Variant rs8400 enhances ALKBH5 expression through disrupting miR-186 binding and promotes neuroblastoma progression
Qian Guan ,Huiran Lin ,Wenfeng Hua ,Lei Lin ,Jiabin Liu ,Linqing Deng ,Jiao Zhang,Jiwen Cheng,Zhonghua Yang,Yong Li,Jun Bian,Haixia Zhou0,Suhong Li,Li Li2,Lei Miao,Huimin Xia,Jing He,2,Zhenjian Zhuo,2,3
1School of Medicine,South China University of Technology,Guangzhou 510006,China;2Department of Pediatric Surgery,Guangzhou Institute of Pediatrics,Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease,Guangdong Provincial Clinical Research Center for Child Health,Guangzhou Women and Children’s Medical Center,Guangzhou Medical University,Guangzhou 510623,China;3Faculty of Medicine,Macau University of Science and Technology,Macau 999078,China;4Research Institute for Maternal and Child Health,Guangdong Second Provincial General Hospital,Guangzhou 510317,China;5 Department of Pediatric Surgery,the First Affiliated Hospital of Zhengzhou University,Zhengzhou 450052,China;6 Department of Pediatric Surgery,the Second Affiliated Hospital of Xi’an Jiaotong University,Xi’an 710004,China;7 Department of Pediatric Surgery,Shengjing Hospital of China Medical University,Shenyang 110004,China;8 Department of Pediatric Surgery,Hunan Children’s Hospital,Changsha 410004,China;9 Department of General Surgery,Xi’an Children’s Hospital,Xi’an Jiaotong University Affiliated Children’s Hospital,Xi’an 710003,China;10Department of Hematology,The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University,Wenzhou 325027,China;11Department of Pathology,Children Hospital and Women Health Center of Shanxi,Taiyuan 030013,China;12Kunming Key Laboratory of Children Infection and Immunity,Yunnan Key Laboratory of Children’s Major Disease Research,Yunnan Institute of Pediatrics Research,Yunnan Medical Center for Pediatric Diseases,Kunming Children’s Hospital,Kunming 650228,China;13 Laboratory Animal Center,School of Chemical Biology and Biotechnology,Peking University Shenzhen Graduate School,Shenzhen 518055,China
Abstract Objective:AlkB homolog 5 (ALKBH5) has been proven to be closely related to tumors.However,the role and molecular mechanism of ALKBH5 in neuroblastomas have rarely been reported.Methods:The potential functional single-nucleotide polymorphisms (SNPs) in ALKBH5 were identified by National Center for Biotechnology Information (NCBI) dbSNP screening and SNPinfo software.TaqMan probes were used for genotyping.A multiple logistic regression model was used to evaluate the effects of different SNP loci on the risk of neuroblastoma.The expression of ALKBH5 in neuroblastoma was evaluated by Western blotting and immunohistochemistry (IHC).Cell counting kit-8 (CCK-8),plate colony formation and 5-ethynyl-2’-deoxyuridine(EdU) incorporation assays were used to evaluate cell proliferation.Wound healing and Transwell assays were used to compare cell migration and invasion.Thermodynamic modelling was performed to predict the ability of miRNAs to bind to ALKBH5 with the rs8400 G/A polymorphism.RNA sequencing,N6-methyladenosine (m6A)sequencing,m6A methylated RNA immunoprecipitation (MeRIP) and a luciferase assay were used to identify the targeting effect of ALKBH5 on SPP1.Results:ALKBH5 was highly expressed in neuroblastoma.Knocking down ALKBH5 inhibited the proliferation,migration and invasion of cancer cells.miR-186-3p negatively regulates the expression of ALKBH5,and this ability is affected by the rs8400 polymorphism.When the G nucleotide was mutated to A,the ability of miR-186-3p to bind to the 3’-UTR of ALKBH5 decreased,resulting in upregulation of ALKBH5.SPP1 is the downstream target gene of the ALKBH5 oncogene.Knocking down SPP1 partially restored the inhibitory effect of ALKBH5 downregulation on neuroblastoma.Downregulation of ALKBH5 can improve the therapeutic efficacy of carboplatin and etoposide in neuroblastoma.Conclusions:We first found that the rs8400 G>A polymorphism in the m6A demethylase-encoding gene ALKBH5 increases neuroblastoma susceptibility and determines the related mechanisms.The aberrant regulation of ALKBH5 by miR-186-3p caused by this genetic variation in ALKBH5 promotes the occurrence and development of neuroblastoma through the ALKBH5-SPP1 axis.
Keywords: Neuroblastoma;risk;ALKBH5;polymorphism;progression
Introduction
Neuroblastoma is a solid tumor of the sympathetic nervous system (1).It accounts for 7% of all childhood cancers and 15% of childhood cancer-related deaths (2).Neuroblastoma is a heterogeneous malignancy with prognoses ranging from spontaneous regression with a good outcome to metastatic disease with a poor outcome(3,4).Despite advances in the treatment of paediatric cancers,the survival rates of patients with high-risk neuroblastomas are still less than 50% (5,6).
With the development of whole-genome sequencing techniques,increasing attention has been paid to the important roles of genetic variations in neuroblastoma.Genetic markers help us to understand the genetic aetiology and guide the treatment of neuroblastoma (7-9).Neuroblastomas are frequently associated with mutations in thePHOX2B(10) andALK(11,12) genes,amplification of theMYCNgene,and loss of heterozygosity of chromosome 1p.The oncogenic effects of mutations inPTPN11,ATRX,TERT,LMO1andNRASpreviously found in neuroblastoma have also been investigated (13-19).Multiple neuroblastoma risk-associated single-nucleotide polymorphisms (SNPs) have been identified by genomewide association studies (GWASs) (20-24).Mariset al.first demonstrated that the rs9295536,rs4712653 and rs6939340 polymorphisms are associated with neuroblastoma susceptibility (25).Interestingly,Oldridgeet al.demonstrated that the SNP rs2168101 polymorphism affects GATA transcription factor binding sites within enhancer elements,resulting in altered LMO1 expression that affects neuroblastoma progression (24).However,the set of currently identified genetic mutations or variants is not sufficient to predict neuroblastoma risk.
With the development of methylated transcriptome analysis approaches,N6-methyladenosine (m6A) has been proven to be the most common posttranscriptional modification of messenger RNA (mRNA) in eukaryotes(26,27).The key regulators of m6A modification include mainly the following three protein complexes: RNA methyltransferases (METTL3/14,WTAP,RBM15/15B and VIRMA,termed “writers”),demethylases (FTO and ALKBH5,termed “erasers”),and m6A-binding proteins(YTHDC1/2,YTHDF1/2/3,IGF2BP1/2/3,HNRNP and eIF3,termed “readers”) (28).Human AlkB homolog H5(ALKBH5),a member of the ALKB family,is a critical m6A demethylase that catalyses the direct removal of m6A(29).AberrantALKBH5expression affects RNA metabolism,RNA nuclear export,and gene expression (30).ALKBH5 is critically implicated in the occurrence and progression of human malignancies.ALKBH5 plays oncogenic or tumor-suppressive roles in different types of cancer (31-35).One study showed that ALKBH5 reduced PER1 degradation in an m6A-YTHDF2-dependent manner via m6A demethylation and that posttranscriptional upregulation of PER1 resulted in inhibition of pancreatic cancer cell growth by activating ATM-CHK2-P53/CDC25C signalling (36).Recent studies show that ALKBH5 can promote tumor progression in head and neck squamous cell carcinoma by inhibiting interferon-alpha production and RIG-I expression (35).Tanet al.established and validated a prognostic glucose metabolism model based on the overlap between differentially expressed genes related toALKBH5and glucose metabolism-related genes in an analysis based on the GSE62564 dataset,and the results suggested thatALKBH5was related to the process of glucose metabolism (37).However,the roles of theALKBH5gene and its SNPs in neuroblastoma remain unclear.
Here we report the identification of the rs8400 polymorphism in theALKBH5gene as a new locus for susceptibility to neuroblastoma in the Chinese Han population.Biochemical analysis showed that the rs8400 polymorphism could affect the expression of the host geneALKBH5to promote neuroblastoma progression.Mechanistically,the rs8400 G>A SNP (located in a microRNA binding site) leads to the failure of miR-186-3p to negatively regulate ALKBH5 expression,leading to upregulation of ALKBH5.In addition,we functionally validated the oncogenic effects of the rs8400 SNP and ALKBH5 expression in neuroblastoma.Subsequently,we further identified SPP1 as the downstream target gene ofALKBH5.Therefore,our findings indicate that the miR-186-3p-ALKBH5-SPP1 axis is closely related to the occurrence and development of neuroblastoma.
Materials and methods
Sample selection
Participants for this study were pooled from 9 hospitals across China.We defined cases as patients with a histologically verified first-time diagnosis of neuroblastoma.Controls who had no diagnosis of any cancers were randomly selected from the same hospital and individually matched to the cases in age and sex.Patients’clinical characteristics are described inSupplementary Table S1.Participants provided written informed consent at screening.Detailed descriptions of the study participants have been published in our previous work (38,39).
Neuroblastoma tissue samples
Frozen neuroblastoma tissue samples were collected from the Guangzhou Women and Children’s Medical Center,and the neuroblastoma samples were categorized into two groups: the early-stage (stages I and II,n=10) and advanced-stage (stages III and IV,n=10) groups,based on the clinicopathological information.The study was approved by the Ethics Committee of Guangzhou Women and Children’s Medical Center and conducted in accordance with the ethical standards of the World Medical Association Declaration of Helsinki.All patients provided written informed consent before enrolment.
Polymorphism selection and genotyping
Potential functional SNPs in theALKBH5gene were retrieved from the dbSNP database and with SNPinfo software (40).DNA was extracted from blood using the QIAamp DNA Blood Mini Kit (QIAGEN Inc.,Valencia,USA) and further genotyped using the standard TaqMan Genotyping Array (Applied Biosystems,Foster City,USA).On each 384-well plate,blind duplicate samples (10%samples) were included for quality control.A concordance rate of 100% was achieved.The genotyping completion rate for all SNPs was >95%.
