Lysobacter hymeniacidonis sp. nov., Isolated from a Crude Oil-Contaminated Marine Sponge
XIN Yanjuan, QU Junge, XU Junyi, WU Peichun, CAO Xupeng, and XUE Song, *
sp. nov., Isolated from a Crude Oil-Contaminated Marine Sponge
XIN Yanjuan1), QU Junge2), XU Junyi3), WU Peichun1), CAO Xupeng1), and XUE Song1), *
1) Marine Bioproducts Engineering Group, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, P.R. China 2)Department of Biology and Pharmacy, Zhejiang Pharmaceutical College, Ningbo315100, P.R. China 3)Liaoning Entry-Exit Inspection and Quarantine Bureau, Dalian 116001,P.R. China
An aerobic, Gram-negative bacterium, strain 2-5T, was isolated from a crude oil-contaminated marine sponge collected near Dalian Bay, China, and subjected to a polyphasic taxonomic investigation. Cells of strain 2-5Twere non-spore forming, non-motile, rods 0.2–0.3µm wide and 1.1–1.2µm long. Strain 2-5Tgrew well on nutrient agar, TSA, R2A agar and LB agar. Colonies of strain 2-5Ton LB agar were circular, smooth with entire margins, non-transparent and pale yellow after 3d of incubation at 30℃. Growth of strain 2-5Toccurred in LN medium with 0–6% NaCl; no growth occurred in the presence of 8.0% NaCl. Strain 2-5Tgrew at 15–42℃ and at pH 6.0–8.0. Comparative 16S rRNA gene sequence analysis showed that strain 2-5Tclustered with the species of the genus. Its closet neighbors were the type strains ofKCTC 12205T(97% similarity),ZS79T(96%), andAPB-9T(96%). The value for DNA-DNA relatedness between strain 2-5TandKCTC 12205Twas 23%. Branched fatty acids iso-C16: 0, iso-C15: 0, iso-C11: 03-OH, iso-C17: 1ω9and iso-C11: 0were found to be predominant. The major polar lipids were diphosphatidylglycerol, phosphatidylethanolamine and phosphatidylglycerol. Strain 2-5Thad a DNA G+C content of 63.8mol%. On the basis of the phenotypic, chemotaxonomic, DNA-DNA hybridization and phylogenetic data, strain 2-5Trepresents a novel species of the genus, for which the namesp. nov. is proposed. The type strain is 2-5T(=CGMCC 1.12190T= JCM 18137T).
sp. nov.; taxonomy; phylogenetic analysis
1 Introduction
The genuswas first described by Christensen and Cook (1978), and the description was emended by Park. (2008). The genus, grouped in the family, is classified in the class(Christensen and Cook, 1978) based on non-fruiting bodies, lack of flagella, gliding nature and high genomic DNA G+C content (typically ranging between 65.4 and 70.1mol%) (Aslam., 2009). At the time of writing, the genuscomprises 27 species with validly published names (http: //www. bacterio. cict. fr/). Recently, three novel species of the genus,ZS79Tfrom iron-mined soil (Luo., 2012),107-E2Tfrom an Antarctic freshwater lake (Fukuda., 2013) andCJ29Tfrom ginsengsoil (Choi., 2014) have been described. Members of the genus were of great potential for the de-velopment of biocontrol agents against plant fungal pathogens (Islam., 2005; Park., 2008) and antibiotic bioactivities against human pathogens (Ahmed., 2003; Hashizume., 2004). Here we report the characterization of a novel marine bacterium, strain 2-5T, of the genus,which was isolated from a crude oil-contaminated marine sponge.
2 Materials and Methods
2.1 Bacterial Strains
Strain 2-5Twas isolated from a crude oil-contaminated sponge specimen (), collected at the inter-tidal beach of Dalian, on the Chinese Yellow Sea, located in northern China (38˚52´N 121˚41´E). Freshly collected sponge specimens were rinsed five times in sterile seawater to remove unassociated bacteria and then thoroughly homogenized in a sterile mortar. A 10-fold dilution series of sponge homogenate was made and plated on 2216E plates (Difco, USA) in triplicate. After incubating the plates at 28℃ for 7d, an isolate, designated 2-5T, was picked and sub-cultured on 2216E plates, repeating the above steps until a pure culture was obtained. The novel strain was deposited into the CGM CC (China General Microbiological Culture Collection Center) as CGMCC 1.12190Tand the JCM (Japan Collection of Microorganisms) as JCM 18137T. The reference strain used for the DNA-DNA homology tests wasKCTC 12205T, obtained from the KCTC (Korean Collection for Type Cultures).
