Single-cell RNA-sequencing and subcellular spatial transcriptomics facilitate the translation of liver microphysiological systems for regulatory application
Reducing the use of animal models in drug development and safety assessment has long been supported by the U.S.Food and Drug Administration(FDA).The report by Royal Society for the Prevention of Cruelty to Animals indicates that in 2020,experiments involved the use of over 100 million animals,with the United States leading the list by utilizing 20 million animals.Beyond ethical considerations associated with animal testing and the costs in terms of time and money,animal models are not always effective in predicting human reactions to drug exposure.While animal testing has been the traditional method for assessing the safety and efficacy of drugs,the focus has been shifting towards developing alternative methods such as in vitro testing,in silico modeling,and tissue engineering,which can provide valuable insights into drug performance without the use of animals.For instance,organ chip technology and 3D cell cluster and organoid models are becoming increasingly attractive and are being rapidly commercialized by companies.
Microphysiological systems (MPS),including organs-on-chips,are designed to overcome the drawbacks of conventional cell culture techniques,such as lack of perfusion,fluid shear stress,or cell-cell interactions.MPS have been developed for almost all tissues,but liver MPS are the most advanced and most mature,likely because in vitro liver models are widely used not only for drug induced hepatotoxicity,which is a leading cause of drug nonapprovals and postmarking withdrawal,but also for drug metabolism and drug-drug interactions (Fig.1A).The most significant challenge regarding liver MPS is the culture time-dependent loss of key hepatic functions,particularly the expression and activities of drug metabolism enzymes and transporters.Other issues to be addressed by liver MPS include the spatial organization and complex interactions between hepatocytes and hepatic nonparenchymal cells,which accounts for 20% of liver weight.Traditional measurements,such as biochemical assays of albumin and urea production,have been used to characterize liver MPS as compared to conventional cell culture,but these techniques fail to provide a comprehensive picture of the complex network of liver biology involving multiple cell types interacting in spatially defined 3D environments.Therefore,how closely MPS platform resembles the in vivo liver organ remains unclear and this is one of the major obstacles to adopting liver MPS in regulatory decision-making.
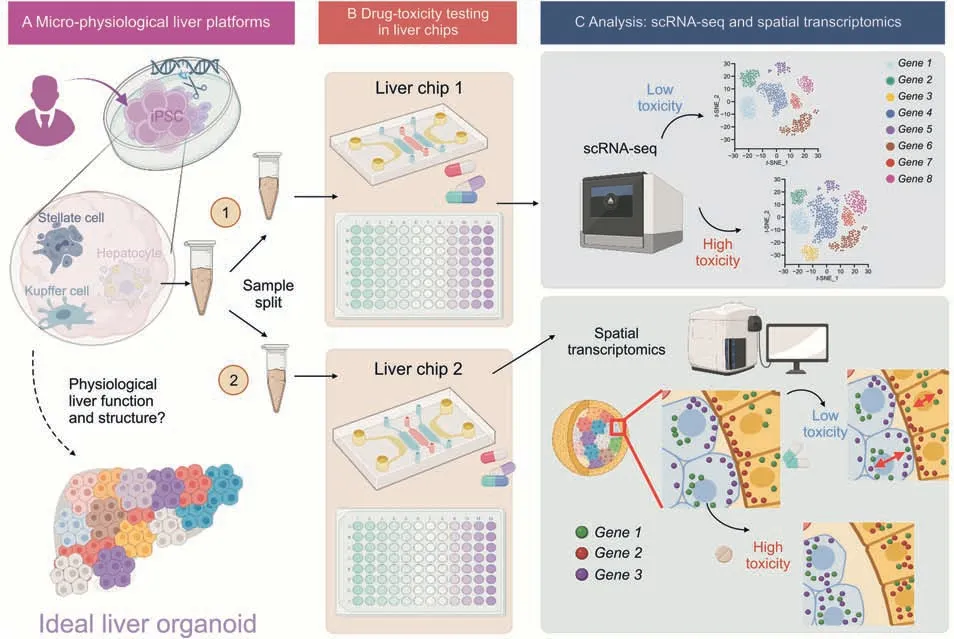
Fig.1.Single-cell RNA sequencing (scRNA-seq)and spatial transcriptomics have great potential to distinguish low and high-toxicity conditions of high-throughput drug screening assays in liver chips and wells.(A)Induced pluripotent stem cells(iPSC)-derived liver organoids are split into replicates.(B)Liver organoids and wells are used for high-content drug toxicity assay.(C) scRNA-seq and spatial transcriptomics are used for downstream analysis of gene distributions and networks associated with low and high toxicity outcomes of drug testing assays. t-SNE: t-distributed stochastic neighbor embedding.Created with BioRender.com.