Immunohistochemistry (IHC)
Neuroblastoma tissue was fixed with 10% formaldehyde by a professional pathologist,and paraffin-embedded tissue sections were generated.After 1 h of baking,the sections were sequentially dewaxed,subjected to antigen retrieval,blocked with 5% goat serum for 30 min,and then incubated with an anti-ALKBH5 rabbit primary antibody at 4 °C overnight.The next day,an anti-rabbit secondary antibody was applied for 30 min after rewarming.After that,3,3’-diaminobenzidine (DAB) chromogenic reagent was used for color development to visualize staining.After confirming staining,the sections were incubated with haematoxylin for 5 s to stain nuclei and differentiated with 1% hydrochloric acid for 10 s;finally,cover slips were mounted for observation.The whole process of immunohistochemical staining was carried out by trained professional laboratory technicians who are not familiar with experiments.In immunohistochemical staining,the ALKBH5 signals of different stages and genotypes were compared and analyzed according to the number of positive signals and the intensity of positive signals (color depth) (41).
Cell culture
The neuroblastoma cell lines SK-N-SH,SK-N-BE(2),SKN-BE(2)-C and SK-N-AS were obtained from the American Type Culture Collection (ATCC,Manassas,VA,USA).The cells were cultured in EMEM,RPMI-1640 medium,and F12 medium (Thermo Fisher Scientific,Inc.,Waltham,MA,USA) containing 10% foetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C in a humidified incubator with 5% CO2.All the cells were authenticated by short-tandem repeat genetic profiling and found to be free of mycoplasma contamination by testing.
Transfection
Production of the ALKBH5 knockdown and overexpression lentiviruses was performed by OBiO (OBiO Technology Corp.,China).Neuroblastoma cells were plated in 6-wells dishes at 50% confluence and infected with ALKBH5 knockdown lentivirus (termed shALKBH5-1) or scramble control lentivirus (termed shNC).The shALKBH5 sequence was 5’-GAAGCTTCAATGGTC TCCTTA-3’.The shNC sequence was 5’-TTCTCCGA ACGTGTCACGT-3’.Pools of stable transductants were generated by selection using puromycin (4 μ g/mL) for 2 weeks.The miR-186 mimics and inhibitors used for transfection were purchased from GenePharma (Shanghai,China).Transfection was performed using Opti-MEM and an IMAX kit (Invitrogen,USA) according to the manufacturer’s instructions.
Cell proliferation
The cell proliferation rate was determined with a cell counting kit (CCK-8,Dojindo,Japan) according to the manufacturer’s instructions.Briefly,3×103cells/well were seeded in 96-well culture plates and incubated for the indicated time.Then,CCK-8 solution (100 μL) was added,and the absorbance at 450 nm was measured.A colony formation assay (plate colony formation assay) was performed to evaluate the long-term proliferation of neuroblastoma cells with knockdown of ALKBH5 expression.
Cell cycle assay
A total of 1×106cells were collected,washed with phosphate-buffered saline (PBS),and fixed with 75% cold ethanol for 24 h at -20 °C.After being washed with PBS twice and stained with propidium iodide from a Cycletest Plus DNA Reagent Kit (BD Biosciences,USA) at room temperature for 30 min,cells were examined by flow cytometry (Becton Dickinson,USA),and the cell cycle distribution was analyzed by ModFit LT software (Version 4.1.7;Verity Software House,Topsham,ME,USA).
Wound healing assay
Cells were seeded in 6-well plates at 5×105cells per well.Then,a wound was made in the cell monolayer by scratching with a 200 μ L pipette tip.The detached cells were then removed by washing with PBS.Cell motility in each group was assessed by visualizing the speed of wound closure at intervals.
Cell migration assay
A Transwell (Costar,New York,NY,USA) assay was used to evaluate the migration capacity.Cells (1×105cells/well)in a serum-free medium were placed in the upper chamber,and medium containing 10% FBS was added to the lower chamber as a chemoattractant.After incubation for the indicated time,cells on the upper surface of the membrane were removed.The migrated cells were fixed with formaldehyde and stained using 0.5% crystal violet(Sigma).The migrated cells were counted under a microscope.
Western blotting analysis
Protein extracts were lysed on ice with RIPA buffer containing both protease and phosphatase inhibitors(Thermo Fisher Scientific,Inc.).Protein concentrations were determined using a BCA protein assay kit (Thermo Fisher Scientific) following the manufacturer’s protocol.Equal amount of protein was then loaded on a gel and finally transferred onto a PVDF membrane.The membrane was blocked using 5% BSA and incubated overnight at 4 °C with primary antibodies diluted 1:1,000.HRP-conjugated anti-mouse (1:5,000) and anti-rabbit(1:2,000) secondary antibodies were used.The membrane was incubated with enhanced chemiluminescence reagents,and band densities were quantified by ImageJ analysis software (Version 1.43;National Institutes of Health,USA).
RNA extraction and reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using an RNA extraction kit (#RN001,ESscience,Shanghai,China) and converted to cDNA using a reverse transcription kit(#RR036A-1,TAKARA,Japan).Quantitative mRNA expression levels were determined using real-time fluorescence quantitative PCR instrument (ABI Q6 System,Applied Biosystems,USA).GAPDH was used as the internal reference gene.Relative mRNA expression was calculated using the 2-ΔΔCTmethod.Specific primer sequences are shown inSupplementary Table S2.
RNA sequencing
mRNA sequencing was performed on ALKBH5-knockout(KO-ALKBH5) and control BxPC cells (Vector) using the HiSeq 2000 platform to generate single-end reads with a read length of 50 bp (PE50) (Illumina Inc.,San Diego,CA,USA).The sequencing data for each gene were then converted to FPKM (fragments per kilobase mRNA sequence per million mapped reads) values by TopHat-Cufflinks (v2.2.1).Gene Ontology (GO) and signalling pathway enrichment analyses were subsequently performed using DAVID (www.david.niaid.nih.gov) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database(http://www.genome.jp/kegg/),respectively.
Methylated RNA immunoprecipitation (MeRIP) and m6A sequencing
Total RNA was extracted from stable KO-ALKBH5 SKN-AS cells and control cells with TRIzol (Thermo Fisher).According to the instructions of the MeRIP kit (C11051-1,Magneto,USA),RNA was fragmented,immunoprecipitated with an anti-m6A antibody,and finally eluted and purified.The concentration of m6A-modified mRNA was then analyzed by RT-qPCR or highthroughput RNA sequencing using the primers listed inSupplementary Table S2.Sequencing was carried out on the Illumina HiSeq X Ten platform,and each group was analyzed in duplicate.
Thermodynamic modelling of miRNA-target interactions
We compared the ability of miRNA-186-3p to bind to the rs8400 G allele and A allele using a secondary structurebased nonparametric thermodynamic modelling study.ETargetrepresents the dissociation energy of the targeted region of the mRNA with no interaction with the microRNA.EIntermediaterepresents the energy required to make the target region available for microRNA binding,and EComplexrepresents the free energy of the final microRNA-target complex.To avoid sequence length dependence,we determined the final target sequence length by increasing number of nucleotides surrounding the region consisting of the target seed region and the SNP site (Supplementary Figure S1).We calculated the minimum free energy (MFE) of the target sequence (G or A) and the free energy of the partition function folding by RNAfold,and we took the MFE as the ETargetafter considering the consistency of the centroid structure free energy and MFE.When computing EComplex,we input the miRNA and target sequences into RNAcofold.Considering the different structures (including suboptimal structures) that the miRNAs and target sequences actually form,we assigned the ensemble energies of the allocation function folding rather than just the MFE as EComplex.We calculated EIntermediateby constraining all possible binding sites of the miRNA and the target sequence to an unpaired state.
Luciferase reporter assay
The ALKBH5 3’-UTR (rs8400) and SPP1 overexpression reporter plasmids (purchased from Promega) were constructed by using the pGL3 and PNL3.1 luciferase reporter plasmids,respectively.The constructed wild-type and mutant plasmids were transfected into both SK-N-AS and SK-N-BE(2)-C cells.After 48 h,the cells were treated according to the instructions of the dual luciferase reporter detection kit (E1910,N1610),and the ratio of firefly luciferase to Renilla luciferase activity was determined.The experiment was performed in triplicate,and the data are expressed as.
Statistical analysis
The distributions of demographic and clinical characteristics were compared between the cases and controls using Student’st-test (for continuous variables)and the Chi-square test (for categorical variables).We first applied a goodness-of-fit Chi-square test to assess whether SNPs in the controls were in Hardy-Weinberg equilibrium(HWE).An odds ratio (OR) and the corresponding 95%confidence interval (95% CI) were estimated for each SNP for neuroblastoma using logistic regression models.An estimation of haplotype frequency and an analysis of the effect of haplotype on neuroblastoma risk were performed(42,43).We also applied false-positive report probability(FPRP) analysis to evaluate notable associations by a method described elsewhere (44,45).Expression quantitative trait loci (eQTL) analysis using the online Genotype-Tissue Expression (GTEx) portal (http://www.gtexportal.org/home/) was adopted to determine the correlations between the SNPs and expression levels of nearby genes (46).P<0.05 indicates statistical significance.Statistical analyses were conducted using SAS software(Version 9.4;SAS Institute Inc.,Cary,NC,USA)
Results
ALKBH5 rs8400 is a new neuroblastoma susceptibility locus in Chinese Han population
We successfully genotyped the twoALKBH5gene SNPs(rs1378602 and rs8400) in 967 cases and 1,813 controls(Table 1).First,we tested whether the two SNPs were in HWE in the control group by Chi-square test,and results showed that the two SNPs were not statistically significant in HWE (HWE P>0.05).We then performed multivariate logistic regression analysis to explore the relationships between the genotypes of these twoALKBH5SNPs(rs1378602 and rs8400) and susceptibility to neuroblastoma.We then performed multivariate logistic regression analysis to explore the relationships between rs1378602 and rs8400 genotypes and susceptibility to neuroblastoma.The results showed that the relationship between rs1378602 genotype and neuroblastoma risk was not statistically significant.Notably,the rs8400 mutant A allele was significantly associated with increased susceptibility to neuroblastoma (AAvs.GG: adjusted OR=1.28,95% CI=1.02-1.61,P=0.030;GGvs.GAvs.AA: adjusted OR=1.12,95% CI=1.004-1.26,P=0.042;AAvs.GG/GA: adjusted OR=1.25,95% CI=1.03-1.52,P=0.028).We further observed that participants with 1 risk genotype (adjusted OR=1.23,95% CI=1.02-1.48,P=0.031) or 1-2 risk genotypes (adjusted OR=1.22,95%CI=1.03-1.44,P=0.022) exhibited an increased risk of neuroblastoma in comparison to that of participants with 0 risk genotypes.