2.2 Morphology and Physiological Characteristics
To investigate the morphological and physiological characteristics, cell grown on R2A agar plates at 30℃ for 2d. Cell morphology and motility were examined using light microscopy (Olympus; ×1000) and transmission electron microscopy (H-7650; Hitachi, Japan) using cells from an exponentially growing culture. The strain Gram straining reaction was carried out according to the classical procedure described by Doetsch (1981). Gliding motility was determined as described by Bowman (2000). The physiological properties of strain 2-5Twere determined by the CGMCC using established procedures described by Gordon. (1974) and Yokota. (1993). Catalase activity, oxidase activity, enzyme activity and acid production from different carbohydrates were determined by the CGMCC with Biolog GN2, API 20E, API 20NE kits according to the manufacturer’s instructions. The assimilation of single carbon substrates was determined by the CGMCC with Biolog GN2 and API 20NE strips cultured at 28℃ for 24 h. Hydrolysis of casein and chitin was determined using previously described test (Smibert and Krieg, 1994; Brown, 2007). Growth at 4, 10, 15, 25, 30, 37, 42 and 45℃ and at pH 4.0–10.0 (at intervals of 1.0pH unit) was assessed after 5d of incubation on 2216E agar. Growth on nutrient agar, trypticase soy agar (TSA; Difco), R2A agar (Difco, USA) and LB agar (Difco, USA) was also evaluated at 28℃. Salt tolerance was tested in LN medium (LB without NaCl) supplemented with 0–10% NaCl after 10d of incubation.
2.3 G+C Content and Analysis of Cellular Fatty Acids
Cell grown on R2A agar plates at 30℃ for 2d were used for the analysis of cellular fatty acid and polar lipid. The fatty acids were extracted, methylated and analysed using the standard Sherlock MIDI (Microbial Identification) system (Sasser, 1990; Kämpfer and Kroppenstedt, 1996). Polar lipids were extracted and analyzed as described by Tindall (1990). A 6.75mL portion of chloroform/methanol/0.3% aqueous NaCl (1: 2: 0.8) was added to 100 mg freeze-dried cell material. The preparation was stirred overnight and cell debris was pelleted by centrifugation. Polar lipids were recovered into the chloroform phase by adjusting the chloroform/methanol/0.3% aqueous NaCl mixture to a ratio of 1: 1: 0.9 and then dried under nitrogen. The dried polar lipids were resuspended in chloroform/methanol (2:1) and separated by two-dimensional TLC.
The G+C content of the genomic DNA was determined by the CGMCC by thermal denaturation (Mandel and Marmur, 1968), for whichK-12 (CGMCC 1.365) was used as a standard.
2.4 Phylogenetic Analysis and DNA-DNA Hybridization
Genomic DNA from strain 2-5Twas extracted and purified according to standard procedures (Sambrook and Russell, 2001). The 16S rRNA gene cloned into pMD- 18T (Takara, Japan) was sequenced using an automated sequencer (Applied Biosystems model 3730). The 16S rRNA gene sequence of strain 2-5Twas compared with known sequences found in the GenBank database using the BLAST program (http://blast.ncbi.nlm.nih.gov/ Blast.cgi). Phylogenetic analysis was performed with MEGA5 program (Tamura., 2011) after multiple alignments of the data via CLUSTAL_X program (Thom- pson., 1997). A distance matrix method (distance options according to the Kimura two-parameter model), including clustering using the neighbor-joining and maximum-likelihood (Kluge and Farris, 1969) method. In each case, bootstrap values were calculated based on 1000 replications (Felsenstein, 1985). The taxonomic relationship between strain 2-5Tand its phylogenetic relative was further examined using DNA-DNA hybridization. DNA–DNA hybridization values between 2-5TandKCTC 12205Twas performed fluorometrically, according to the method developed by Ezaki. (1989) using photobiotin-labelled DNA probes and micro dilution wells. Hybridization was conducted in five replications for each sample. The highest and lowest values obtained for each sample were excluded, and the remaining three values were utilized in the calculation of hybridization values.
