Demonstration of safety characteristics and effects on gut microbiota of Lactobacillus gasseri HMV18
Xianxian Jia,Miaomiao Jia,Xiang Gao,Xiang Li,Mengyuan Wang,Shengqiang Du,Rui Huang,Xiaotong Li,Jun Zhang,Shujin Li,Chunling Ma,Yan Zhang,Bin Cong,
a Department of Pathogen Biology, Institute of basic Medicine, Hebei Medical University, Shijiazhuang 050017, China
b Research Unit of Digestive Tract Microecosystem Pharmacology and Toxicology, Chinese Academy of Medical Sciences, Shijiazhuang 050017, China
c Institute of Basic Medicine, Hebei Medical University, Shijiazhuang 050017, China
d College of Forensic Medicine, Hebei Key Laboratory of Forensic Medicine, Collaborative Innovation Center of Forensic Medical Molecular Identif ication,Hebei Medical University, Shijiazhuang 050017, China
e Hebei Food Safety Key Laboratory, Hebei Food Inspection and Research Institute, Shijiazhuang 050227, China
Keywords: Probiotics Lactobacillus gasseri Safety assessment Gut microbiota
ABSTRACT Human normal flora is a source of probiotics.The safety characteristics of a specific isolate determine its application in foods or drugs.The food-borne-pathogen antagonist strain Lactobacillus gasseri HMV18 is one of the isolates from normal human f lora.In this work,we assessed the in vitro pH tolerance,bile tolerance,biogenic amine production,mucin utilization,and safety of in vivo administration to mice to evaluate general health,organ-body weight index,organ histopathological change,whether L.gasseri HMV18 can colonize in the gut or modulate the gut microbiota after oral administration.The results suggest that L.gasseri HMV18 can tolerate pH 3 for 2 h,3% bile for 3 h,biogenic amine negative,mucin usage negative,does not encode verif ied toxins,and cause no visible change in mice’s organs.L.gasseri HMV18 might not colonize in mice’s gut,but can signif icantly affect the structure of gut microbiota.A bibliographical survey suggested that there were as few as 8 opportunistic infection cases from 1984 to 2022 and that the possibility for L.gasseri to cause infection is relatively low.Therefore,this work provides a basis for the foods or drugs application of L.gasseri HMV18 and gives a map of experiments for the safety assessment of probiotics.
1.Introduction
Normal f lora benef its human health through mucosal homeostasis maintenance,promoting immune status,and being reactive in drug metabolism and metabolite utilization[1-2].Multiple probiotic strains have been isolated and identif ied from human f lora[3].Probiotics are live microbes,when administered in an adequate dose,confer health benefits to the host.The genusLactobacillusis belonged to phyla Firmicutes,and it is an important constituent of the gut microbiome[4].These gut-residingLactobacillusspecies not only contribute to microflora but also improve the gut barrier integrity[5].Some strains ofLactobacillusgasseriwere proven to be benefit to lipid metabolism[6-7],decreased the Irritable Bowel Syndrome symptom score[8]and changed the composition of intestinal microbiota[9].A new strainL.gasseriHMV18 was reported to inhibit the growth of pathogenic bacteria such asEscherichia coli,Staphylococcus aureus,andKlebsiella oxytocaby our research group[10].AlthoughL.gasseriis one of the strains used in foods and approved by the National Health Commission of the People’s Republic of China [2010] No.65,the safety of new probiotic strains needs to be studied as probiotic bacteria are strain-specific.Therefore,it is necessary to investigate whetherL.gasseriHMV18 is safe to be an additive in foods or used as a drug to prevent these food-borne pathogens.
To evaluate probiotic safety is to assess the potential hazards of a certain probiotic strain,such as pathogenicity,toxicity,antibiotic resistance,which ensures the safe use of probiotic products.However,the characteristics of different strains make the testing experiments various among different research group.Such as genomic analysis to detect antibiotic resistant genes or toxin related genes[11-13],invivoanimal acute toxicology evaluation[11,14-15],subacute oral toxicity study[15],chronic toxicology test[14,16],and the potential migration to cause bacteremia[17].Some research interested in the evaluation of adhesion of host cells or inhibitory effect ofL.gasserion pathogenic bacteria[18-19].According to theGuidelines for the Evaluation of Probiotics in Food(FAO/WHO,2002),the criteria and methodology to systematically evaluate probiotics in food are: 1) assess antibiotic resistance profiles,2) assess certain metabolic activities,3) assess side-effects in human studies,4) post-market surveillance of adverse incidents,5) if the strain is a known mammalian toxin producer,the test for toxin production is recommended,6) if the strain has hemolytic potential,hemolytic activity is required.We considered all described criteria to design and recruit experiments to demonstrate the safety ofL.gasseriHMV18.The antibiotic resistance and hemolytic potential have been conducted in our previous study.According to the results of screening the antibiotic-resistant gene and Kirby-Bauer Disk Diffusion Susceptibility Test,L.gasseriHMV18 is sensitive to various antibiotics such as penicillin,ampicillin,erythromycin,tetracycline,and chloramphenicol[10].Although it is resistant to several antibiotics,the resistant genes are on the genome,not plasmid,which suggests the possibility is limited for the antibiotic gene to transfer to another bacterium[20].
In this article,we aim to investigate metabolic activities,the possible mammalian toxin,the clinical case report of bacteremia and effects on gut microbiota caused byL.gasseri.Notably,human side effects and post-market surveillance should be further evaluated.
2.Material and methods
2.1 Bacterial strains and culture conditions
L.gasseriHMV18 andL.caseiHMV1803 were kept in the Pathogenic Biology Laboratory,Hebei Medical University (China).Isolates were routinely grown anaerobically at 37 °C in Rogosa and Sharpe (MRS) broth or MRS agar using the methods described before[10].The MRS broth recipe includes 10 g/L tryptone,10 g/L beef extract,5 g/L yeast extract,20 g/L glucose,1 mL/L Tween 80,2 g/L K2HPO4,5 g/L sodium acetate,2 g/L ammonium citrate dibasic,0.2 g/L MgSO4·7H2O,0.05 g/L MnSO4·4H2O,and a final pH 7.0.Notably,the recipe of MRS agar is the same as MRS broth,with 1.5%agarose.TheLactobacillusstrains were recovered by streaking on MRS agar and incubated at 37 °C under anaerobic conditions for 72 h.Finally,grey colonies were picked and inoculated into screw-capped tubes filled with MRS broth for anaerobic growth and incubated at 37 °C for 48 h.