The MPS liver models need to be rigorously validated to ensure the in vitro model matches the organ's physiological features.Cell types,cell communities,cell-to-cell communication,subcellular RNA organization,and RNA-RNA proximities are major features that could impact model performance and need to be evaluated in MPS.Single-cell RNA sequencing (scRNA-seq) provides the cell distributions,abundance,and identities but lacks the “spatial”context of cell communities and networks.Emerging spatial transcriptomics techniques allow the study of multiplexed transcriptomic profiles using two mainstream approaches: image-based spatial transcriptomics and sequencing-based spatial transcriptomics.While both approaches resolve the spatial context,the main differences between the two are the transcriptome coverage and spatial resolution.Imaging-based spatial transcriptomics techniques such as sequential fluorescent in situ hybridization (seq-FISH) detect multiple RNA (10-10,000 transcript types) molecules at 0.1-1 μm resolution.Sequencing-based spatial transcriptomics techniques provide a spatial resolution of 10-100 μm while maintaining high genome coverage.The advancement of machine learning (ML) and artificial intelligence(AI) enables the extraction of more information from complex spatial transcriptomics datasets.For example,a recently published spatial transcriptomics analysis pipeline entitled spatial gene neighborhood network (SpaGNN)can study the spatial proximity between genes and organelles using multiplexed seqFISH in distinct subcellular regions of a cell.Pipelines such as SpaGNN can be used to examine any alterations to the proximity due to drug toxicity in liver MPS.
scRNA-seq analysis and subcellular spatial transcriptomics will be powerful tools to characterize hepatic cells maintained in live MPS compared to those in conventional culture and the intact cells under in vivo conditions.Such comparative analysis will provide a more comprehensive characterization of liver MPS than conventional approaches and help identify factors and pathways that are important to improve the performance of liver MPS.For example,scRNA-seq of normal human liver identified a subgroup of cells that have a strong potential to form organoids[1],and the characteristics of these cells may vary among individuals due to genetic factors or disease states,which may explain why primary liver cells from only about 30%of human donors are suitable for organoid culture[2],and help improve the likelihood of success in liver MPS by characterizing predictive markers before use.
When combined with automated microfluidics,termed “liver chips”,and microwell arrays,“liver wells”,MPS growth condition factors can be tightly controlled in terms of pH,cell shear stress,temperature,and media(Fig.1B).Custom materials such as polydimethylsiloxane and glass bottom high content microwell arrays create flexibility for perfusion of drugs in toxicity testing,enabling the design of different perfusion (oxygen and media) setups in rapid drug screening with microarrays.For example,the blood flow rate in human liver is about 60 mL/h/g tissue mass.Each gram of human liver contains around 100 million hepatocytes.This suggests that the blood flow rate normalized to the number of hepatocytes would be 0.6 mL/million cells/h.Therefore,for a liverchip with about 0.05 million hepatocytes per chip,the flow rate of the perfusing cell culture medium is usually set at 0.03 mL/h to mimic the liver blood flow rate in a healthy human subject.
Recently,induced pluripotent stem cells(iPSC)-derived 3D liver organoids have emerged as liver development and disease models.Hepatocytes combined with iPSCs have been demonstrated to form functional,3D clusters that successfully recapitulate liver biology.Following differentiation,liver organoids contain diverse cell types such as mature hepatocytes,hepatic stellate,Kupffer,and endothelial cells.Such advantageous in vitro models can successfully screen drug efficacy and toxicity on a personalized basis.
Such 3D cell clusters represent more complex and realistic models of cellular organization and interactions within tissues that can be used for a wide range of applications,such as drug discovery,toxicity testing,and tissue engineering [3].Spatial transcriptomics techniques can be used to study the spatial organization of cells and molecules within the 3D clusters,which can provide insights into cellular behavior and tissue function.3D cell clusters can be preserved and sliced into sections,and 3D spatial transcriptomics profiles can be reconstructed.Organ-chip models can be examined as a whole or deconstructed.For example,spatial transcriptomics can be used to analyze gene expression patterns within different regions of 3D cell clusters,providing a more detailed understanding of how cells within the cluster are interacting with each other and responding to external stimuli.