3 Results and Discussion
3.1 Morphological and Physiological Characteristics
Cells of strain 2-5Twere Gram-negative, non-spore forming, non-motile (but showing gliding activity), aerobic rods 0.2–0.3µm wide and 1.1–1.2µm long (Supplementary Fig.S1-2). Strain 2-5Tgrew well on nutrient agar, TSA, R2A agar and LB agar. Colonies of strain 2-5Ton LB agar were circular, smooth with entire margins, non- transparent and pale yellow after 3d of incubation at 30℃. Growth of strain 2-5Toccurred in LN medium with 0–6% NaCl; no growth occurred in the presence of 8.0% NaCl. Strain 2-5Tgrew at 15–42℃ (optimal 28–30℃) and at pH 6.0–8.0 (optimum pH 7.0). In the Biolog GN2, API 20E and API 20NE kits, strain 2-5Tassimilated glycogen,-acetyl-D-galactosamine,-acetylglucosamine, ribitol, L-arabinose, D-arabitol, D-cellobiose,D-fructose, alpha-D-glucose, M-inositol,D-galactonic acid lactone,D-galacturonic acid, D, L-lactic acid, malonic acid, propionic acid, quinic acid, D-saccharic acid, succinamic acid, glucuronamide, L-ala- ninamide, L-alanine, L-alanyl-glycine, L-asparagine, L- aspartic acid, glycyl-L-aspartic acid, glycyl-L-glutamic acid, hydroxy-L-proline.The phenotypic characteristics of strain 2-5Tare summarized in the species description and a comparison of selective characteristics with refe- rence strains are summarized in Table 1.
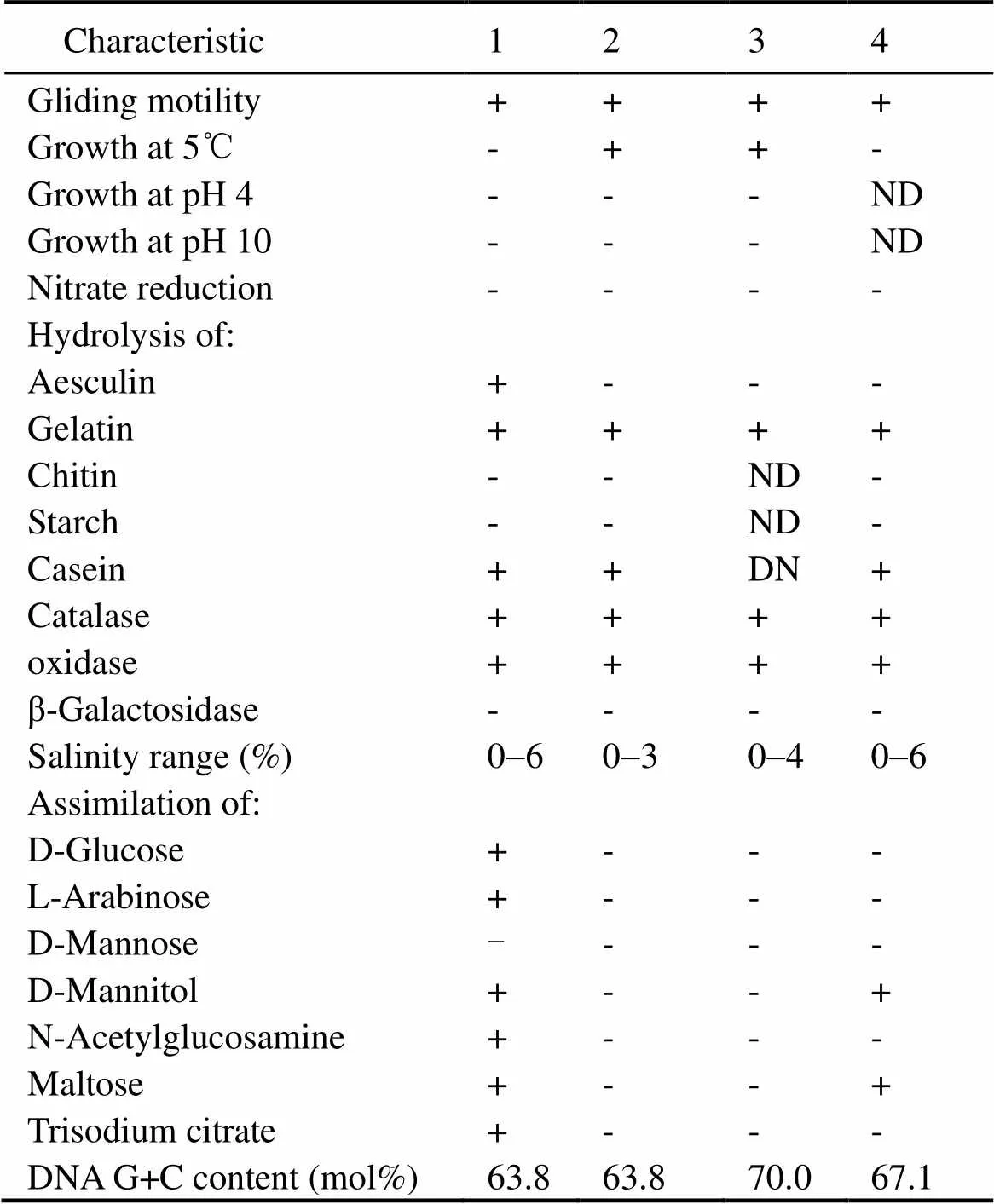
Table 1 Physiological and biochemical characteristics that differentiate strain 2-5T from related type strains of the genus Lysobacter
Notes: Strains: 1, 2-5T(data from this study); 2,KCTC 12205T(Bae., 2005); 3,ZS79T(Lue., 2012); 4,DSM 18482T(Yassin., 2007). +, positive; -, negative; ND, no data available.
3.2 G+C content and Analysis of Cellar Fatty Acids

Table 2 Cellular fatty acid compositions (%) of strain 2-5T and related type strains of the genus Lysobacter.
Notes: Strains: sames as those in Table 1. -, <1% or not detected.