2.2 Resistance to GI stress (pH and bile)
The pH of the human gastrointestinal (GI) tract is 1.0-8.0[21],the stomach pH can reach 0.9-1.5,and the intestinal pH is 7.6[22].The pH levels 1.0,2.0,and 3.0 were tested in the acid tolerance ofLactobacillus.The culture ofL.gasseriHMV18 in MRS can reach pH 4.0,so we wanted to assess whether it can tolerate this pH from the beginning of growth.To test the pH tolerance characteristic ofL.gasseriHMV18,we prepared media with pH 1.0,2.0,3.0,4.0,7.0,and 8.0.The pH was adjusted by 10% (V/V) HCl or 10% NaOH.
The 15 mL of an overnight culture ofL.gasseriHMV18 was normalized to 1 at OD600nmby Macy UV-1700PC spectrophotometer(Shanghai,China).Bacterial cells were concentrated by centrifugation (Centrifuge TDL-5-A,Shanghai Anting Scientific Instrument Factory,China) at 5 000 r/min for 10 min and resuspended in 1/10 volume of MRS broth so that the final bacteria concentration was 1010CFU/mL in each pH solution.0.5 mL of each solution was immediately collected at times 0 and after 0.5 h,and 2 h.The 10-fold serial dilutions were made with sterile saline,and viable-cell numbers were determined by the pour-plate method.
According to the instruction,an 8%-10% solution of the dehydrated product is equivalent to fresh bile.Therefore,we tested the bile tolerance in solutions with 3.0%,6.0%,and 8.0% of dehydrated ox bile (Hefei Qiansheng Biotechnology Co.,Ltd.,China) in MRS,pH 6.4,and MRS with 0 ox bile was used as control.The inoculation process was the same as the pH tolerance test.0.5 mL of each solution was immediately taken at times 0 and after 3,6,12,and 24 h.Similar dilutions were made,and viable-cell numbers were determined,as mentioned above.Finally,the plates were incubated at 37 °C for 24 h under anaerobic conditions.
These experiments were repeated independently 3 times.
2.3 Bibliographical survey (opportunistic infection case reports)
Lactobacilliare the residents of the normal human flora,and the bacteremia caused byLactobacilliis only occasionally reported in clinical infection cases.The literature review is based on the open access database using the keywords: “L.gasseri” and “infection” or“abscess” or “sepsis” or “septic” or “bacteremia” or “bacteraemia”or “toxin” for surveillance of sepsis,“endocarditis” or “abscess” or“meningitis” for surveillance of identified disease.The references used in the article were further searched for other case reports,as some of the cases are unavailable on PUBMED or not using the above mentioned keywords.An additional search was performed on Baidu Scholar to search for non-PUBMED references.This “multi-layered”web search assisted us in finding more cases than reported in the previous review published onL.gassericase reports.
2.4 In vitro toxicity study
2.4.1 Biogenic amine production
We adopted the qualitative testing method described by Bover-Cid to confirm the biogenic amine production.This test involved two steps: activating decarboxylase,and screening the amine-forming strains.The culture media used in the two steps are slightly different.
The activation culture medium includes 0.05 g/L pyridoxal-5-phosphate (Sinopharm Chemical Reagents Co.,Ltd.,Shanghai,China),1 g/L either one of the precursors amino acids:L-tyrosine,L-histamine,L-lysine,L-ornithine monohydrochloride,andL-arginine(Solarbio®Life sciences,Beijing,China) in MRS broth.
The screening medium includes 5 g/L tryptone,5 g/L beef extract,5 g/L yeast extract,2.5 g/L sodium chloride,0.5 g/L glucose,2 g/L diammonium hydrogen Citrate,0.4 g/L MgSO4·7H2O,0.055 g/L MnSO4·4H2O,0.04 g/L FeSO4,0.01 g/L thiamine,0.1 g/L CaCO3,2 g/L K2HPO4,0.06 g/L bromocresol purple,0.05 g/L pyridoxal-5-phosphate,10 g/L precursor amino acid,1 mL/L Tween 80,pH 5.3.The medium was autoclaved at 121 °C for 10 min to avoid excessive hydrolysis of the agar at low pH value.The prepared plates were light purple.
The recovered HMV18 over-night culture was inoculated at 1% (V/V) into the activation medium and sub-cultured for 5 times.Subsequently,the culture was grown at 37 °C,24 h under anaerobic conditions for one time.The activated HMV18 was concentrated by centrifugation and streaked on the screening medium,and the streaked plates were incubated at 37 °C under anaerobic conditions for 4 days.Furthermore,both strains (L.gasseriHMV18 andL.caseiHMV1803)were streaked in duplicate with and without amino acids.
2.4.2 Mucin degradation
Kim’s approach[23]was adopted to evaluate whetherL.gasseriHMV18 can utilize mucin as a carbon source.In this experiment,glucose was supplemented as a positive control.1% (V/V) of the o/n culture ofL.gasseriHMV18 was inoculated in four kinds of MRS broth: no glucose (the basal medium,MRS broth without any glucose),0.5% glucose in base medium,1% glucose in basal medium,0.5% mucin in basal medium,and 1% mucin in basal medium.Mucin from the bovine submaxillary gland was purchased from Shanghai Zeye Biotechnology Co.,Ltd.,China.These cultures were incubated anaerobically at 37 °C.Bacterial growth was monitored by measuring OD600nmat 0,12,24,36,and 48 h.Each group was repeated three times independently in parallel.
2.5 Virulence factors
The genomic data ofL.gasseriHMV18 were uploaded to the virulence factor Database (VFDB,http://www.mgc.ac.cn/VFs/search_VFs.htm),and the potential virulence genes were further searched on the NCBI gene bank for their function and experimental verification.
