Mobile genetic elements facilitate the transmission of antibiotic resistance genes in multidrug-resistant Enterobacteriaceae from duck farms
Xin’er Zheng,Dingting Xu,Jinhng Yn,Min Qin,Peng Wng,Dvood Zeim,Jinzhong Hn,Dofeng Qu,
a School of Food Science and Biotechnology, Zhejiang Gongshang University, Hangzhou 310035, China
b The Second Aff iliated Hospital, School of Medicine, Zhejiang University, Hangzhou 311100, China
c Agricultural and rural Bureau of Wangdian Town, Xiuzhou District, Jiaxing 314011, China
Keywords: Duck farm Mobile genetic element Antibiotic resistance gene Plasmid Food safety
ABSTRACT Multidrug-resistant (MDR) Enterobacteriaceae critically threaten duck farming and public health.The phenotypes,genotypes,and associated mobile genetic elements (MGEs) of MDR Enterobacteriaceae isolated from 6 duck farms in Zhejiang Province,China,were investigated.A total of 215 isolates were identif ied as Escherichia coli (64.65%),Klebsiella pneumoniae (12.09%),Proteus mirabilis (10.23%),Salmonella (8.84%),and Enterobacter cloacae (4.19%).Meanwhile,all isolates were resistant to at least two antibiotics.Most isolates carried tet(A) (85.12%),blaTEM (78.60%) and sul1 (67.44%) resistance genes.Gene co-occurrence analysis showed that the resistance genes were associated with IS26 and integrons.A conjugative IncFII plasmid pSDM004 containing all the above MGEs was detected in Proteus mirabilis isolate SDM004.This isolate was resistant to 18 antibiotics and carried the blaNDM-5 gene.MGEs,especially plasmids,are the primary antibiotic resistance gene transmission route in duck farms.These f indings provide a theoretical basis for the rational use of antibiotics in farms which are substantial for evaluating public health and food safety.
1.Introduction
Duck meat is an essential source of proteins and other nutrients commonly consumed worldwide[1].China is one of the largest producers and consumers of duck meat;thus,duck breeding is a considerable part of its agriculture.However,industrial-scale production and dense stocking have led to the frequent emergence of contagious diseases,such as H9 avian influenza virus (AIV)and avian pathogenicEscherichiacoli(APEC).Although the extensive application of conventional antibiotics initially controlled such contagious diseases,the unmonitored and continued use of antibiotics has led to contamination of diversified environments,results in selective pressure on bacteria,and subsequently increases the prevalence of multidrug-resistant (MDR) strains,which are resistant to at least three different antimicrobial categories[2-3].These strains restrict the eff iciency of antibiotics,a critical challenge to the duck breeding industry.Most notably,the transmissibility of drugresistant agents from one pathogenic strain to others may lead to superbugs and intensify lethal bacterial infections.Thus,it is essential to monitor MDR strains in duck farms to prevent the spreading of bacterial diseases.Furthermore,there is an urgent need to reduce the anthropogenic burden of antibiotics in the environment and encourage their judicious use[4].
Nowadays,infections caused by antibiotic-resistant Gramnegative bacteria are emerging as a greater threat[5].Most MDR strains belong to conditional pathogenic bacteria,of which Gramnegative bacteria account for 71%,and Enterobacteriaceae make up more than 53% of them[6].Moreover,MDRs have been increasingly observed in Enterobacteriaceae[7-9].E.coli(a common Enterobacteriaceae) has the most significant probability of animalhuman transmission and is a crucial microorganism of communityassociated antibiotic resistance[10].The prevalence of antibioticresistantE.coliin duck farms has been previously reported[11-13].Another common Enterobacteriaceae isProteusmirabilis,which can cause opportunistic infections.P.mirabilisis generally found in natural environments like soil and water[14].
MDR variants are usually created through horizontal gene transfer.Antibiotic resistance genes (ARGs) are sometimes accumulated by mobile genetic elements (MGEs) in one cell[15].MGEs contain insertion sequences (IS),transposons (Tn),plasmids,integrons,etc.For example,ISAba1withblaOXA-51-like genes inAcinetobacter baumanniirepresents carbapenem resistance[16].Plasmids are the primary vehicles for carrying ARGs.For instance,Inc18-like mosaic plasmid pEF-01,the first identified inEnterococcusfaecalisto carry thecfrgene,can confer resistance to multiple antimicrobial classes,including oxazolidinones,phenicols,and lincosamides[17].Integrons are another type of MGEs containing multiple ARGs.The integrase gene (int) is one of the critical elements of integrons,which are the major vehicle for spreading multi-drug resistance and are often carried by plasmids[18].Classification of integrons into classes 1,2,and 3 are based on the integrase genesintI1,intI2,andintI3[19].