The major cellular fatty acids of strain 2-5Twere iso- C16: 0(15.1%), iso-C15: 0(14.4%), iso-C17: 1ω9(9.9%), iso-C11: 03-OH (8.4%) and iso-C11: 0(7.7%) (Table 2). The presence of branched fatty acids, namely iso-C16:0, iso-C15: 0, iso-C17:1ω19and iso-C17: 0, was consistent with the placement of strain 2-5Twithin the genus(Bae., 2005; Weon., 2006, 2007; Romanenko., 2008). However, strain 2-5Tcontained C16:0ω7/16:1 ω6and C19:0cyclo ω8, which differences with the reference strains. The predominant polar lipid of strain 2-5Twere phosphatidylethanolamine (PE), phosphatidylglycerol (PG) and diphosphatidylglyceroal (DPG) (Supplementary Fig.S3). The DNA G+C content of strain 2-5Twas 63.8 mol%. This value is within the range reported for the genus(Christensen and Cook, 1978; Aslam., 2009).
3.3 Phylogenetic Analysis and DNA-DNA Hybridization
An almost-complete 16S rRNA gene sequence of strain 2-5Tconsisting of 1436bp was obtained. The phylogenetic tree shows that strain 2-5Tclusters within the genusin the class Gammaproteobacteria(Fig.1). 16S rRNA gene sequence analysis revealed that strain 2-5Twas related most closely to.Ko07T(97%similarity),.ZS79T(96%), and.APB-9T(96%). Based on the above results, DNA- DNA hybridization experiments were carried out between strain 2-5TandKo07T. The value obtained was 23%, which is significantly below the value of 70% proposed by Wayne. (1987) for species discrimination.
3.4 Toxonomic Conclusion
It is clear from the 16S rRNA gene sequence comparison and DNA–DNA hybridization data that strain 2-5Trepresents a novel species of the genus(Wayne., 1987). In addition, strain 2-5Twas positive for L-Arabinose, D-Mannitol, N-Acetylglucosamine, Maltose, and Trisodium citrate, differs from the type strain.Ko07T. Based on the phenotypic, phylogenetic and genomic evidence, strain 2-5Twas identified as a novel species of the genus, for which the name.sp. nov. is proposed.
3.5 Description ofsp. nov.
(hy.me.ni.a.ci΄do.nis. N.L. gen. n.of, the generic name of the marine sponge, the source of the type strain).