2.6 In vivo toxicology studies
2.6.1 Animals
The animal protocol listed below was reviewed and approved by the Laboratory Animal Ethical and Welfare Committee of Hebei Medical University (No.20190051).20 Kunming mice (male,8 weeks old) were purchased from the Experimental Animal Center of Hebei Medical University.The mice were acclimated for 3 days at room temperature ((22 ± 2) °C) and humidity ((55 ± 2)%) with a 12 h-12 h light-dark alteration.The mice were randomly grouped: the control group and the HMV18 gavaged group.Both groups were fed sufficient food and sterile water.
2.6.2 Oral toxicity study
L.gasseriHMV18 was subcultured to MRS broth at 1% (V/V).After 12 h of incubation at 37 °C,bacteria were centrifuged at 5 000 r/min for 10 min.The cell pellet was harvested and washed with sterilized phosphate buffer solution (PBS) three times and resuspended in PBS to reach a final concentration of 109CFU/mL.
Each mouse in the HMV18 group was gavaged 200 µL of the HMV18 suspension (109CFU/mL) once per day,while the mice in the control group were gavaged 200 µL PBS.The oral administration lasted for 7 days.The mice’s activity,general health,food intake (FI),and body weight were observed and measured daily.On the 8thday,the mice were euthanized by cervical dislocation.
2.6.3 Organ index and histopathological examination
The overall anatomy of each mouse’s organs was examined.The heart,liver,spleen,lungs,and kidney were quickly removed with a flame-sterilized scalpel,placed in a petri dish,washed with normal saline,dried with filter paper,and weighed.The small intestine was removed,washed,and dried.The equation calculated the organ index (OI).
where,mOwas actual organ weight (mg),andmWwas mouse’s body weight before anatomy (g).
The organs were fixed in 10% formaldehyde,dehydrated with a step-wise gradient of ethanol,embedded in paraffin,cut into 5 μm sections,stained with hematoxylin-eosin (H&E),examined histologically,and analyzed with Olympus light microscope (Tokyo,Japan).The magnification is 200,and the images were captured using a Cannon digital CCD camera (Tokyo,Japan).
2.6.4 Gut microbiota
Five mice from each group were randomly selected for 16S rRNA sequencing.Total genomic DNA samples were extracted using the OMEGA Soil DNA Kit (M5635-02) (Omega Bio-Tek,Norcross,GA,USA),following the manufacturer’s instructions.The quantity and quality of extracted DNAs were measured using a NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific,Waltham,MA,USA).PCR amplification of the bacterial 16S rRNA genes V3-V4 region was performed using the forward primer 338F(5’-ACTCCTACGGGAGGCAGCA-3’) and the reverse primer 806R(5’-GGACTACHVGGGTWTCTAAT-3’).PCR amplicons were purified with Vazyme VAHTSTM DNA Clean Beads (Vazyme,Nanjing,China) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen,Carlsbad,CA,USA).After the individual quantification step,amplicons were pooled in equal amounts,and pair-end 2 × 250 bp sequencing was performed using the Illlumina NovaSeq platform with NovaSeq 6000 SP Reagent Kit (500 cycles) at Shanghai Personal Biotechnology Co.,Ltd.(Shanghai,China).
Microbiome bioinformatics were performed with QIIME 2 2021.8[24].Raw sequence data were demultiplexed and quality filtered using the q2-demux plugin followed by denoising with DADA2.All amplicon sequence variants (ASVs) were aligned with mafft and used to construct a phylogeny with fasttree2.Theα-diversity metrics(observed features and Faith’s Phylogenetic Diversity,which was abbreviated as faith_pd),β-diversity metrics (weighted UniFrac),and Principle Coordinate Analysis (PCoA) were estimated using q2-diversity after samples were rarefied (subsampled without replacement) to 900 sequences per sample.Taxonomy was assigned to ASVs using the q2-feature-classifier classify-sklearn naïve Bayes taxonomy classifier against the Greengenes 13_8 99% OTUs reference sequences.
2.7 Statistical analysis
The results are expressed as mean ± standard deviation.The comparisons of body weight and OI were performed by ANOVA comparison of multiple samples.Statistical Package for Social Science (SPSS) version 26.0 (SPSS Inc.,USA) was used for all statistical analyses,and the difference was considered significant whenP< 0.05.
3.Results and discussion
3.1 Resistance to GI stress (pH)
Six pH values were selected to simulate the human digestive tract’s internal environment,and the pH 7 group was set as the control.As shown in Table 1,there is no significant difference in the viable cell number ofL.gasseriHMV18 in pH 3,4,and 8 compared with that in pH 7.The viable cell number in the pH 2 group significantly different at 2 h compared with that in pH 7.In the extremely low pH(pH 1),the viable cell number significantly differed from that in pH 7 at 0.5 and 2 h.Notably,the viable cell number significantly decreased to 8.64 (lg (CFU/mL)) at 2 h from 9.48 (lg (CFU/mL)) in pH 7(P< 0.05).Although we normalized the starting bacterial cell number,there still might be some difference among different pHs,and pH 3 shows a significant difference.Notably,L.gasseriHMV18 could resist pH 3,4,and 8 for 2 h treatment.
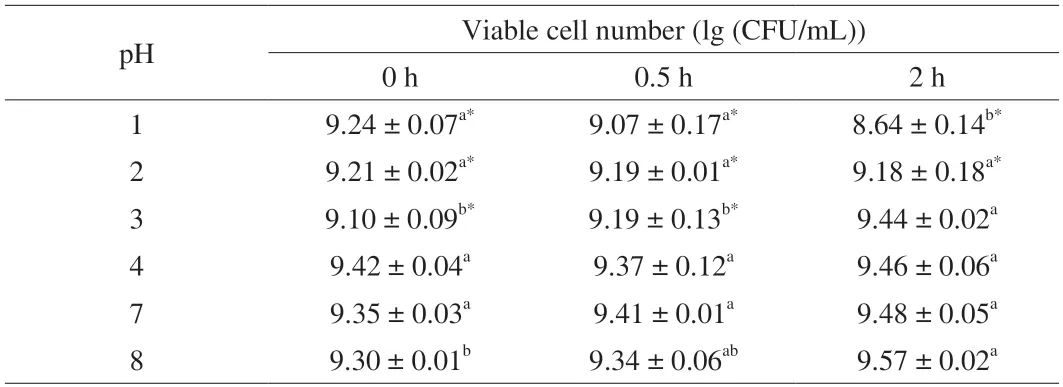
Table 1 L.gasseri HMV18 resistance to pH.
