit. Many studies have confirmed that transcription factors play an important regulatory role in the development and maturation of horticultural plants. Through transcriptome sequencing, YE found that there are 37 transcription factors and five structural genes of carotenoid metabolism pathways that are significantly related. In tomato crops, the AP2/ERF transcription factor family plays a key role in the accumulation of carotenoids. Recent studies have found that the AP2/ERF transcription factor SleRF6 plays an important negative regulatory role on carotenoids during tomato fruit ripening. also identified two transcription factors (RAP2.2 and PIFs), a bHLH family transcription factor, which blocks gene expression through the PSY promoter cassette. In pepper, recent studies have also found that the expression patterns of ER transcription factor family genes, such as CAERF82, CAERF97, CAERF66, CAERF107, and CAERF10, are consistent with the accumulation of carotenoids, which may have regulatory effects. MYB transcription factor is also widely involved in the biosynthetic pathway of carotenoids. In maize, PBF and GAMYB independently activate the expression of BCH2. In kiwifruit, the transcription factor MYB7 regulates carotenoid anabolism by transcriptionally activating the LCYB gene.
The genes or proteins that regulate the color of pepper fruit are affected by many factors, such as light temperature, adversity stress, and hormones. In this study, the WGCNA analysis suggested that 29 transcription factors such as Pkinase, F-box, and zf-MYND may be involved in the regulation of pepper fruit color change. However, the regulatory mechanism of these transcription factors is unclear and needs to be further clarified.
- LI Q H, YANG S P, YU Y N, KHAN A, FENG P L, ALI M, SHAO D K, WANG Y Y, ZHANG R X, GAI W X, HAN R, MA X, HOU Q G, GONG Z H. Comprehensive transcriptome-based characterization of differentially expressed genes involved in carotenoid biosynthesis of different ripening stages of [J]. Scientia Horticulturae, 2021, 288: 110311.
- HURTADO HERNANDEZ H, SMITH P G. Inheritance of mature fruit color in L.[J]. Journal of Heredity, 1985, 76(3): 211-213.
- MATSUFUJI H, ISHIKAWA K, NUNOMURA O, CHINO M, TAKEDA M. Anti-oxidant content of different coloured sweet peppers, white, green, yellow, orange and red (L.)[J]. International Journal of Food Science & Technology, 2007, 42(12): 1482-1488.
- ARI E, BEDIR H, YILDIRIM S, YILDIRIM T. Androgenic responses of 64 ornamental pepper (L.) genotypes to shed-microspore culture in autumn season[J]. Turkish Journal of Biology, 2016, 40: 706-717.
- LIGHTBOURN G J, GRIESBACH R J, NOVOTNY J A, CLEVIDENCE B A, RAO D D, STOMMEL J R. Effects of anthocyanin and carotenoid combinations on foliage and immature fruit color of L.[J]. The Journal of Heredity, 2008, 99(2): 105-111.
- BOROVSKY Y, OREN SHAMIR M, OVADIA R, DE JONG W, PARAN I. The a locus that controls anthocyanin accumulation in pepper encodes a MYB transcription factor homologous to Anthocyanin2 of [J]. Theoretical and Applied Genetics, 2004, 109(1): 23-29.
- CHAIM A B, BOROVSKY Y, DE JONG W, PARAN I. Linkage of the a locus for the presence of anthocyanin and, a major fruit-shape QTL in pepper[J]. Theoretical and Applied Genetics, 2003, 106(5): 889-894.
- PAZ-ARES J, GHOSAL D, WIENAND U, PETERSON P A, SAEDLER H. The regulatory c1 locus of encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators[J]. The EMBO Journal, 1987, 6(12): 3553-3558.
- DUBOS C, STRACKE R, GROTEWOLD E, WEISSHAAR B, MARTIN C, LEPINIEC L. MYB transcription factors in [J]. Trends in Plant Science, 2010, 15(10): 573- 581.