Stratification analysis of identified SNPs
To reduce the influence of other factors,we further explored the associations betweenALKBH5gene polymorphisms and neuroblastoma susceptibility in certain groups stratified by age,sex,sites of origin and clinical stages (Table 2).Compared with the rs8400 GG/GA genotype,the rs8400 AA genotype increased susceptibility to neuroblastoma,and this association remained statistically significant in the subgroup with mediastinal tumor origin(adjusted OR=1.43,95% CI=1.03-1.98,P=0.034) and the subgroup with clinical stage I+II+4s (adjusted OR=1.29,95% CI=1.01-1.64,P=0.041).The combined analysisshowed that participants with 1-2 risk genotypes in the following subgroups had a significantly increased neuroblastoma risk: age >18 months (adjusted OR=1.27,95% CI=1.02-1.58,P=0.033),males (adjusted OR=1.26,95% CI=1.00-1.58,P=0.049),retroperitoneal tumor origin (adjusted OR=1.29,95% CI=1.004-1.65,P=0.046),and clinical stage I+II+4s (adjusted OR=1.29,95%CI=1.04-1.59,P=0.018).
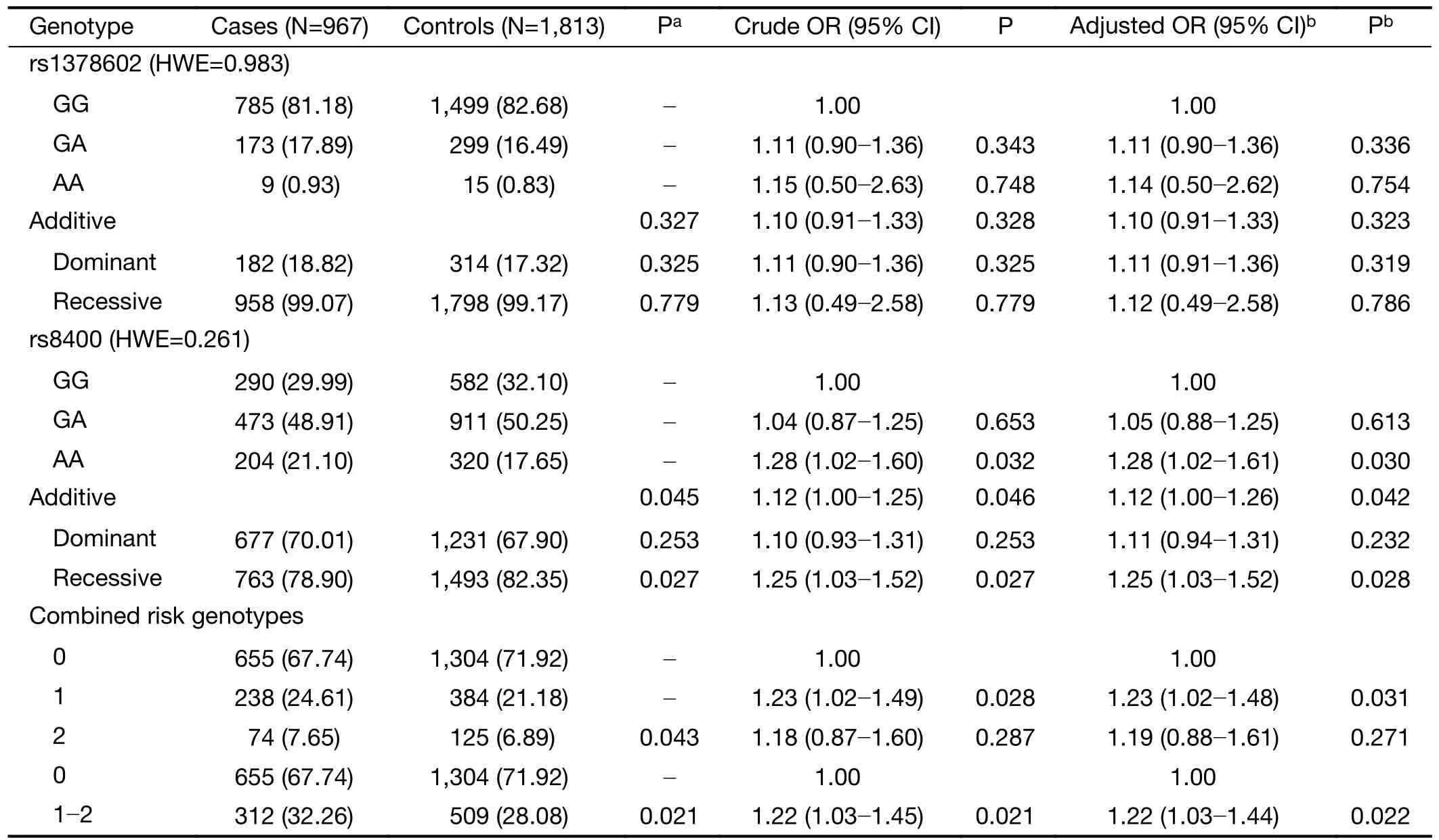
Table 1 Logistic regression analysis of associations between ALKBH5 gene polymorphisms and neuroblastoma susceptibility
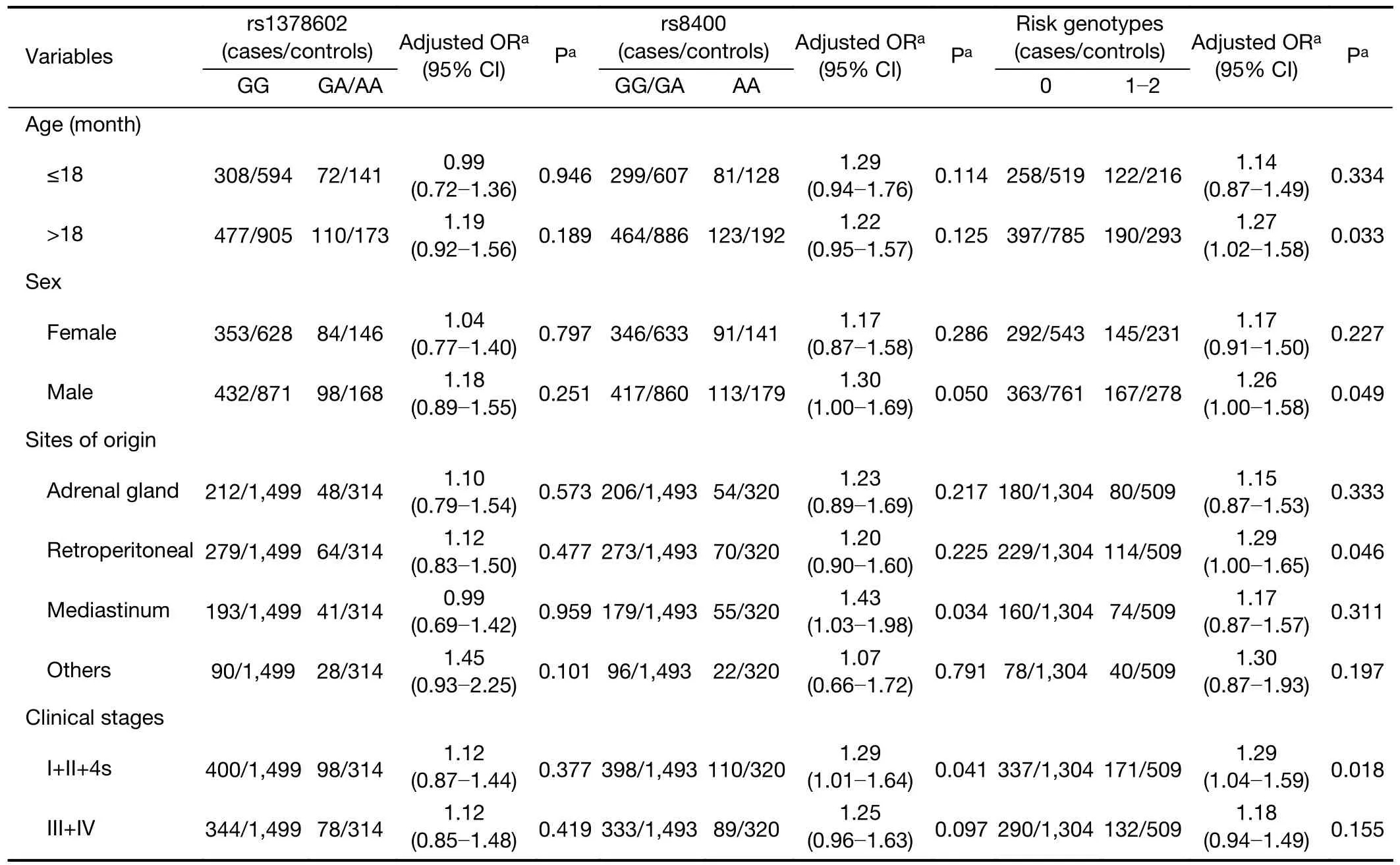
Table 2 Stratification analysis of ALKBH5 gene polymorphisms with neuroblastoma susceptibility
Characterization of ALKBH5 expression in neuroblastoma
To explore the expression of ALKBH5 in neuroblastoma,we first analyzed the publicly accessible data in a GEO dataset (GSE49710) and GTEx.The results showed that ALKBH5 was highly expressed in neuroblastoma tissues(P<0.0001) (Figure 1A).We then measured and compared the expression of ALKBH5 in neuroblastoma cell lines[(SK-N-SH,SK-N-BE(2),SK-N-AS,SK-N-BE(2)-C,and KELLY] and non-neuroblastoma cell lines (NT22 and HELS) by Western blotting.The results showed that the expression of ALKBH5 protein in neuroblastoma cells was significantly higher than that in non-neuroblastoma cells(Figure 1B).In addition,neuroblastoma occurred more frequently at 18 months of age,and clinical correlation analysis showed that ALKBH5 expression was increased in individuals younger than 18 months (P<0.0001) (Figure 1C).Additionally,m6A levels were elevated in ALKBH5 knockdown (si-ALKBH5) neuroblastoma cells compared with control cells (P<0.05) (Figure 1D).The IHC results showed that ALKBH5 expression was higher in tissues from patients with advanced neuroblastoma (stage 3 and stage 4) than in tissues from patients with early neuroblastoma (stage 1 and stage 2) (Figure 1G).These results suggest that ALKBH5 is significantly upregulated in neuroblastoma and that ALKBH5 upregulation may be related to the poor progression of neuroblastoma.The tissue samples of neuroblastoma patients were further analyzed by RT-qPCR and Western blotting (Figure 1E,F),and the results showed that the mRNA and protein expression of ALKBH5 in late neuroblastoma (stage III and IV) tissues was significantly upregulated compared with that in early neuroblastoma (stage I and II) tissues.