3.2 Resistance to bile
We used a maximum concentration of 8% ox bile to mimic the fresh bile.The results of bile tolerance ofL.gasseriHMV18 are shown in Table 2.When the HMV18 strain incubates with 3%,6% and 8% concentrations of bile for 3 h,the viable bacteria decrease as the concentration increases.The same trend can also be observed as the incubation time is prolonged.When the HMV18 strain was incubated with 3%,6%,and 8% concentrations of bile for 24 h,the viable bacteria were 8.28,7.36,and 6.92 (lg (CFU/mL)) compared with the control,respectively.Therefore,bile salt can be considered one of the stress conditions forL.gasseriHMV18 in the GI tract.As reported by Liu et al.[17],we also performed experiments to test the resistance to 0.15%,0.3%,and 0.5% bile salts.However,the results showed thatL.gasseriHMV18 was sensitive to these concentrations of sodium cholate (data not shown).Considering the Grading Specification for Probiotic Viable Rate in Probiotic Foods (T/CNHFA 006-2022),whenL.gasseriHMV18 was added to foods,the viable cell number should be above 106CFU/g.To get 106CFU/g viableL.gasseriHMV18 after passing through the GI tract,the cell number should be above 109CFU/g.
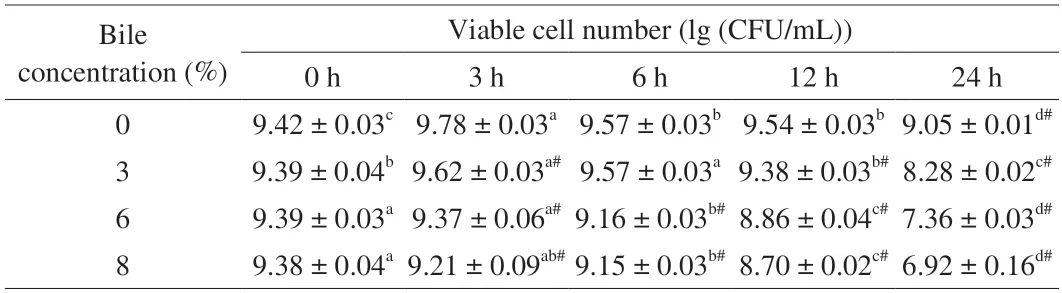
Table 2 L.gasseri HMV18 resistance to bile.
3.3 Opportunistic infections
When we performed the literature review,we foundL.gasserionly caused 8 cases (Table 3).The frequency of bacteremia caused byL.gasseriwas much less than bacteremia caused by the genusLactobacillus(3 cases in 86 bacteremia patients with a lactobacillus isolate) or the species likeL.caseiandL.rhamnosus[25].One case report ofL.gasseriempyema was recovered and was seen for 4 months follow-up[26].One case of Fournier gangrene due to the monomicrobial etiology byL.gasseri[27].One case of septicemia following a urinary tract infection was noted due toL.gasseri[28].One case ofL.gasseri-associated urinary and wound infection was reported in a female renal transplant patient[29].One case of pneumonia and empyema was due to a mixed infection ofL.delbrueckiiandL.gasseri.After the condition had been successfully cured,the patient developed esophageal malignancy 5 months later[30].One case ofL.gasseriendocarditis on the aortic valve bioprosthesis was successfully treated with amoxicillin/clavulanate acid(6 weeks) and with gentamycin (2 weeks)[31].In a case,a 59-year-old man with a history of diabetes mellitus and multiple abdominal surgeries developed liver abscesses and bacteremia caused byL.gasseri[32].All of these indicated that the prevalence of infection over the last 30 years was low: all 8 reported cases were after 1984.
3.4 Biogenic amine production
Excessive amounts of biogenic amines can cause adverse effects in humans,such as headaches,blood pressure changes,breathing disturbances,heart palpitation,and vomiting[33-34].If nitrites arepresent,biogenic amines could be converted to nitrosamines,which are suggested to cause cancer[35].Tyramine,histamine,cadaverine,putrescine,and spermine are common biogenic amines tested for safety and toxicity evaluation[34-35].Furthermore,pyridoxal-5-phosphate was a coenzyme of aminotransferase and decarboxylase in amino acid metabolism[36],which activates decarboxylase.
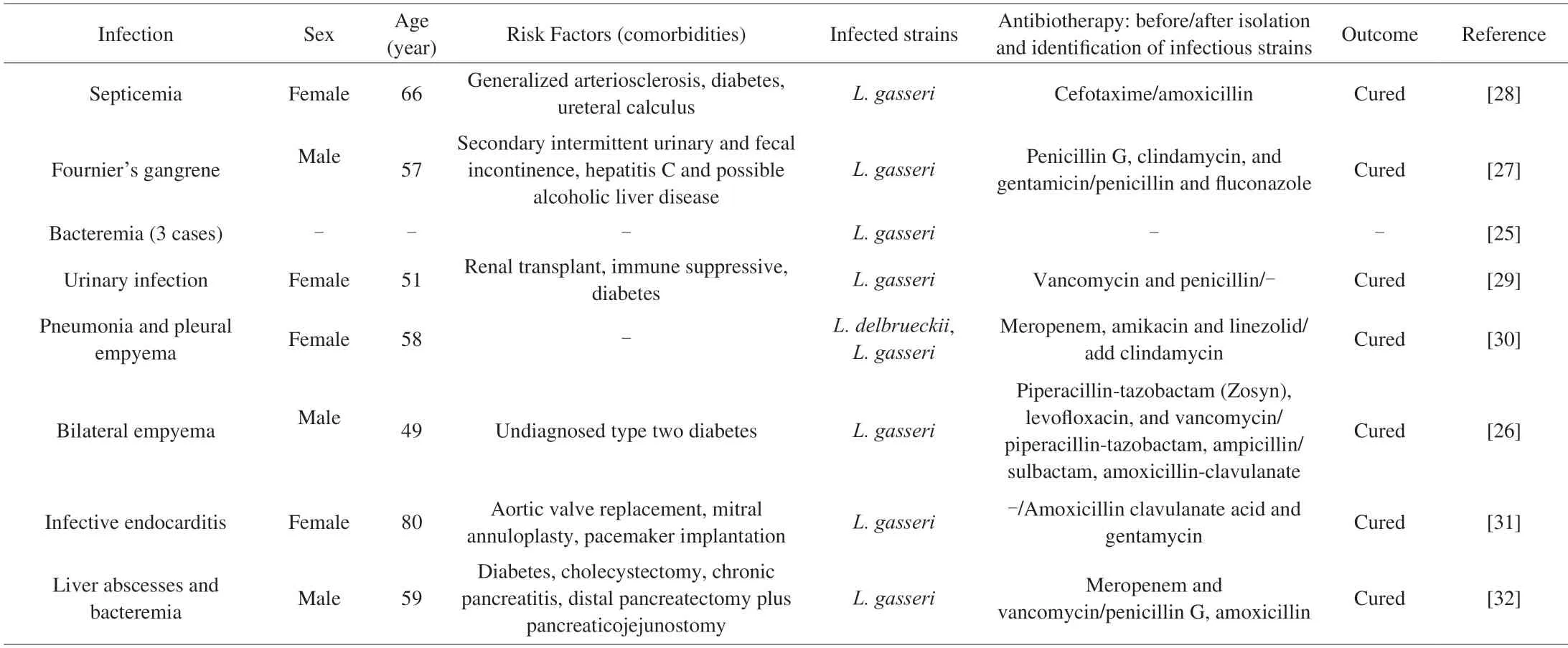
Table 3 Opportunistic infection case reports for L.gasseri isolates.