However,challenges remain regarding 3D cell cluster heterogeneity,scalability,and stability.For example,hepatocytes may vary in maturity levels and arrangement within liver organoids,thus decreasing the reproducibility and reliability of toxicity tests.Microwells and microfluidics are poised to combat this issue by systematically controlling organoid size,uniformity,and growth conditions.Besides,3D bioprinting has emerged as another method to precisely control 3D cell cluster patterns and architecture,but vascularization issues within the organoid remain difficult to address.
With the growing interest in the use of in vitro models such as organ chips and 3D cell cluster models as alternatives to animal models,it is becoming increasingly important to ensure the quality of the outcomes.scRNA-seq and spatial transcriptomics play an essential role in studying cellular heterogeneity and organization in these in vitro models,making it necessary to ensure their reproducibility and accuracy across different methods and tools.This is particularly crucial for drug development and safety assessment,where reliable and consistent results are vital.Therefore,continued efforts are needed to develop and improve quality control standards and guidelines for scRNA-seq and spatial transcriptomics.This will ensure their reliability and reproducibility in the context of in vitro models.
Comparative studies of scRNA-seq and spatial transcriptomics in liver chips composed of multiple cell types of present opportunities to resolve the differences between the technical variability and biological heterogeneity (Fig.1C).scRNA-seq achieves genome-wide detection but the sensitivity is much lower compared to image-based spatial transcriptomics that is inherently capable of capturing >90% RNA molecules owing to the omission of complementary DNA conversion steps.Image-based spatial transcriptomics then enhances the specificity of RNA type resolvability,but the limited coverage of a targeted number of markers (20-100) complicates the cell type resolvability despite its high sensitivity and molecular specificity.However,sequencing-based spatial transcriptomics still suffers from low sensitivity and specificity as in scRNA-seq.Therefore,the technical variations of scRNA-seq and image-based spatial transcriptomics may benefit from standardization efforts to establish the quality control metrics in drug toxicity studies using emerging MPS platforms,including in vitro 3D clusters,liver organoids,and chips.A sample split strategy from biological replications would be used to quantitatively evaluate the differences in separately performed scRNA-seq and spatial transcriptomics measurements on the identical drug-treated 3D cell clusters.Spatial proteomics assays complement such sample split strategies to further benchmark standardization efforts.
The FDA-led SEQC2 project conducted a study [4] to compare diverse scRNA-seq datasets from different laboratories using different technologies.The project also provided guidance on algorithms to generate accurate biological interpretations.Despite the efforts made by the project,the results indicated that obtaining high-quality and reproducible results through an optimized pipeline is still quite challenging.This is due to the many contributing elements involved,such as preprocessing,normalization,batch effect correction,and bioinformatic pipelines.The complexity of single-cell sequencing technology adds to the difficulty of ensuring consistency and reproducibility of the results.Modern AI/ML methods have also been developed to better analyze the scRNAseq data to address batch effects,implement simultaneous measurement of gene expression and chromatin accessibility in the same cell,and reduce the dimension for downstream analyses and visualization [5].
In conclusion,scRNA-seq and spatial transcriptomics have become increasingly valuable tools in understanding cellular heterogeneity and organization in a wide range of contexts,including in vitro models and disease states.However,these technologies also present significant challenges,particularly in terms of quality assurance.In addition,MPS is still in its infancy and the technology is rapid evolving.It is unlikely that MPS can completely replace animal tests in the near future.Despite these challenges,the continued development and improvement of quality control standards and guidelines for single-cell sequencing and spatial transcriptomics are necessary to ensure reliable and reproducible results in future research.With the growing interest and investment in these fields,there are numerous opportunities for advancements in technology and data analysis,as well as exciting prospects for new discoveries and therapeutic interventions.
Disclaimer
The views presented in this article do not necessarily reflect those of the U.S.Food and Drug Administration.Any mention of commercial products is for clarification and is not intended as an endorsement.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
 Journal of Pharmaceutical Analysis2023年7期
Journal of Pharmaceutical Analysis2023年7期
- Journal of Pharmaceutical Analysis的其它文章
- Dissecting the brain with spatially resolved multi-omics
- New discoveries in the field of metabolism by applying single-cell and spatial omics
- Single-cell analyses reveal cannabidiol rewires tumor microenvironment via inhibiting alternative activation of macrophage and synergizes with anti-PD-1 in colon cancer
- Cux1+ proliferative basal cells promote epidermal hyperplasia in chronic dry skin disease identified by single-cell RNA transcriptomics
- Single-cell analysis of cellular heterogeneity and interactions in the ischemia-reperfusion injured mouse intestine
- Discovering metabolic vulnerability using spatially resolved metabolomics for antitumor small molecule-drug conjugates development as a precise cancer therapy strategy