Figure 1 Downregulation of m6A demethylase gene ALKBH5 represents a characterization of neuroblastoma.(A) ALKBH5 is highly expressed in neuroblastoma tissues compared to normal tissues;(B) Five neuroblastoma cell lines [(SK-N-SH,SK-N-BE(2),SK-N-AS,SKN-BE(2)-C and KELLY] ALKBH5 protein expression was higher compared to non-neuroblastoma cell lines (NT22 and HELS);(C)ALKBH5 expression increases at less than 18 months of age;(D) m6A expression levels are elevated in ALKBH5 knockdown neuroblastoma(si-ALKBH5) cells compared to controls;(E,F) mRNA and protein expression of ALKBH5 was significantly increased in late neuroblastoma (stages III and IV) tissues compared with early neuroblastoma (stages I and II) tissues;(G) Immunohistochemistry results show that ALKBH5 expression is higher in tissues of patients with advanced neuroblastoma (Magnification 10×).*,P<0.05;**,P<0.01;***,P<0.001;****,P<0.0001.
Effect of ALKBH5 on neuroblastoma cell transcriptome
To demonstrate the oncogenic effect of ALKBH5 expression on neuroblastoma,we first investigated the effect of ALKBH5 on the transcriptome of neuroblastoma cells.We used CRISPR-Cas9 genome editing to generate neuroblastoma cell lines with stable ALKBH5 knockout(Supplementary Figure S2A).The control cell line was left untreated,and the cells were collected for RNA-seq analysis.Volcano plots generated from the RNA-seq data showed that neuroblastoma cells with knockout of ALKBH5 expression exhibited 34 differentially expressed genes (28 upregulated genes and 6 downregulated genes)(Figure 2A,Supplementary Figure S2B).Cluster analysis of the differentially expressed genes revealed thatSPP1,MIR6859-3,ANKRD1,andADAMTS12were up-regulated in neuroblastoma cell lines with knockout of ALKBH5 compared to the corresponding control cells (Figure 2B).GO enrichment analysis showed that knockout of ALKBH5 resulted in significant downregulation of genes related to RNA trafficking,RNA localization,ribonucleic acid export,and DNA biosynthesis processes,and upregulation of genes related to the cell response to tumor necrosis factor and interleukin 1 (Figure 2C,D).KEGG enrichment analysis of the differentially expressed genes indicated that the upregulated genes were involved in pathways such as transcriptional dysregulation in cancer,NOD-like receptor signalling pathway,and NF-kappa B signalling pathway (Figure 2E).In conclusion,ALKBH5 showed neuroblastoma-promoting activity based on transcriptome regulation.
ALKBH5 promotes neuroblastoma cell proliferation and invasion
Due to the high expression of ALKBH5 in patients with advanced neuroblastoma,we investigated the relationship between ALKBH5 expression and neuroblastoma cell invasiveness.We first determined the interference efficiency of the siRNA targeting ALKBH5.The results of RT-qPCR and Western blotting analysis consistently showed that ALKBH5 mRNA and protein expression were significantly decreased in neuroblastoma cell lines with ALKBH5 knockdown compared with the corresponding control cells (Supplementary Figure S3A,B).Cell proliferation assays using the CCK-8 kit showed that two si-ALKBH5 neuroblastoma cell lines (SK-N-SH,SK-NAS) had significantly reduced numbers of viable cells compared to the corresponding control neuroblastoma cells(Figure 3A).The effect of ALKBH5 expression on longterm neuroblastoma cell proliferation was determined by a colony formation assay.After two weeks,we observed that ALKBH5 knockdown significantly reduced the efficiency of clone formation (Figure 3B).The EdU incorporation assay also showed that the proliferation of neuroblastoma cell lines with knockdown of ALKBH5 was inhibited(Supplementary Figure S3C).Flow cytometric analysis further demonstrated that the proportion of cells in G2/M phase were increased after perturbing ALKBH5 expression(Figure 3C).Inhibition of ALKBH5 reduced the expression of CDK2,CDK4 and cyclinD3 and increased the expression of p27 (Figure 3D).The wound closure ability of the two cell lines with inhibited ALKBH expression [SKN-SH,SK-N-BE(2)] was decreased compared to that of the corresponding control cells (Figure 3E).Consistent with these findings,these ALKBH5-knockdown neuroblastoma cells had decreased migratory and invasive abilities (Figure 3F).Taken together,these findings suggest that ALKBH5 exerts tumorigenic effects.
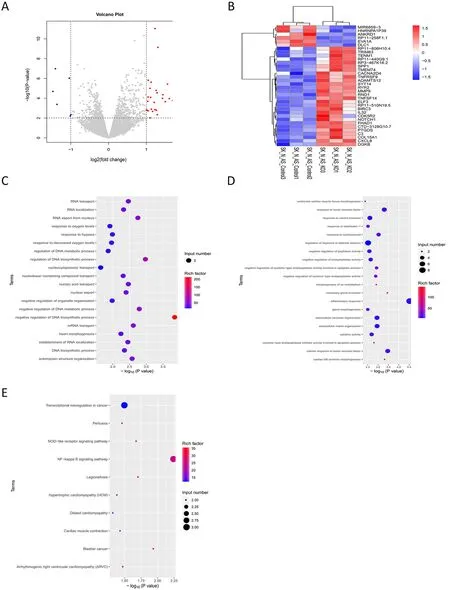
Figure 2 Effects of ALKBH5 on transcriptome of neuroblastoma cells.(A) Volcano plots showing up-regulated genes (red) and downregulated genes (blue) displayed by ALKBH5 knockout neuroblastoma cells;(B) Cluster analysis shows that SPP1,MIR6859-3,ANKRD1 and ADAMTS12 genes are up-regulated in neuroblastoma cell lines with down-regulated ALKBH5 expression;(C,D) GO enrichment analysis showed that genes related to RNA trafficking,localization,ribonucleic acid export,and DNA biosynthesis processes were down-regulated in ALKBH5 knockdown neuroblastoma cells (C),and genes related to the cellular response to tumor necrosis factor are up-regulated (D);(E) KEGG enrichment analysis of differential genes revealed that up-regulated genes are involved in pathways including transcriptional dysregulation in cancer,NOD-like receptor signaling and NF-kappa B signaling.GO,Gene Ontology;KEGG,Kyoto Encyclopedia of Genes and Genomes.