TheL.caseiHMV1803 positive control was changed to purple.In contrast,the plates without amino acids were yellow.Notably,the screening plates streaked withL.gasseriHMV18 were yellow.Although the glucose was reduced to 0.5 g/L and the colonies ofL.gasseriHMV18 were small,the culture medium became acidic.Therefore,these results suggest thatL.gasseriHMV18 produces acid and could not produce any of the five biogenic amines under anaerobic conditions.
3.5 Mucin degradation
Goblet cells and enterocytes of the GI tract secrete mucin and mucus,which help comprise the gut’s first line of defense[37].The bacterium is considered pathogenic or toxic if it can degrade mucin and penetrate mucus[38].To test whether HMV18 can degrade mucin,we inoculated it in 5 modified MRS media and monitored the growth by OD600nm.As glucose is the optimized carbon source in MRS for mostLactobacilli,HMV18 grew fastest in the basal medium with 1%glucose,and the growth rate slowed with less glucose,as observed in the basal medium with 0.5% glucose.HMV18 slightly grew in the glucose-free basal medium.The growth rate in 0.5% mucin shows no significant difference from that in the basal medium (P> 0.05)(Fig.1).However,the growth rate in 1% mucin was significantly slower than that in the basal medium (P< 0.05).
These results suggest that HMV18 prefers glucose over mucin as a carbon source in the growth conditions.A high concentration of mucin might even inhibit the growth of HMV18.HMV18 could not destroy the intestinal surface or translocation.
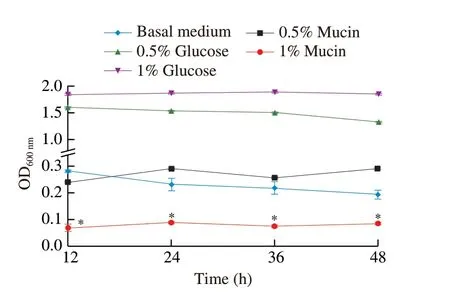
Fig.1 Effect of mucin on the growth of L.gasseri HMV18. L.gasseri HMV18 was inoculated into glucose-free MRS,basal medium with 0.5% glucose,basal medium with 1% glucose,basal medium with 0.5% mucin,basal medium with 1% mucin.*P < 0.01 vs basal medium group.
3.6 Virulence factors
BLASTP comparison was performed between the whole genome sequence ofL.gasseriHMV18 and VFDB 2005,and 269 putative virulence factors were identified (Table S1).When we sorted these factors with the conditions-E < 0.01,coverage > 70%,identity >30%,172 putative pathogenic genes were identified (Table S2),and the annotated virulence factors are summarized in Table 4.These genes include 43 transporter genes,27 of which were related to the transport of iron,magnesium,zinc,and copper.The gene 1849 shows the highest similarity asctpV(identity 44.79%,Table S2),which can maintain copper resistance[39]and is essential to the intracellular pathogenicity ofMycobacterium tuberculosis.
Despite the 172 VFDB genes,the low percentage of coverage and identity suggest they seldom can be true virulence factors.Adhesion and colonization must be the first step for probiotic survival in intestinal mucus.Understandably,Lactobacillusencodes genes related to these functions for colonization but not pathogenicity:adhesion,capsule,polysaccharides,and fibronectin-binding proteins.Probiotics express anti-phagocytosis proteins,stress proteins,transporter components,some enzymes,and virulence-related gene regulators that may help bacteria adapt to new ecological niches for survival in the host gut[40].For example,clpC(51.41%),clpE(57.16%),andclpP(68.59%) encode chaperones and proteases necessary in general stress response and are involved inListeriavirulence[41].The two-component signal transduction systemlisR(68.40%) andlisK(38.02%) associated to sense ethanol,pH,hydrogen peroxide,and resistant antimicrobial and are essential in virulence[42].Additionally,bile salt hydrolases are enzymes for bile salt resistance.The gene 0051 shows 98.42% identity with the gene of LagBSH,a new bile salt-resistant protein reported by Kusada et al.[43].The possible virulence factors ofL.gasseriHMV18 could be cell toxins/hemolysins/invasive factors (25 genes) and secretion systems(8 genes),which account for 19.19% of the virulence factors.However,the similarities are low.Only 4 genes encode proteins with more than 50% similarity: the nonspecific virulence enzyme gene 1348 (Lgt,53.44%),gene 1327 (Eno,69.12%),gene 0798 (Eno,50.84%),and the invasion factor gene 0718 (Cthe_0827,54.29%).