- SAGASSER M, LU G H, HAHLBROCK K, WEISSHAAR B. is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins[J]. Genes & Development, 2002, 16(1): 138-149.
- STOMMEL J R, LIGHTBOURN G J, WINKEL B S, GRIESBACH R. Transcription factor families regulatethe anthocyanin biosynthetic pathway in [J]. Journal of the American Society for Horticultural Science, 2009, 134(2): 244-251
- HUH J H, KANG B C, NAHM S H, KIM S, HA K S, LEE M H, KIM B D. A candidate gene approach identified phytoene synthase as the locus for mature fruit color in red pepper (spp.)[J]. Theoretical and Applied Genetics, 2001, 102(4): 524-530.
- POPOVSKY S, PARAN I. Molecular genetics of the locus in pepper: its relation to capsanthin-capsorubin synthase and to fruit color[J]. Theoretical and Applied Genetics, 2000, 101(1): 86-89.
- FENG C, CHEN M, XU C J, BAI L, YIN X R, LI X, ALLAN A C, FERGUSON I B, CHEN K S. Transcriptomic analysis of Chinese bayberry () fruit development and ripening using RNA-Seq[J]. BMC Genomics, 2012, 13: 19.
- QIN Y L, DJABOU A S M, AN F F, LI K M, LI Z G, YANG L, WANG X J, CHEN S B. Proteomic analysis of injured storage roots in cassava (Crantz) under postharvest physiological deterioration[J]. PLoS One, 2017, 12(3): e0174238.
- CHABIKWA T G, BARBIER F F, TANURDZIC M, BEVERIDGE C A. transcriptome assembly and annotation for gene discovery in avocado, macadamia and mango[J]. Scientific Data, 2020, 7(1): 9.
- TATA S. A comprehensive study on chilli peppers ( L.)[M]. Chisinau: Lambert Academic Publishing, 2016.
- DELI J, MATUS Z, SZABOLCS J. Carotenoid composition in the fruits of black paprika (variety longum nigrum) during ripening[J]. Journal of Agricultural and Food Chemistry, 1992, 40(11): 2072-2076.
- GOODMAN C D, CASATI P, WALBOT V. A multidrug resistance-associated protein involved in anthocyanin transport in [J]. The Plant Cell, 2004, 16(7): 1812-1826.
- GUZMAN I, HAMBY S, ROMERO J, BOSLAND P W, O’CONNELL M A. Variability of carotenoid biosynthesis in orange colored spp.[J]. Plant Science, 2010, 179 (1): 49-59.
- CHUNG M Y, VREBALOV J, ALBA R, LEE J, MCQUINN R, CHUNG J D, KLEIN P, GIOVANNONI J. A tomato () APETALA2/ERF gene, , is a negative regulator of fruit ripening[J]. Plant Journal, 2010, 64(6): 936-947.
- WELSCH R, MAASS D, VOEGEL T, DELLAPENNA D, BEYER P. Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of leaves[J]. Plant Physiology, 2007, 145(3): 1073- 1085.
- SONG J L, CHEN C M, ZHANG S L, WANG J T, HUANG Z B, CHEN M X, CAO B H, ZHU Z S, LEI J J. Systematic analysis of the ERF transcription factor family: identification of regulatory factors involved in the regulation of species-specific metabolites[J]. BMC Genomics, 2020, 21(1): 573-587.
- JIN X, BAI C, BASSIE L, NOGAREDA C, ROMAGOSA I, TWYMAN R M, CHRISTOU P, ZHU C. ZmPBF and ZmGAMYB transcription factors independently transactivate the promoter of the maize () β-carotene hydroxylase 2 gene[J]. New Phytologist, 2019, 222(2): 793-804.
- AMPOMAH DWAMENA C, THRIMAWITHANA A H, DEJNOPRAT S, LEWIS D, ESPLEY R V, ALLAN A C. A kiwifruit () R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation[J]. New Phytologist, 2019, 221(1): 309-325.