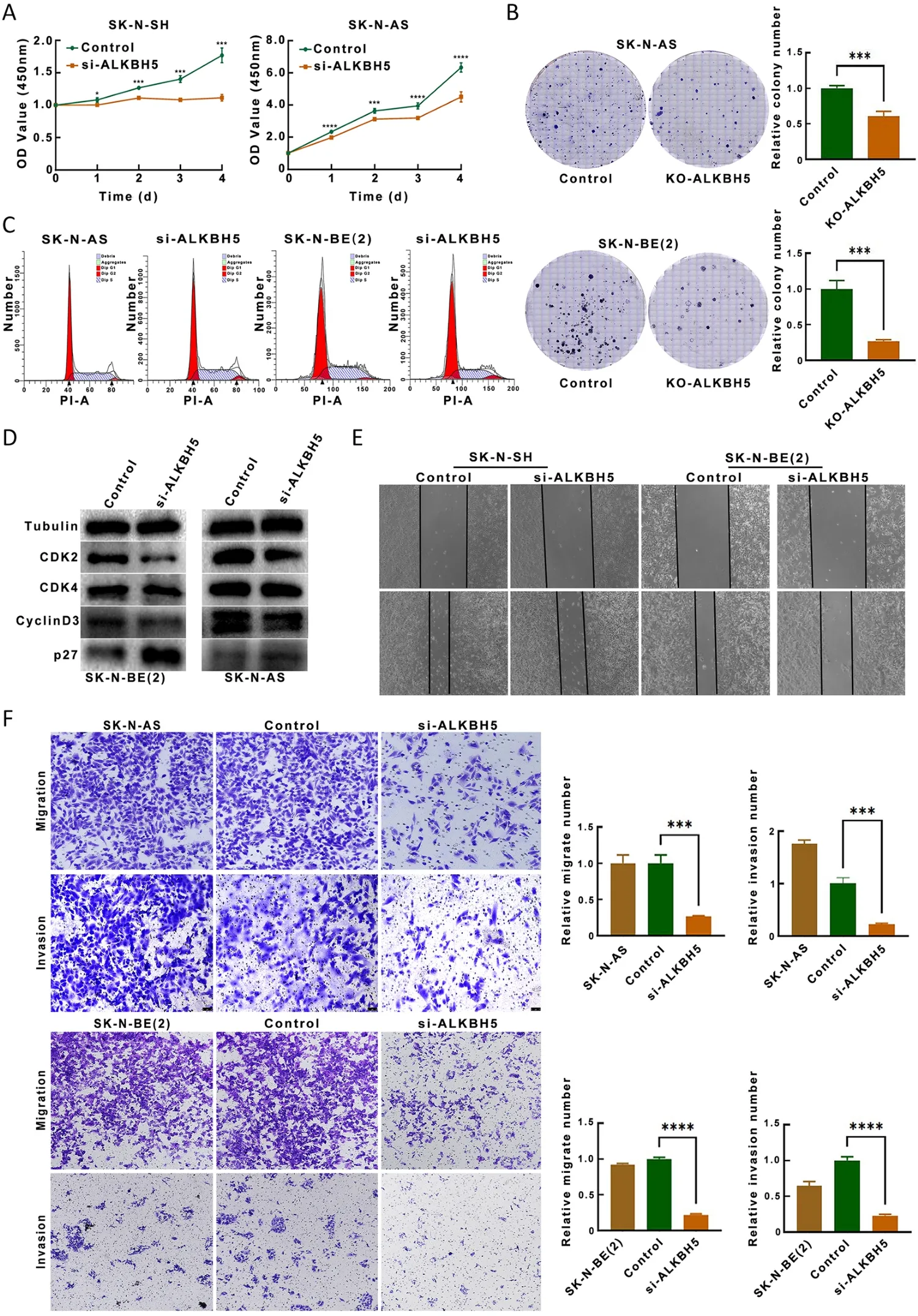
Figure 3 ALKBH5 promotes proliferation,migration and invasion of neuroblastoma cells.(A) CCK-8 assay showed that two neuroblastoma cell lines (si-ALKBH5) that interfered with ALKBH5 expression had significantly reduced rates of viable cell proliferation;(B) Colony formation assay (plate colony formation assay) was performed to evaluate the long-term proliferation of neuroblastoma cells with knockdown of ALKBH5 expression;(C) Flow cytometric analysis revealed an increased proportion of cells in G2/M phase after the downregulation of ALKBH5 expression;(D) Inhibition of ALKBH5 increased the expression of CDK2 and CDK4 and decreased the expression of p27;(E) Two cell lines that inhibited ALKBH expression [SK-N-SH,SK-N-BE(2)] had reduced wound closure ability compared to controls (Magnification 10×);(F) Downregulation of ALKBH5 inhibits neuroblastoma cell migration and invasion(Magnification 10×).CCK-8,cell counting kit-8.*,P<0.05;**,P<0.01;***,P<0.001;****,P<0.0001.
rs8400 A allele increases ALKBH5 level and is an oncogenic variant
To further investigate whether the functional polymorphism rs8400 affects ALKBH5 expression,we performed cis-eQTL analysis using newly published data from Genotype-Tissue Expression (GTEx).The results showed that individuals carrying the rs8400 A allele had significantly higher ALKBH5 mRNA levels in multiple tissue samples compared with individuals carrying the rs8400 G allele (Supplementary Figure S4A).The same results were found in adrenal tissues,which are often the origin of neuroblastoma (Figure4A).The immunohistochemstry results also showed that tissue samples from individuals carrying the rs8400 A allele had higher ALKBH5 expression (Figure 4B).We then transfected the ALKBH5 3’-UTR fragment with the G or A allele into SK-N-SH and SK-N-BE(2) cells and showed that the rs8400 A allele significantly increased luciferase activity compared with the rs8400 G allele.It was proven that the rs8400 A allele resulted in enhanced expression of ALKBH5 (Figure 4C,D).To further characterize functional variants of rs8400,we first generated cell lines overexpressing rs8400 A allele and cell lines overexpressing rs8400 G allele.The expression of ALKBH5 in cell lines overexpressing rs8400 A allele and cell lines overexpressing rs8400 G allele was measured by RT-qPCR and Western blotting.The results showed that the mRNA and protein expression levels of ALKBH5 in the rs8400 A alleleoverexpressing cell lines was higher than that in the rs8400 G allele-overexpressing cell lines or the control vectortransfected cell lines (Supplementary Figure S4B,C).Overexpression of rs8400 A allele significantly promoted the proliferation of neuroblastoma cells compared with overexpression of rs8400 G allele or expression of the control vector (Figure 4E).Flow cytometry revealed a higher proportion of cells in G2/M phase in neuroblastoma cells with rs8400 A allele relative to rs8400 G allele (Figure 4F).The colony formation assay further verified that rs8400 A allele-overexpressing cells formed the most colonies (Figure 4G).The wound closure ability of cells with stable overexpression of rs8400 A allele was increased compared with that of cells with stable overexpression of rs8400 G allele,cells transfected with the control vector or cells in the untreated blank group (Figure 4H).Compared with stable overexpression of rs8400 G allele or transfection of the control vector,stable overexpression of rs8400 A allele also significantly enhanced the migration and invasion abilities of SK-N-AS cells (Figure 4I).In conclusion,these findings suggest that the rs8400 G>A SNP can lead to increased expression of ALKBH5 and thus have tumorigenic effects.
rs8400 G>A SNP,located in miR-186 binding site,is a functional regulatory site
In order to explore the reason that the G>A base substitution in rs8400 leads to a change in ALKBH5 expression,we first used the miRNA target prediction software miRanda (Version 3.3a;Memorial Sloan Kettering,USA) to find that the ALKBH5 3’-UTR contains a hypothetical binding site for miR-186-3p (47)(Figure 5A,B).Through the RNAfold and RNAcofold web servers,we found that the rs8400 G>A substitution changes the local folding structure and free energy of ALKBH5.This means that the G>A substitution in rs8400 may destroy the binding site of miR-186-3p,resulting in a decrease in the ability of miR-186-3p to bind to the 3’-UTR of ALKBH5 (Figure 5C,D).Then,we further verified the effect of miR-186-3p on the expression of ALKBH5in vitro.MiR-186-3p mimics or inhibitors were transfected into SK-N-BE(2) and SK-N-AS cells,and the expression of the ALKBH5 protein was evaluated.The Western blotting results showed that inhibition of miR-186-3p promoted the expression of ALKBH5,while an increase in the miR-186-3p level inhibited the expression of ALKBH5 (Figure 5E).This indicates that miR-186-3p inhibits ALKBH expression.
We further verified the difference in the miRNA binding ability between different genotypes by a luciferase reporter assay.The reporter plasmid containing the G or A allele was transiently transfected with the miR-186 mimic or NC(or inhibitor or NC) into SK-N-AS cells,and the relative activity of firefly luciferase compared with Renilla luciferase was determined.The results showed that the luciferase activity in cells transfected with the miR-186-3p mimics was decreased,and the decrease was more obvious in cells with the G allele than in cells with the A allele(Figure 5F).This proves miR-186-3p has a weaker ability to bind toALKBH5with rs8400 A allele than toALKBH5with rs8400 G allele.This shows that the rs8400 G>A substitution reduces the inhibitory effect of miR-186-3p on ALKBH expression and increases the expression of ALKBH5.
SPP1 was identified as downstream target of ALKBH5
In order to comprehensively explore the methylation mode of ALKBH5,we knocked out ALKBH5 in the SK-N-AS cell line and performed MeRIP-seq analysis on the ALKBH5-knockdown group and the control group.Data analysis showed that the m6A consensus motif (GGAC) was highly abundant in SK-N-AS cells (Figure 6A) and enriched near stop codons (Supplementary Figure S5A,B).Compared with that in the control group,the m6A modification landscape in the ALKBH5 knockdown group showed slight changes in the distribution across different elements of the gene (Supplementary Figure S5C,D).Analysis of m6A peaks showed that there were 10,945 and 12,508 peaks in ALKBH5-knockdown and control cells,respectively,and that 1,500 of them overlapped (Figure 6B).In ALKBH5-knockdown and control cells,there were 7,092 and 7,617 genes associated with peaks,respectively,and 1,931 and 2,465 uniquely associated genes,respectively (Figure 6C).We focused on the genes whose methylation patterns and expression levels were regulated by ALKBH5.Therefore,we further intersected the differential peak-associated genes identified by m6A-seq with the differentially expressed genes identified by RNA-seq to obtain five genes related to the ALKBH5 effect,namely,SPP1,COL15A1,ADAMTS12,SYT14,andRP11-440G9.1(Figure 6D).When ALKBH5 was knocked down,the m6A peaks of these target genes were significantly increased (Figure 6E-I).Moreover,these relationships were initially validated by RT-qPCR in ALKBH5 knockdown cells.We found that SPP1 was negatively regulated by ALKBH5 in the three cell types,with significant and consistent differences (Figure 6J).This was further verified by Western blotting,which showed that the expression of SPP1 increased when ALKBH5 was knockdowned (Figure 6K).In conclusion,SPP1 may be the direct downstream target of ALKBH5 in neuroblastoma.