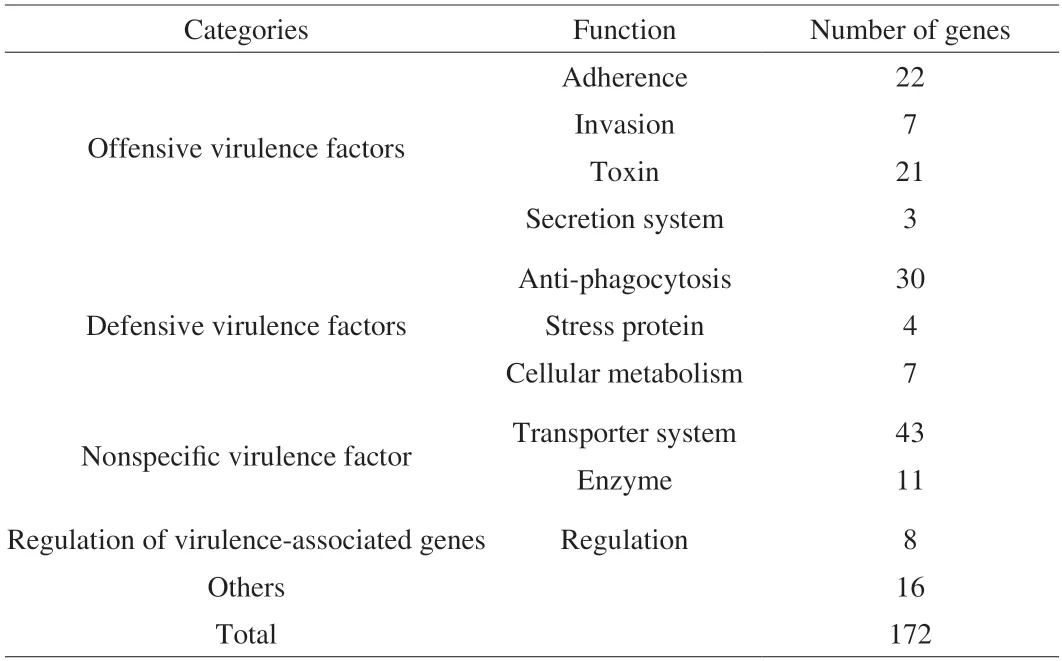
Table 4 Putative virulence genes of L.gasseri HMV18 predicted by VFDB 2005.
3.7 In vivo safety evaluation
All mice survived at the end of the experiment.During the investigation,all the mice in both groups were active.The general health of each mouse was in good condition,including pupil size,respiratory rate,stool color and hardness,skin integrity,eyelid,and eyeball.Each mouse’s average (FI was (5.88 ± 0.46) and (5.03 ± 0.55) g in the HMV18 and control group,respectively.Furthermore,all the mice in both groups showed similar growth patterns (P> 0.05)(Fig.2).
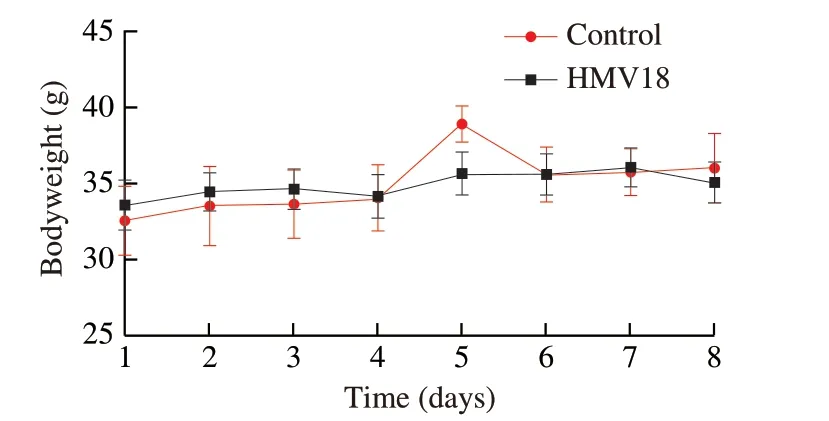
Fig.2 Body weight of mice during the experiment in the L.gasseri HMV18 gavaged group and the PBS control group.
Organ-to-body weight ratios are always used when body weight shifts significantly.It is more reliable than absolute organ weight in toxicity studies and useful in normalizing body weight due to nutritional status.Organs like the liver,kidneys,spleen,heart,and lungs are the most valuable to this testing[15].Notably,an autopsy found no distinct pathological change in the two groups’ overall appearance or size of internal organs.OIs of mice are shown in Table 5,and there was no significant difference between the two groups(P> 0.05).Thus,we conclude that there was no apparent organ toxicity in the mice gavagedL.gasseriHMV18.
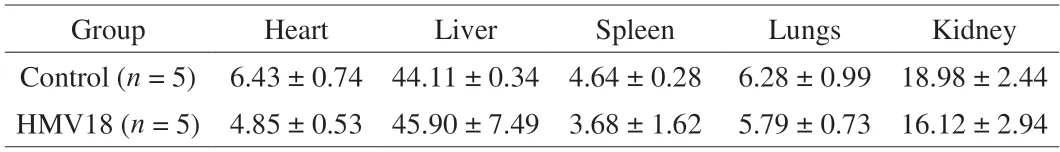
Table 5 OI of micea.
Representative microscopic images of histopathological changes in the heart,liver,spleen,lungs,and kidneys are shown in Fig.3.In the HMV18 treated group (similar with the control group),cell morphology and structure of all the tested organs remained intact,and cytoplasm and nuclei were uniform in appearance.Additionally,the intercellular matrix remains unchanged.There is no apparent inflammation,infiltration,or necrosis.Therefore,oral administration of HMV18 had no observable adverse effects on the organs of mice.
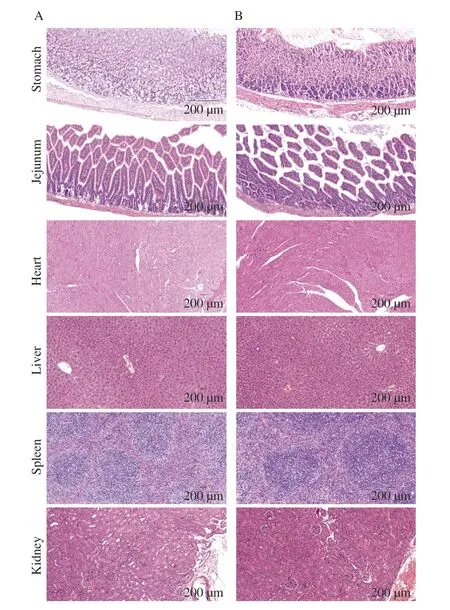
Fig.3 Histopathological images of mice’s organ sections in the control group (A) and HMV18 strain treatment group (B).Magnification: 200×.