Duck farms are a vital upstream stage in the supply chain of duck meat,and antibiotics are prevalently used to control pathogens in them.Overuse of antibiotics may facilitate the evolution of MDR strains.Moreover,domestic ducks can act as potential vehicles for resistant bacteria that may spread ARGs.Duck wastes are also a potential reservoir for antibiotic resistance genes[20-21].Therefore,duck farms may play a significant role in generating and spreading MDR strains.The prevalence and molecular characteristics of Enterobacteriaceae carrying ARGs have been identified in many Chinese duck flocks.Moreover,the characteristics of MDR and ARGs have shown diversity in different places[13,22].There is a significant lack of data on the prevalence and molecular characteristics of Enterobacteriaceae bearing ARGs in Zhejiang duck farms.This study aimed to isolate Enterobacteriaceae strains from duck farms in Zhejiang,China and analysis their drug resistance.In order to investigate the transferability of ARGs,the characteristics and conjugation performance of plasmids extracted from the MDR strains were analyzed.Surging drug resistance in animal pathogens is a big challenge to food safety and public health.This study provides a theoretical basis for detecting and controlling antibiotic resistance.
2.Materials and methods
2.1 Sample collection, isolation, and identification of strains
Ninety-six duck fecal samples were collected from 6 commercial duck farms in the Zhejiang Province of China in 2020.Feces were sampled from individual droppings.These samples were subsequently transported to the laboratory with ice bags within 6 h for bacteriological analyses.Samples were enriched in brainheart infusion broth and streaked on MacConkey plates.Putative colonies,depending on colors and morphological characteristics of Enterobacteriaceae,were sub-cultured on nutrient agar for purification.Then,isolates were identified by 16S rRNA sequencing with universal primers (Table S1).
2.2 Antibiotic susceptibility testing
Kirby-Bauer disc diffusion method was employed to check the susceptibility of isolates to 23 common antibiotics.Mueller-Hinton(MH) agar plates were used to determine antibiotic susceptibility[23].After 18-24 h incubation,antibiotic resistance properties were studied according to Clinical Laboratory Standards Institute(CLSI) breakpoint values[24].The following antibiotics were tested:tetracycline,minocycline,erythromycin,azithromycin,ciprofloxacin,enrofloxacin,ofloxacin,amoxicillin,imipenem,meropenem,cefazolin,aztreonam,cefoxitin,ceftazidime,oxacillin,streptomycin,gentamicin,amikacin,kanamycin,trimethoprim,sulfamethoxazole,chloramphenicol,and fosfomycin.
2.3 Detection and correlation analysis of ARGs and MGEs
Bacterial DNA was extracted using Tianamp Bacteria DNA Kit(Tiangen biochemical technology (Beijing) Co.,Ltd.) and stored at-20 °C.Polymerase chain reaction (PCR) and sequencing were exploited to detect ARGs and MGEs in all isolates.Amplicons of ARGs and MGEs were extracted from the gel using a Gel Extraction Kit (Thermo Fisher Scientific).Sequencing was performed by Sangon Biotech Co.,Ltd.(Shanghai,China).DNA sequences were compared with published DNA sequences using BLAST.The primers are presented in Table S1.Data analysis was carried out by Origin 2021 and SPSS Statistics version 26.0.Co-occurrence and contributor networks of ARGs and MGEs were analyzed by R software(X64 3.5.1) with the package of “psych” and “vegan” based on Spearman’s rank correlations (P≤ 0.05 indicating significant differences) and visualized by Gephi.
2.4 Conjugation assay
The plasmid transferability between an MDRProteusmirabilisstrain (SDM004) and the rifampin-resistantE.coliEC600 was examined by the conjugation assay.TheP.mirabilisstrain was used as a donor in conjugation experiments,which showed high resistance to 18 antibiotics.Briefly,donor and recipient cells were mixed and incubated overnight at 37 °C.Transconjugants were detected on Muller-Hinton (MH) plates containing 1 000 µg/mL rifampin and 20 mg/mL ampicillin,then confirmed by the 16S rRNA assay[25-26].