Figure 4 rs8400 A allele increases ALKBH5 levels and promotes neuroblastoma cancer progression.(A) In adrenal tissues,individuals with rs8400 AA genotype had statistically higher levels of ALKBH5 mRNA than those with rs8400 GG or GA genotype;(B)Immunohistochemistry results show that tissue samples with the rs8400 A allele have higher ALKBH5 expression relative to rs8400 G allele(Magnification 10×);(C,D) Luciferase activity was significantly reduced in SK-N-SH (C),SK-N-BE(2) (D) cells with the rs8400 G allele construct compared with the construct with the rs8400 A allele;(E) Overexpression of rs8400 A significantly promoted neuroblastoma cell proliferation compared to overexpression of rs8400 G or the control vector;(F) Flow cytometry showed a higher proportion of cells in G2/M phase in neuroblastoma cells with rs8400 A allele relative to rs8400 G allele;(G) Colony formation experiments further demonstrated that rs8400 A-overexpressing cells had the most colonies;(H) Wound closure ability of stably overexpressed rs8400 A allele was increased compared with stably overexpressed rs8400 G allele or control vector or blank group without any treatment (Magnification 10×);(I) Stable overexpression of rs8400 A also significantly promoted the migration and invasion abilities of SK-N-AS cells compared with stably overexpressed rs8400G or control vector or blank group (Magnification 10×).*,P<0.05;**,P<0.01;***,P<0.001;****,P<0.0001.

Figure 5 SNP rs8400 A/G causes different binding abilities of miR-186-3p to ALKBH5 3’UTR.(A) We found the binding sites for ALKBH5 3’-UTR and miR-186-3p by prediction software (Bartel,2009);(B) When a miRNA binds to a target,the target first transitions to an intermediate state by unraveling the seed target nucleotides,and then forms a complex upon binding to the miRNA;(C,D) Using RNAfold and RNAcofold WebSever,we found that the rs8400 A allele (D) binds miR-186-3p to a reduced capacity compared with the rs8400 G allele (C);(E) Western blotting results showed that inhibiting miR-186-3p could promote expression of ALKBH5,and increasing miR-186-3p could inhibit the expression of ALKBH5;(F) Luciferase analysis showed that miR-186-3p could bind to the 3’-UTR of ALKBH5 gene,and rs8400 A allele reduced the binding ability of miR-186 to the 3’-UTR.*,P<0.05;**,P<0.01;***,P<0.001.
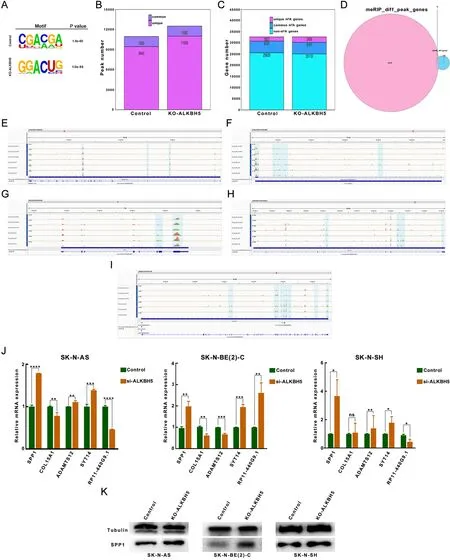
Figure 6 SPP1 is identified as a downstream target of ALKBH5.(A) m6A consensus motif of GGAC is highly concentrated in SK-N-AS cells;(B) The study of m6A peaks showed that there were 12,508 and 10,945 in control cells and ALKBH5 knockdown,respectively,of which 1,500 were common;(C) There were 7,092 and 7,617 genes associated with the peak,with 1,931 and 2,465 uniquely associated genes,respectively;(D) Among the shared genes of differential peak-related genes of m6A-seq and differential genes of RNA-seq,there are 5 genes related to the ALKBH5 effect,namely SPP1,COL15A1,ADAMTS12,SYT14 and RP11-440G9.1;(E-I) When ALKBH5 was knocked down,m6A modification peak of the target gene was significantly increased;(J,K) RT-qPCR (J) and Western blotting (K) analysis demonstrated that SPP1 is inversely regulated by ALKBH5 in three cell types with significant differences.RT-qPCR,reverse transcription-quantitative real-time PCR.*,P <0.05;**,P<0.01;***,P<0.001;****,P<0.0001.
ALKBH5 downregulates SPP1 mRNA expression by m6A modification
We identified the m6A site in theSPP1gene with significantly differential modification through MeRIP-seq.MeRIP-qPCR for this significant site confirmed the demethylation of SPP1 mRNA mediated by ALKBH5.Overexpression of ALKBH5 significantly reduced the m6A level of PER1 in the exon,while knockdown of ALKBH5 resulted in increased m6A modification of PER1(Figure 7A).To further verify the regulatory effect of m6A modification on SPP1 expression,we substituted the adenosine (A) in the significantly differential m6A site in theSPP1gene with cytosine (C) and constructed a mutant SPP1 fragment resistant to m6A modification.The luciferase activity of the wild-type (WT) SPP1 fragment was much lower than that of the mutant (mut) fragment.In addition,luciferase activity was decreased in ALKBH5 overexpressing cells and increased in cells with knockdown of ALKBH5 expression relative to controls (Figure 7B).Therefore,the mRNA level of SPP1 is regulated by m6A modification mediated by ALKBH5.
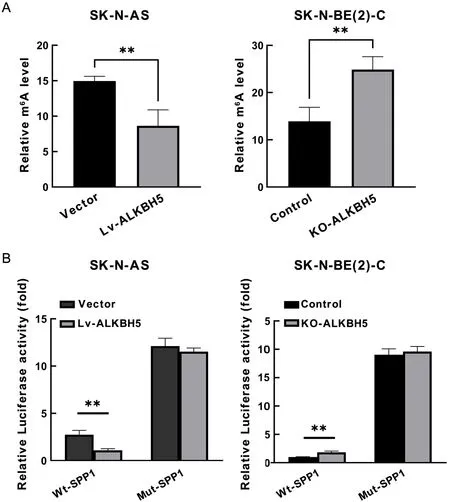
Figure 7 ALKBH5 decreases SPP1 mRNA levels in an m6A manner.(A) MeRIP-qPCR detected m6A modification of SPP1.Compared with the control group,the overexpression of ALKBH5 inhibited m6A abundance of SPP1 mRNA,and increased m6A abundance of SPP1 mRNA after ALKBH5 was knocked out;(B) Detected the luciferase activity of SPP1 wild type and mutant type in overexpressed or knockdown ALKBH5 cells,respectively.MeRIP-qPCR,methylated RNA immunoprecipitation quantitative polymerase chain reaction;**,P<0.01.
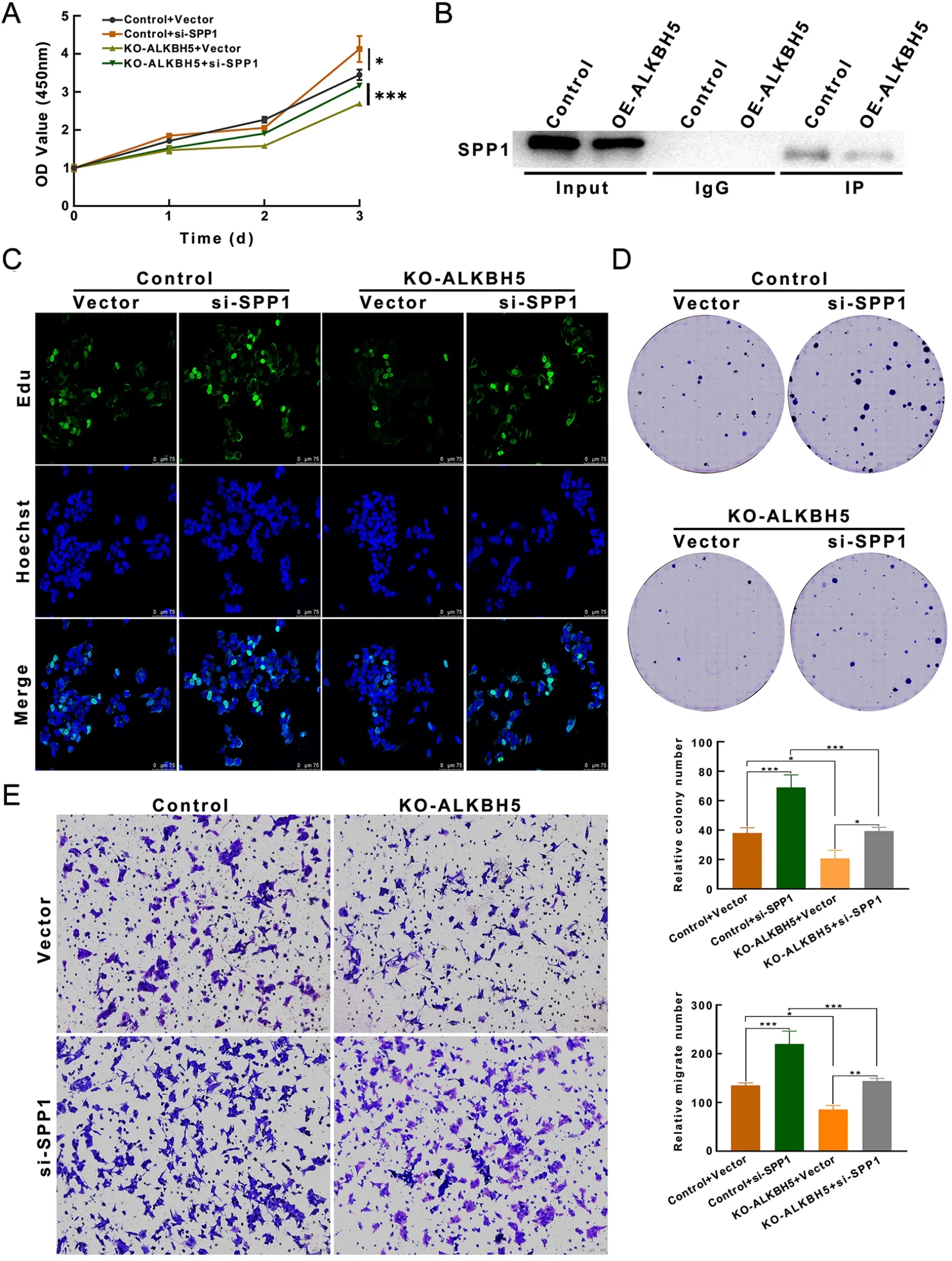
Figure 8 Oncogenic effects of ALKBH5 were reversed by overexpression of SPP1.(A) CCK-8 showed that SPP1 expression inhibition promoted neuroblastoma cell progression and partially reversed ALKBH5 knockdown-induced reduction in neuroblastoma proliferation;(B) Co-precipitation results in Co-IP experiments indicated that ALKBH5 interacts with SPP1;(C,D) EdU (C) and colony (D) assays showed that inhibition of SPP1 expression promoted neuroblastoma cell proliferation and could partially reverse the decreased proliferation of neuroblastoma cells caused by ALKBH5 knockdown;(E) Knockdown of SPP1 also significantly abolished the reduced invasiveness caused by ALKBH5 deletion (Magnification 20×).CCK-8,cell counting kit-8;Co-IP,coimmunoprecipitation.*,P<0.05;**,P<0.01;***,P<0.001.