Cells are Gram-negative, aerobic, non-spore forming, non-motile (but show gliding activity) and rod-shaped,about 0.2–0.3µm wide and 1.1–1.2µmlong. Colonies grown on LB agar are convex, circular, smooth, non- transparent and pale yellow after 3d of incubation at 30℃. Grows at 15–37℃ (optimal 28–30℃); no growth occurs below 4℃ or above 45℃. The pH range for growth is 6.0–8.0 (optimal pH 7.0). Growth occurs in the absence of NaCl and no growth occurs in 8.0% NaCl. Oxidase and catalase-positive. Positive for acid production from glucose, but negative for reduction of nitrates to nitrites. Hydrolyses aesculin, gelatin and casein, but not starch and chitin. Positive for citric acid utilization andacetoin production (Voges–Proskauer reaction). Activities of beta-galactosidase, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase, urease and tryptophan deaminase are negative (API 20 E).Negative for assimilation of dextrin, tween40, tween80, I-erythritol, L-fruc- tose, D-galactose, dextrinose, D-mannose, D-allulose, sucrose, methyl pyruvate, succinic acid methyl, betaphenylglycollic acid, itaconic acid, alpha-oxo acid, sebacic acid, succinic acid, bromosuccinic acid, D-alanine, L-histidine, D-serine, gamma-amino butyric acid, rocanic acid, thymidine, phenyl ethylamine, putrescine, 2-amino ethanol, 2, 3-butanediol, glycerol, glycerol phosphate, D-glucose-1-phosphate, D-glucose-6-phosphate. Utilizes alpha-Cyclodextrin, glycogen, N-acetylgalactosamine, N- acetylglucosamine, ribitol, L-arabinose, D-arabitol, D- cellobiose, D-fructose, D-glucose, m-inositol,alpha-D- lactose, lactulose, maltose, D-mannitol, D-melibiose, beta-methyl-D-glucoside, D-raffinose, L-rhamnose, D- sorbitol, D-trehalose, turanose, xylitol, acetic acid, cis-aconitic acid, citric acid, formic acid, D-galactose acid lactone, D-galacturonic acid, D-gluconic acid, beta- methyl glucoside, D-glucuronic acid, alpha-hydroxy butyric acid, beta-hydroxy butyric acid, gamma-hydroxy butyric acid,alpha-oxo-butanoic acid,alpha-oxo-penta- nedioic acid,D,L-lactic acid, malonic acid, propionic acid, quinic acid, D-saccharinic acid, succinamic acid, glucuronamide, L-alanyl amine, L-alanine, L-alanylglycine, L-asparagine, glycyl aspartic acid, glycyl glutamic acid, hydroxy proline, L-leucine, L-ornithine, L-phenylalanine, L-proline, L-pyroglutamic acid, L-serine, L-threonine, D, L-carnitine, inosine, uridine(API 20 NE and Biolog GN2).

Fig.1 Phylogenetic tree of strain 2-5T and the type strains of related taxa based on 16S rRNA gene sequences. The tree wasreconstructed using the neighbor-joining (NJ) and maximum-likelihood (ML) methods and numbers at nodes representbootstrap percentages (NJ/ML, based on 1000 resamplings). Filled circles indicate genetic branches that were present in boththe NJ and ML trees. GenBank accession numbers are given in parentheses. Bootstrap values over 50% are shown at branching points. Bar, 0.005 substitutions per nucleotide position.
The major cellular fatty acids of strain 2-5Twere iso- C16:0, iso-C15:0, iso-C17:1ω9and iso-C11:03-OH; detailed fatty acid compositions are given in Table 2. The polar lipids consist of phosphatidylethanolamine, phosphatidylglycerol and diphosphatidylglyceroal. The genomic DNA G+C content of the type strain is 63.8 mol%. The type strain 2-5T(=CGMCC 1.12190T= JCM 18137T), was isolated from a crude oil-contaminated marine spon- ge in Dalian, China.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31100092), the Hundred Talent Program of the Chinese Academy of Sciences (No. A1097) and the Ningbo Natural Science Foundation of China (2011A610028).
Supplementary

Fig.S1 Transmission electron micrograph of strain 2-5.
Fig.S2 Scanning electron micrograph of strain 2-5.