2.5 Plasmid sequencing
Plasmids were isolated from transconjugants with a plasmid extraction kit (Axygen,Hangzhou,China).Sequencing and assembly were performed by Sangon Biotech Co.,Ltd.(Shanghai,China).Nucleotide sequencing reactions were done with a paired-end library with an average insert size of 300 bp on a HiSeq sequencer(Illumina,San Diego,CA,USA).The filtered clean reads were assembled using Velvet 1.2.10[27].Contig assembly and predicted gaps were confirmed and filled by PCR amplification and Sanger sequencing[28-29].The GapCloser module from SOAPdenovo software was then used for the gap closure[30].
2.6 Sequence annotation and comparison
BLASTn was used to identify similar plasmid sequences.Furthermore,open reading frames and pseudogenes were predicted using RAST 2.0 with default parameters and verified with BLASTP/BLASTN searches against the UniProtKB/Swiss-Prot and RefSeq databases[31-34].Resistance genes were annotated using Resfinder 4.0[33,35].Mobile elements were annotated with ISfinder[36],INTEGRALL 1.2,and Tn Number Registry[37-38].The Inc group was classified by Plasmidfinder[39].Results were graphically depicted by SnapGene (http://www.snapgene.com/),Adobe Illustrator (https://www.adobe.com/it/) and BRIG (http://brig.sourceforge.net/).Plasmid sequences were then submitted to the NCBI database with accession numbers: ON843497.The control plasmids were pHKU49_CIP (GenBank No.MN543570) and pKPC2_095132(GenBank No.CP028389),which have the highest similarities in comparison with others.
3.Results
3.1 Antibiotic resistance phenotypes of Enterobacteriaceae
In total,215 Enterobacteriaceae isolates were identified in duck fecal samples.All 215 isolates were resistant to at least two antibiotics,and 186 (86.51%) isolates were MDR.PCR identification of 215 presumptive isolates resulted in a higher detection frequency forE.coli(64.65%),followed byKlebsiellapneumoniae(12.09%),Proteusmirabilis(10.23%),Salmonella(8.84%),andEnterobacter cloacae(4.19%).Notably,P.mirabilisSDM004 recovered from feces was resistant to 18 antibiotics,including ciprofloxacin,enrofloxacin,erythromycin,azithromycin,tetracycline,minocycline,gentamicin,streptomycin,amikacin,kanamycin,trimethoprim,sulfamethoxazole,amoxicillin,cefazolin,cefoxitin,ceftazidime,oxacillin,and chloramphenicol.Moreover,Enterobacteriaceae isolates were mainly resistant to 6 antibiotics: tetracycline (86.05%),oxacillin (80.93%),amoxicillin (73.49%),trimethoprim (70.23%),sulfamethoxazole(73.95%),and chloramphenicol (70.70%).Low resistance to imipenem (3.26%) and meropenem (4.19%) was detected (Fig.1).

Fig.1 The resistance rate to 23 antibiotics of the Enterobacteriaceae strains isolated from duck farms.
3.2 Antibiotic resistance genotypes of Enterobacteriaceae
Twenty-nine ARGs were identified in 215 Enterobacteriaceae isolates.Except formcr-2,mcr-3,andmcr-4genes,all the rest ARGs tested were detected.Overall,the detection rates of resistance genestet(A) (85.12%),blaTEM(78.60%),sul1(67.44%) were the highest in 215 isolates (Fig.2).On the contrary,carbapenems resistance geneblaNDM-5andblaNDM-1displayed a lower detection rate,with 1.86%and 0.93%,respectively.The colistin resistance genemcr-1was also exhibited low prevalence,detected in only 2.79% of isolates(Fig.2).Other detection rates of common ARGs to tetracyclines,β-lactams,sulfonamides,quinolones,macrolides,chloramphenicol,and fosfomycin were ranged from 11.16% to 67.44%.The detection rates for others were as follow:fosX(4.65%),erm(B) (7.91%),erm(C) (4.65%),tet(D) (7.91%),tet(E) (4.65%),andqnrB(9.30%)(Fig.2).Multiple ARGs were identified in SDM004 strain includingβ-lactams (blaTEM,blaNDM-5,blaCTX-M),tetracyclines (tet(A),tet(B),tet(C)),quinolones (qnrA,qnrS),aminoglycosides (aadA1,aph(3’)-Ia,aac(6’)-Ib-cr),sulfonamides (sul1,sul2),chloramphenicol (floR),and macrolides (erm(A),erm(B)).

Fig.2 Distribution of 29 ARGs of the Enterobacteriaceae strains isolated from duck farms.