3.8 Effects of HMV18 on gut microbiota
We sought to determine whether HMV18 modulated the structure and composition of the gut microbiota in mice by 16S rRNA sequence analysis.Theα-diversity,β-diversity and species composition were analyzed to explore the pattern of microbial variation.In this experiment,267 000 valid reads were obtained from 20 samples.The rarefaction curve and Shannon index curve were balanced as the number of reads increases,which indicated that enough sequencing data were obtained and the sequencing depth were reasonable.There were no significant differences between the control and HMV18 group with respect to theα-diversity of the microbiota as indicated by the observed_features,faith_pd,and evenness (Fig.4A).HMV18 administration by intragastrical gavage resulted in a modest increase inα-diversity as indicated by the faith_pd,in comparison to the control group,suggesting that HMV18 gavaged can boost the species richness and diversity of gut microbiota.
Next,the relative similarity of gut microbiota was visualized by PCoA followingβ-diversity analysis.As shown in Fig.4B,the PCoA plot of weighted UniFrac distance showed that the same treatment was clustered.PCoA1 and PCoA2 axis in PCoA diagram explained 58.96% of the overall difference after HMV18 gavaged (R=0.496,P=0.014).Overall,an apparent separation between control and HMV18 gavaged group was observed,which suggests that HMV18 shifted the structure of the microbial community.
To further understand the impact of HMV18 administration on the composition of gut microbiota,we examined the relative abundance of the predominant taxa within and among groups.The distribution of gut microbiota at the phylum,family and genus level in different treatments were analyzed.At the phylum level (Fig.4C),the gut microbiota was mainly composed of Bacteroidetes,Firmicutes,Campilobacterota,Verrucomicrobiota,Patescibacteria,Actinobacteria,Cyanobacteria,Proteobacteria and Desulfobacterota.Compared with the control group briefly,the abundance of Firmicutes,Campilobacterota,and Patescibacteria in all HMV18 treatments were increased,and the Firmicutes/Bacteroidetes (F/B)ratio (a key indicator of microbiota imbalance)[44]of the HMV18 group was significantly raised (P< 0.05).This variation of F/B ratio might be induced byL.gasseriHMV18 gavaged,which belongs to Firmicutes.At the genus level (Fig.4D),HMV18 treatment increased the abundance ofHelicobacterand Lachnospiraceae_NK4 A136_group,etc.,while decreased the abundance of Muribaculaceae andClostridia_UCG-014,etc.To research the key gut strains,the Lefse analysis was further conducted.ASVs with relative abundance greater than 0.5% were screened for analysis based on linear discriminant analysis (LDA) score.As shown in Fig.4E,there were 19 higher ASVs in HMV18-day7 (HMV18-d7) group,8 higher ASVs in HMV18-day0 (HMV18-d0) group,18 higher ASVs in control-day7(Con-d7) group and 9 higher ASVs in control-day0 (Con-d0) group.Bifidobacteriaceae,Coriobacteriales and Helicobacteraceae which belong to Actinobacteria and Oscillospirales,Monoglobaceae,Anaerovoracaceae,Peptostreptococcales,Clostridia_UCG_014andEubacterium_coprostanoligenes_group which belong to Firmicutes were key family in HMV18-day7 group.Besides,Rikenellaceae which belong to Bacteroidetes,and Helicobacteraceae which belong to Proteobacteria also enriched in HMV18-day7 group.However,after 7 days of intragastric administration of HMV18,no advantage ofLactobacillusabundance was observed in HMV18-day7 group,which was consistent with the results of other studies[16].TheLactobacilluscount was lowered in HMV18-day7 group,which was probably owing to the presence of antimicrobial agents,in particularly bacteriocins,in the intestine.Although HMV18 colonization was not observed,it modulated the structure of gut microbiota effectively.

Fig.4 Structure and composition of gut microbiota analysis by 16S rRNA sequence.(A) α-Diversity (observed_feature,shannon_entropy,faith_pd,pielou_evenness).(B) PCoA analysis,using the weighted normalized UniFrac metric,at the genus level.(C) Bacterial taxonomic profiling at the phylum level and the variation of F/B ratio.(D) Bacterial taxonomic profiling at genus level.(E) LEfSe analysis at family levels.Significantly different species among groups with LDA values greater than 2 are displayed.Data presented as mean ± SD.*P < 0.05 vs HMV18-d0 group.

Fig.4 (Continued)
3.9 A route map for safety assessment of probiotics for human food and medicine
To assess whether a new isolate is safe for probiotic application,we recruit 4 sections and list the detailed experiments in Fig.5.Section 1 is to identify the genus,species,and strain of the new isolate by 16S rRNA gene sequencing and compare its phylogenetic relationship with known isolates.We also verify its identity with biochemical tests,Gram staining and microscopy.The isolate is then deposited in the state strain collection center.Section 2 is to test the interaction between the isolate and the human host.We want to answer two questions: what is the beneficial effect of the isolate,and can it survive in the human GI tract? We artificial gastric and bile juices to test the bacterial survival count.Section 3 is the core of safety evaluation,which includesinvitroexperiments like hemolysis,antibiotic resistance,and its potential for horizontal transferring of antibiotic resistant gene,invivoanimal toxicity studies,genome-wide screening and validation of toxin genes,and literature review to check for any reported bacteremia cases.Section 4 provides some criteria for food and drug probiotics evaluation,including clinical trials and post-market feedback.