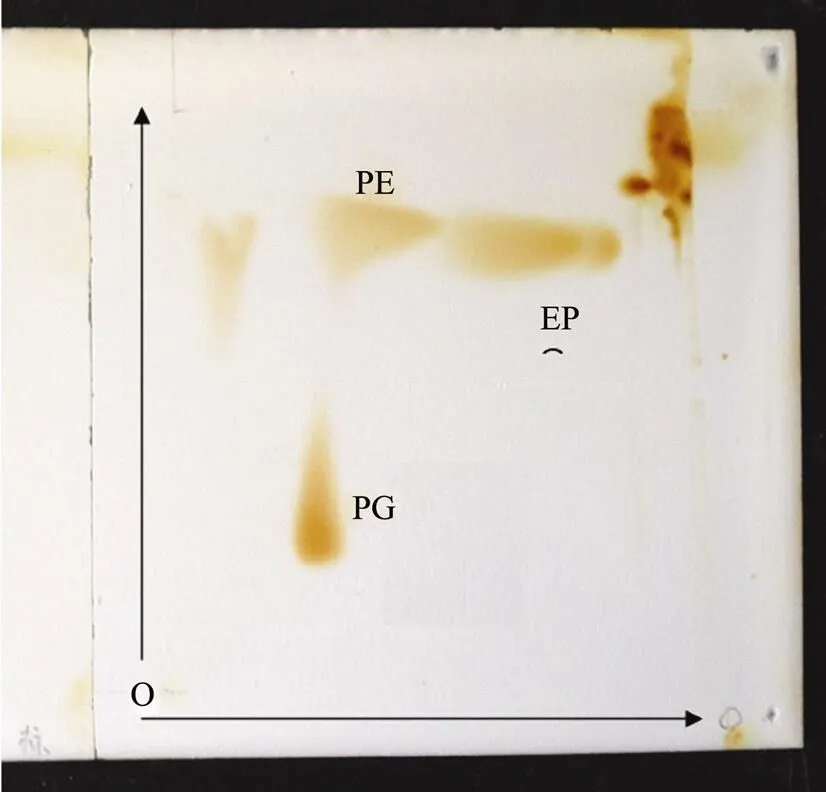
Fig.S3 Two-dimensional TLC of polar lipids of strains 2-5Tstained with 5% ethanolic molybdophosphoric acid. PE, phosphatidylethanolamine; PG, phosphatidylglycerol; DPG, diphosphatidylglycerol.
Ahmed, K., Chohnan, S., Ohashi, H., Hirata, T., Masaki, T., and Sakiyama, F., 2003. Purification, bacteriolytic activity, and specificity of β-lytic protease fromsp. IB-9374., 95: 27-34.
Aslam, Z., Yasir, M., Jeon, C. O., and Chung, Y. R., 2009.sp. nov., isolated from the rhizosphere of rice (L.)., 59: 675-680.
Bae, H.-S., Im, W.-T., and Lee, S.-T. 2005.sp. nov., isolated from anaerobic granules in an upflow anaerobic sludge blanket reactor., 55: 1155- 1161.
Bowman, J. P., 2000. Description ofsp. nov., isolated from the surfaces of Antarctic algae, and reclassification of(ZoBell and Upham 1944) Reichenbach 1989 ascomb. nov., 50: 1861-1868.
Brown, A. E., 2007.. New York, McGraw-Hill, 20-42.
Christensen, P., and Cook, F. D., 1978., a new genus of nonfruiting, gliding bacteria with a high base ratio., 28: 367-393.
Choi, J. H., Seok, J, H., Cha, J. H., and Cha, C. J., 2014.sp. nov., isolated fromginseng soil., 64: 2193-2197
Doetsch, R. N., 1981. Determinative methods of light microscopy. In:, Gerhardt, P.,., eds., American Society for Microbiology, Washington, DC, 21-33.
Ezaki, T., Hashimoto, Y., and Yabuuchi, E., 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains., 39: 224-229.
Felsenstein, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap., 39: 783-791.
Fukuda, W., Kimura, T., Araki, S., Miyoshi, Y., Atomi, H., and Imanaka, T., 2013.sp. nov., isolated from an Antarctic freshwater lake in Antarctica., 63: 3313-3318.
Gordon, R. E., Barnett, D. A., Handerhan, J. E., and Pang, C. H. N., 1974.,, and the nocardin strain., 24: 54-63.
Kämpfer, P., and Kroppenstedt, R. M., 1996. Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa., 42: 989-1005.
Kluge, A. G.,and Farris, F. S., 1969. Quantitative phyletics and theevolution of anurans., 18: 1-32.
Hashizume, H., Hattori, S., Igarashi, M., and Akamatsu, Y., 2004. Tripropeptin E, a new tripropeptin group antibiotic produced bysp. BMK333-48F3., () 57: 394-399.