Oncogenic effect of ALKBH5 is reversed by overexpression of SPP1
To confirm that the observed phenotype is mediated by dysregulation of the ALKBH5-SPP1 axis,we performed several functional rescue assays.CCK-8,EdU,and colony formation assays showed that SPP1 silencing promoted the malignant progression of neuroblastoma cells and partially reversed the reduction in neuroblastoma cell proliferation induced by ALKBH5 knockdown (Figure 8A,C,D).Knockdown of SPP1 also significantly abolished the attenuation of motility and invasion induced by ALKBH5 loss (Figure 8E).In addition,the coprecipitation of ALKBH5 and SPP1 in coimmunoprecipitation (Co-IP)experiments fully demonstrated their interaction(Figure 8B).In conclusion,abnormal SPP1 expression may be responsible for ALKBH5-mediated proliferation or migration of neuroblastoma cells.
ALKBH5 inhibition potentiates effect of chemotherapeutic drugs on neuroblastoma cells
To further investigate the clinical significance of ALKBH5 in neuroblastoma treatment,we explored whether ALKBH5 inhibition affects the therapeutic effect of chemotherapeutic drugs on neuroblastoma cells.We hypothesized that inhibition of ALKBH5 expression would affect the sensitivity of neuroblastoma cells to chemotherapeutic drugs.We studied six chemotherapeutic drugs (cyclophosphamide,carboplatin,etoposide,vincristine,doxorubicin and cisplatin) commonly used in patients with neuroblastoma (5,47,48).We performedin vitrodrug susceptibility experiments for verification.We added different concentrations of the chemotherapeutic drugs to the experimental group of neuroblastoma cells with knockdown of ALKBH5 expression and the untreated control group of neuroblastoma cells,respectively.Cell viability was measured by a CCK-8 assay after 72 h of incubation.It was observed that the number of viable cells decreased significantly in all neuroblastoma cell groups with increasing concentrations of the chemotherapeutic drugs.Compared with the control groups,the number of viable cells in the ALKBH5 knockout groups was significantly reduced after incubation with the same low concentration of two chemotherapeutic drugs (carboplatin and etoposide) (Figure 9).That is,the sensitivity of ALKBH5 knockout neuroblastoma cells to carboplatin and etoposide was significantly increased.These results suggest that ALKBH5 inhibition can enhance the anticancer effects of some chemotherapeutic drugs.
Discussion
Neuroblastoma is the most common tumor in infants and young children,accounting for about 15% of all childhood cancers (49).In recent years,with the in-depth study of neuroblastoma,the prognosis of patients has improved to some extent,and the five-year survival rate has increased from 52% to 74% (1).However,this has been attributed to increased cure rates for patients with benign neuroblastoma.However,there has been little effect on the cure rate of patients with high-risk neuroblastoma.Therefore,understanding the molecular features and genetic variants involved in the pathogenesis of neuroblastoma is essential for the development of tools for early diagnosis and precision treatment.In our study,we identified anALKBH5SNP,rs8400,associated with neuroblastoma risk.It is in the microRNA-binding region on chromosome 17q11.2,and the risk of rs8400 A allele corresponds to higher ALKBH5 gene expression.We proved that the rs8400 G>A SNP resulted in decreased binding of a microRNA to ALKBH5 and promoted its expression,leading to accumulation of ALKBH5.ALKBH5 upregulation resulted in enhancement of the proliferation,migration and invasion of cancer cells.SPP1 was selected as the downstream target gene by RNA-seq combined with MeRIP-seq,and downregulation of SPP1 was found to restore the tumor-suppressive effect of ALKBH5 downregulation.In addition,we further investigated the therapeutic effect of ALKBH5 knockdown combined with common chemotherapeutic drugs on neuroblastomain vitro,and the results showed that the effects of carboplatin and etoposide were enhanced in ALKBH5 knockdown cells.Our workflow is shown inSupplementary Figure S6.This study has greatly expanded the understanding of neuroblastoma mechanisms by revealing that genetic variants located in microRNA binding sites contribute to neuroblastoma-associated phenotypes by affecting the expression of their host genes.ALKBH5gene may also be a potential new therapeutic target for neuroblastoma(Figure 10).
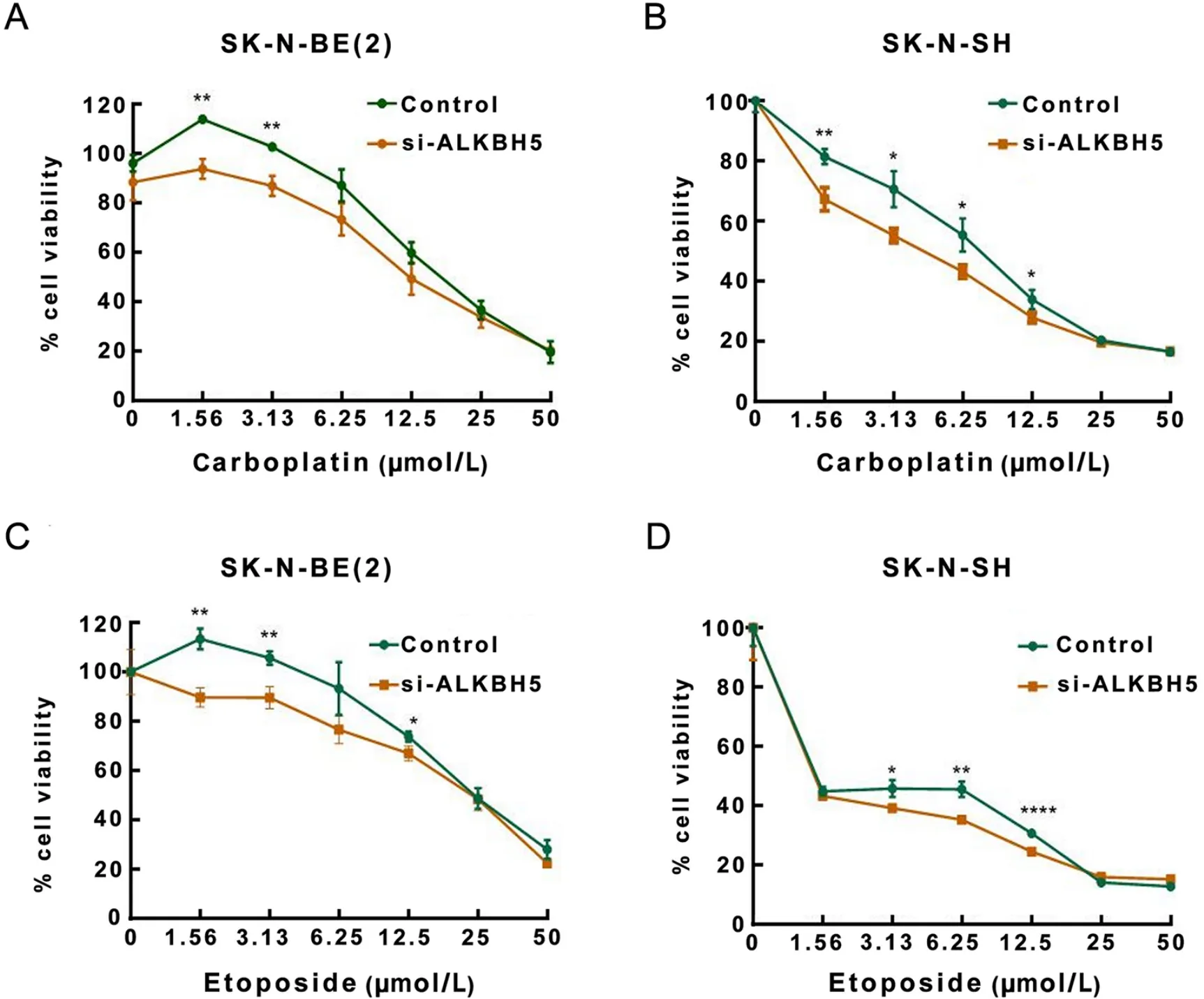
Figure 9 ALKBH5 inhibition promotes therapeutic effects of chemotherapeutic drugs on neuroblastoma cells.(A,B) SK-N-BE(2) (A) and SK-N-SH (B) neuroblastoma cell lines with ALKBH5 knockdown were significantly more sensitive to low doses of carboplatin compared to controls;(C,D) SK-N-BE(2) (C) and SK-N-SH (D) cell lines with ALKBH5 knockdown were significantly more sensitive to low doses of etoposide compared to control.*,P<0.05;**,P<0.01;****,P<0.0001.