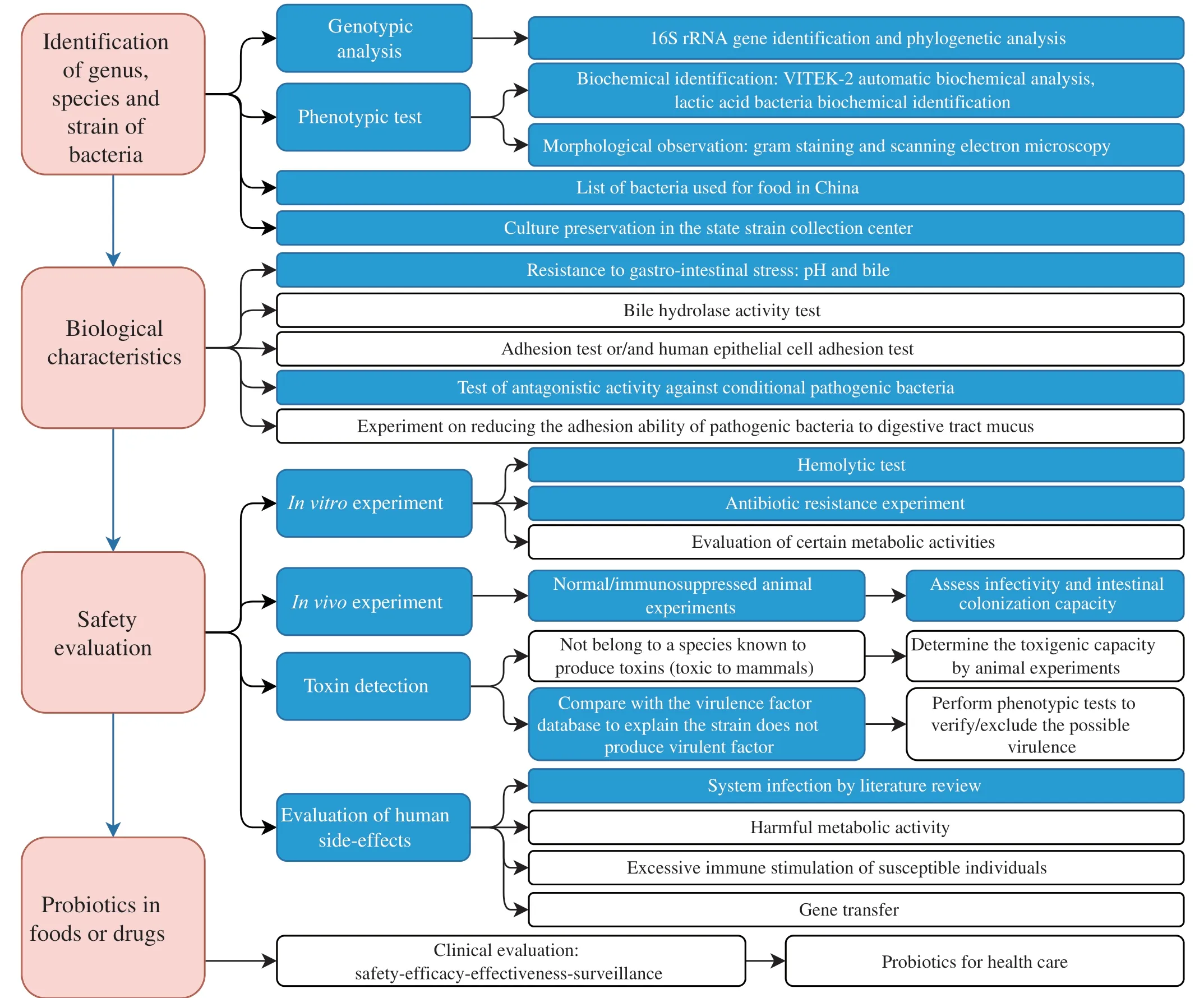
Fig.5 A summary for evaluating probiotics in foods and drugs for human use.Blue blocks indicate this publication’s completed experiments for the safety evaluation of L.gasseri HMV18.
4.Conclusion
L.gasseriHMV18 is one of the dominant species isolated from normal human flora.The pathogens inhibited byL.gasseriHMV18 are also common food-borne pathogens,suggesting the potential application ofL.gasseriHMV18 in foods.In reference to the FAO/WHO guidelines (FAO/WHO,2002) and published articles,we summarized the core experiments for the safety evaluation of probiotics in foods or drugs (Fig.5).In this study,we assessed thatL.gasseriHMV18 could resist the 2 h treatment of pH 3,4,and 8.Additionally,L.gasseriHMV18 resisted 3%,6%,and 8% of bile incubation for 3 h,was unable to produce biogenic amine,did not utilize mucin as a carbon source,did not encode verified toxins,and caused no significant change in the organs of mice.There were rarely clinical infections investigated and as few as 8 cases of were reported during 1984-2022.L.gasseriHMV18 administration by intragastrical gavage could modulate the structure and composition of gut microbiota.Therefore,this work provides evidence for the safe oral application ofL.gasseriHMV18 and lays the foundation for future research to assess the safety of probiotics.
Declaration of interests
Bin Cong is editor-in-chief and Yan Zhang is an associate editor forFood Science and Human Wellnessand were not involved in the editorial review or the decision to publish this article.The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was financially supported by postdoctoral funding of Hebei Medical University,Hebei Province Postdoctoral Research Project Funding (B2022003035),Natural Science Foundation of Hebei Province (H2020206579),CAMS Innovation Found for Medical Sciences (2019-I2M-5-055),2023 Scientific Research Projects of Colleges and Universities in Hebei Province (QN2023131),S &T Program of Hebei (18277743D),Undergraduate Innovation Experiment Project from Hebei Medical University (USIP2019008),and Spring rain project of Hebei Medical University (CYCZ201906).
Appendix A.Supplementary data
Supplementary data associated with this article can be found,in the online version,at http://doi.org/10.26599/FSHW.2022.9250052.
- 食品科学与人类健康(英文)的其它文章
- Protective effects of oleic acid and polyphenols in extra virgin olive oil on cardiovascular diseases
- Diet and physical activity inf luence the composition of gut microbiota,benef it on Alzheimer’s disease
- Inf luence of nitrogen status on fermentation performances ofnon-Saccharomyces yeasts: a review
- Ganoderma lucidum: a comprehensive review of phytochemistry,eff icacy,safety and clinical study
- Resveratrol combats chronic diseases through enhancing mitochondrial quality
- Cyanidin-3-glucoside protects the photooxidative damage of retinal pigment epithelium cells by regulating sphingolipid signaling and inhibiting MAPK pathway