Islam, M. T., Hashidoko, Y., Deora, A., Ito, T. and Tahara, S., 2005. Suppression of damping-off disease in host plants by the rhizoplane bacteriumsp. strain SB-K88 is linked to plant colonization and antibiosis against soilborne peronosporomycetes., 71: 3786-3796.
Luo, G., Shi, Z.,and Wang, G., 2012.sp. nov.,an arsenite-resistant bacterium isolated from iron-mined soil., 62: 1659-1665.
Mandel, M., and Marmur, J., 1968. Use of ultraviolet absorbancetemperature profile for determining the guanine plus cytosine content of DNA., 12B: 195-206.
Park, J. H., Kim, R., Aslam, Z., Jeon, C. O., and Chung, Y. R., 2008.sp. nov., with antimicrobial activity, isolated from the rhizosphere of pepper, and emended description of the genus., 58: 387-392.
Romanenko, L. A., Uchino, M., Tanaka, N., Frolova, G. M., and Mikhailov, V. V., 2008.sp. nov., isolated from a deep-sea sponge., 58: 370-374.
Sasser, M., 1990. Identification of bacteria by gas chromatography of cellular fatty acids. Technical Note 101. Newark, DE: MIDI.
Sambrook, J., and Russell, D. W., 2001.:, 3rd edtion. Cold Spring Harbor, Cold Spring Harbor Laboratory, 12-29.
Smibert, R. M., and Krieg, N. R., 1994. Phenotypic characterization. In:, Gerhardt, P.,., eds., American Society for Microbiology, Washington, DC, 607-655
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar,S., 2011. MEGA5: Molecular evolutionary genetics analysis usingmaximum likelihood, evolutionary distance, and maximum parsimonymethods., 28: 2731-2739.
Tindall, B. J., 1990. A comparative study of the lipid composition offrom various sources., 13: 128-130.
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G., 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools., 25: 4876- 4882.
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D.,Kandler, O., Krichevsky, M. I., Moore, L. H., Moore, W. E. C., Murray,R. G. E., 1987. International Committee onSystematic Bacteriology. Report of the ad hoc committee onreconciliation of approaches to bacterial systematics., 37: 463- 464.
Weon, H. Y., Kim, B. Y., Baek, Y. K., Yoo, S. H., Kwon, S. W.,Stackebrandt, E., and Go, S. J., 2006. Two novel species,sp. nov. andsp. nov., isolatedfrom Korean greenhouse soils., 56: 947-951.
Weon, H. Y., Kim, B. Y., Kim, M. K., Yoo, S. H., Kwon, S. W., Go, S. J., and Stackebrandt, E., 2007.sp. nov. andsp. nov., isolated from greenhouse soils inKorea., 57: 548-551.
Yassin, A. F., Chen, W. M., Hupfer, H., Siering, C., Krop- penstedt,R. M., Arun, A. B., Lai, W. A., Shen, F. T., Rekha, P. D., and Young, C. C., 2007.sp. nov., isolated from municipal solidwaste., 57: 1131-1136.
Yokota, A., Tamura, T., Hasegawa, T., and Huang, L. H., 1993.gen. nov., sp. nov., nom. rev., a new genus of the order Actinomycetales., 43: 805-812.
(Edited by Ji Dechun)
DOI 10.1007/s11802-015-2789-4
ISSN 1672-5182, 2015 14 (6): 1019-1024
© Ocean University of China, Science Press and Springer-Verlag Berlin Heidelberg 2015
(October 27, 2014; revised August 14, 2015; accepted August 30, 2015)
* Corresponding author. Tel: 0086-411-84379069 E-mail:xuesong@dicp.ac.cn
 Journal of Ocean University of China2015年6期
Journal of Ocean University of China2015年6期
- Journal of Ocean University of China的其它文章
- Inversion Study on Pollutant Discharges in the Bohai Sea withthe Adjoint Method
- Different Responses of Sea Surface Temperature in the North Pacific to Greenhouse Gas and Aerosol Forcing
- The Change Features of the West Boundary Bifurcation Line of the North Equatorial Current in the Pacific
- Wave Pressure Acting on V-Shaped Floating Breakwater in Random Seas
- Dynamic Response of a Riser Under Excitation of Internal Waves
- An Effective Method of UV-Oxidation of Dissolved Organic Carbon in Natural Waters for Radiocarbon Analysis by Accelerator Mass Spectrometry