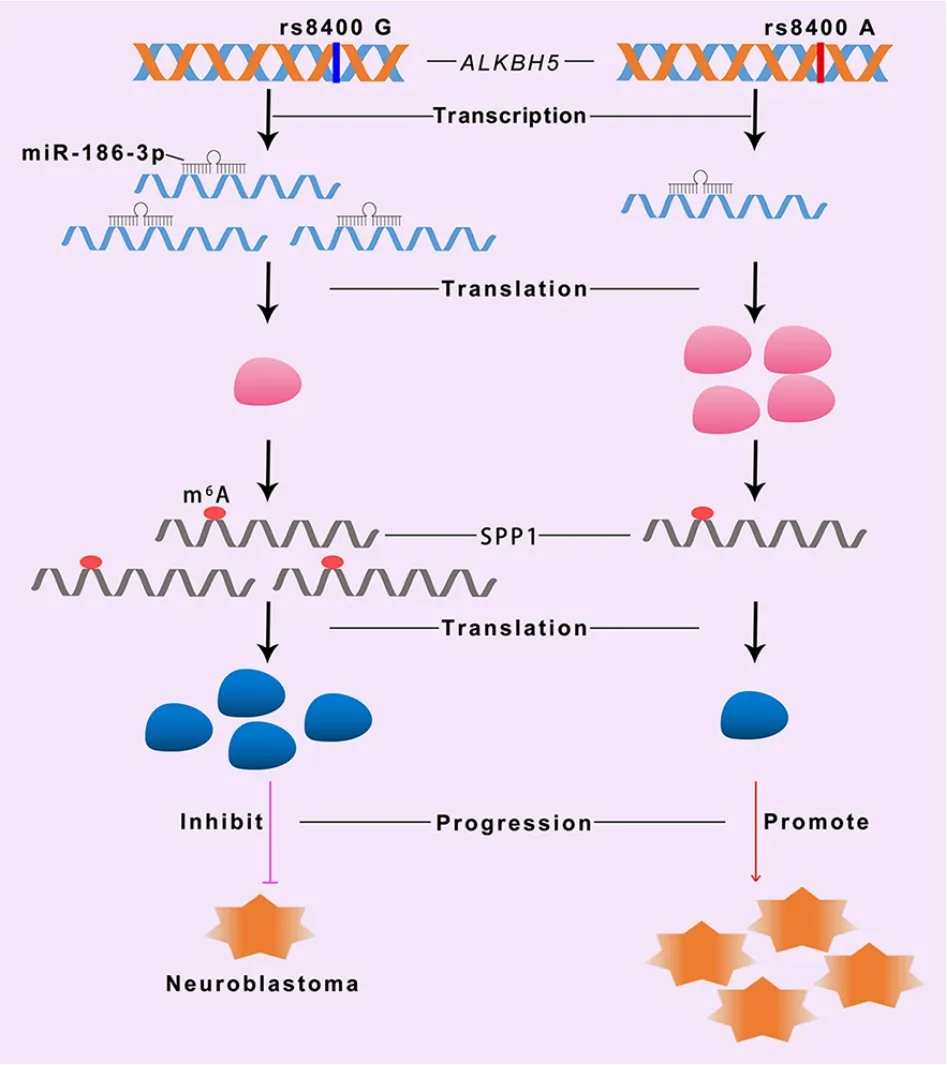
Figure 10 rs8400 G>A of ALKBH5 gene affects binding of miR-186-3p to the ALKBH5 binding site,making miR-186-3p unable to inhibit expression of ALKBH5,resulting in upregulation of its mRNA and protein levels.Upregulation of ALKBH5 further inhibited downregulation of tumor suppressor gene SPP1 and promoted progression of neuroblastoma.
m6A modification regulates a variety of cellular processes,including RNA stability,splicing,translation and biogenesis (50,51).Accumulating evidence suggests that aberrant m6A modification is closely related to the pathogenesis of many cancers (52-54).Although the importance of m6A modification-related genes in cancer is highly appreciated,the study of SNPs in m6A modificationrelated genes is a newly emerging field.However,the information regarding the SNPs inALKBH5gene is still limited.Only recently has it been recognized that SNPs in theALKBH5gene contribute to genetic susceptibility to cancer.In 2019,Menget al.published the first case-control study on SNPs in m6A modification-related genes and colorectal cancer risk.They comprehensively genotyped 240 SNPs in 20 m6A modification-related genes in the Chinese population.However,all the analyzedALKBH5SNPs (rs2124370,rs8400,rs9899249,rs9913266 and rs2925137) could not predispose individuals to colorectal cancer (55).We previously genotyped two SNPs(rs1378602 and rs8400) in theALKBH5gene to determine whether they are associated with Wilms tumor susceptibility in a large multi-center case-control study.No SNP contributed to Wilms tumor susceptibility.Stratification analysis did reveal some significant relationships between these SNPs and Wilms tumor risk in certain subgroups,indicating a weak influence ofALKBH5gene SNPs on susceptibility to Wilms tumor (56).In this study,we identified rs8400 A allele as being significantly associated with the risk of neuroblastoma and as the firstALKBH5locus robustly associated with the risk of neuroblastoma in individuals of Chinese ancestry,and we further explored the biological significance and molecular mechanism of the SNP rs8400.
miRNAs have crucial effects on mRNA deadenylation,mRNA decay,and mRNA translation inhibition by binding to the 3’-UTRs of most protein-coding genes (57).Previous studies have demonstrated that miRNAs are involved in tumorigenesis and tumor development by affecting tumor cell proliferation,apoptosis,migration,invasion and angiogenesis (58).Nevianiet al.previously demonstrated that natural killer cell-derived exosomal miR-186 is downregulated in patients with high-risk neuroblastoma and that its low expression is a poor prognostic factor.They further demonstrated that miR-186 could inhibit the growth of neuroblastoma by targeting MYCN and AURKA (59).miR-186 also exerts inhibitory effects on neuroblastoma by targeting the 3’-UTR of Eg5(60) and MAP3K2 (61).As predicted by miRBase and TargetScan,miR-186-3p was identified as a potential miRNA targeting rs8400 ofALKBH5.miR-186-3p negatively regulated the expression of theALKBH5gene,but this regulatory effect disappeared when the G nucleotide in rs8400 was mutated to A.We describe a possible mechanism of abnormal oncogene expression caused by genetic variation.
ALKHB5 is a key m6A demethylase that can remove m6A(30).A decreased ALKBH5 level was detected in hepatocellular carcinoma (HCC) and associated with worse survival in HCC patients.ALKBH5 functions as a tumor suppressor in HCC cells by inhibiting LY6/PLAUR Domain Containing 1 (LYPD1) via an m6A-dependent mechanism (62).ALKBH5 was found to impair the progression of non-small cell lung cancer (NSCLC) by reducing YAP expression and activity (63).ALKBH5 also functions as an oncogene in other specific types of cancer.ALKBH5 can upregulateFOXM1gene expression and promote the self-renewal and proliferation of glioblastoma stem-like cells (GSCs) (33).Increased ALKBH5 expression serves as a marker for poor prognosis in acute myeloid leukaemia (AML) patients.Mechanistically,ALKBH5 plays a tumorigenic role in AML by posttranscriptional regulation of critical targets such as TACC3 (64).The dual role of ALKBH5,which has different effects in different tumors,depends on tissue specificity and different downstream molecular mechanisms.In our study,we found that theALKBH5gene was frequently amplified and upregulated in high-grade neuroblastoma and that ALKBH5 promoted the malignant progression of neuroblastoma by inhibiting the expression of SPP1,suggesting thatALKBH5is an important oncogene in neuroblastoma.
Although the downstream target genes ofALKBH5in neuroblastoma have been explored,the detailed mechanism by which SPP1 inhibits the occurrence and development of neuroblastoma remains unknown.We have provided a large amount of evidence on the function of ALKBH5 in tumorigenesis,but our work still has some limitations.For example,ALKBH5 is a methyltransferase;whether it acts through m6A modification,and if so,which reader proteins are involved,are unknown and need further study.
To investigate the clinical significance of ALKBH5 in neuroblastoma,we conducted a preliminary drug sensitivity test to study the therapeutic effects of six common chemotherapeutic drugs on neuroblastoma.Our study found that carboplatin and etoposide combined with ALKBH5 inhibition had a good therapeutic effect on neuroblastoma cells,suggesting that ALKBH5 could be used as a clinical therapeutic target for neuroblastoma.The clinical significance of ALKBH5 needs further study.
Conclusions
Our study demonstrated that a SNP inALKBH5gene(rs8400 G>A) was significantly associated with the risk of neuroblastoma.The rs8400 G>A SNP causes an imbalance in the negative regulatory effect of miR-186-3p and further causes abnormal expression of ALKBH5.Dysregulation of the ALKBH5-SPP1 axis can promote the proliferation,migration,and invasion of neuroblastoma cells.The role of the miR-186-ALKBH5-SPP1 axis in neuroblastoma was revealed.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (No.82002635,82002636 and 82173593);Guangzhou Science and Technology Project (No.202102021227 and 2021 02020421);the Science Technology and Innovation Commission of Shenzhen (No.JCYJ20220531093213030);and Guangzhou Municipal Basic Research Program Joint Funding of City and Hospitals (No.202201020622).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
 Chinese Journal of Cancer Research2023年2期
Chinese Journal of Cancer Research2023年2期
- Chinese Journal of Cancer Research的其它文章
- Stomach cancer burden in China: Epidemiology and prevention
- When immunotherapy meets liver transplantation for hepatocellular carcinoma: A bumpy but promising road
- Transforming cancer cells for long-term living with cancer:An inspiring new approach
- Integrated strategies for chemotherapy cycles in nasopharyngeal carcinoma patients: Real-world data from two epidemic centers guiding decision-making
- Exploration and optimization of surgical techniques for laparoscopic transhiatal lower mediastinal lymph node dissection for adenocarcinoma of esophagogastric junction: A prospective IDEAL 2a study with qualitative design
- Aqueous-soluble components of sporoderm-removed Ganoderma lucidum spore powder promote ferroptosis in oral squamous cell carcinoma