3.3 Co-occurrence analysis of ARGs and MGEs
As shown in Fig.3,the gene co-occurrence analysis of the ARGs network implied a close correlation between ARGs and MGEs (only correlations withP< 0.05 are displayed).The resistance genes were significantly associated with the integrase genesintI1andintI2.TheintI1gene was mostly co-occurred withsul1,tet(A),tet(B),tet(C),tet(D),tet(E),qnrS,aadA1,floR,andblaTEMgenes.These genes respectively conferred resistance to sulfonamides,tetracyclines,quinolones,aminoglycosides,chloramphenicol,andβ-lactams.TheintI2gene mostly co-occurred withsul1,sul2,tet(A),tet(B),tet(C),tet(D),tet(E),floR,andblaTEM.These resistance genes were all detected at relatively higher levels,especiallytet(A),sul1,andblaTEM,which were significantly associated withintI1(represented class 1 integrons) and IS26(IS26belonged to IS6family elements).Besides,mcr-1,andblaNDM-1genes,with a relatively lower detection rate,were associated with MGEs such as IS26.The network demonstrated a complex connection between the ARGs of tetracyclines (tet(A),tet(B),tet(C),tet(D)),β-lactams (blaCTX-M,blaTEM),sulfonamides (sul1,sul2),chloramphenicol (floR),aminoglycosides (aadA1,aph(3’)-Ia),and quinolones (qnrS,qnrA,andqnrB).

Fig.3 Co-occurrence gene network of ARG subtypes and MGEs in MDR isolates collected.11 different colors represent 9 kinds of antibiotics and mobile elements.Nodes belonging to the same kinds of MGEs or resistant to the same class of antibiotics are presented in the same color.Only correlations with P < 0.05 are displayed.
3.4 Overview of a representative conjugative plasmid pSDM004 containing a large variety of ARGs and MGEs
Conjugation assays confirmed that the plasmid with antibioticresistance genes could be transferred fromP.mirabilisSDM004 toE.coliEC600.Notably,the plasmid type was identified as IncFII by the plasmid replicon analysis.This conjugative plasmid,called pSDM004,was further analyzed.The plasmid pSDM004 was 135 720 bp in size with a backbone composed of conjugal transfer,maintenance,and replication regions (Fig.4).The accessory modules that acquire DNA regions associated with the adjacent mobile elements were inserted at different backbone locations.A pairwise sequence comparison using BLASTN showed that the pSDM004 exhibited the highest sequence identity to the plasmid pHKU49_CIP and pKPC2_095132 (Fig.4).pHKU49_CIP was isolated fromK.pneumoniae,and pKPC2_095132 was obtained from a hospital.A comparative analysis was conducted among pSDM004,pHKU49_CIP,and pKPC2_095132.Plasmid pHKU49_CIP and pKPC2_095132 are circular DNA sequences of 140.57 and 166.03 kb in size,respectively (Fig.S1a).According to Fig.S1b,the backbones of pSDM004,pHKU49_CIP,and pKPC2_095132 are almost identical (97% BLAST coverage and 99% nucleotide identity).In addition,3 plasmids share the major resistance genes:floR,arr-3,tet(A),mph(A),dfrA27,aac(6’)-Ib-cr,qnrB2,blaTEM-1b,sul1,aac(3)-IId,aadA16,andaph(3’)-Ia.The plasmid pSDM004 possessed an approximately 46 kb MDR region,which contained 5 major MGEs modules,including ISKpn19-aac(3)-IIdunit,qnrB2-sul1-IS6100unit,ΔTn2-related region,In1021region,and IS26repeat region (Fig.4).The MDR region of pHKU49_CIP contains the ISKpn19-qnrS1unit,ΔISEcp1-CTX-M unit,ΔTn2-related region,In1021,and IS26repeat region.pKPC2_095132 harbors ΔISKpn6-KPC-2unit,qnrB2-sul1-IS6100unit,ΔTn2-related region,In1021region,and IS26repeat region (Fig.S1a).
3.5 Major modular differences among pSDM004, pHKU49_CIP, and pKPC2_095132

Fig.4 DNA alignment of plasmids compared in the study.The concentric circles represent BLAST comparisons of pHKU49_CIP (light green) and pKPC2_095132(dark blue) against the pSDM004 (pink).The annotated coding DNA sequence (CDS) of the pSDM004 is shown in the outer circle,with the plasmid maintenance(red),replication initiation encoding genes (teal),conjugal transfer module (blue),resistance genes (purple),and mobile genetic elements (gray) highlighted.Color codes for DNA identity range from 100% to 50% as indicated.
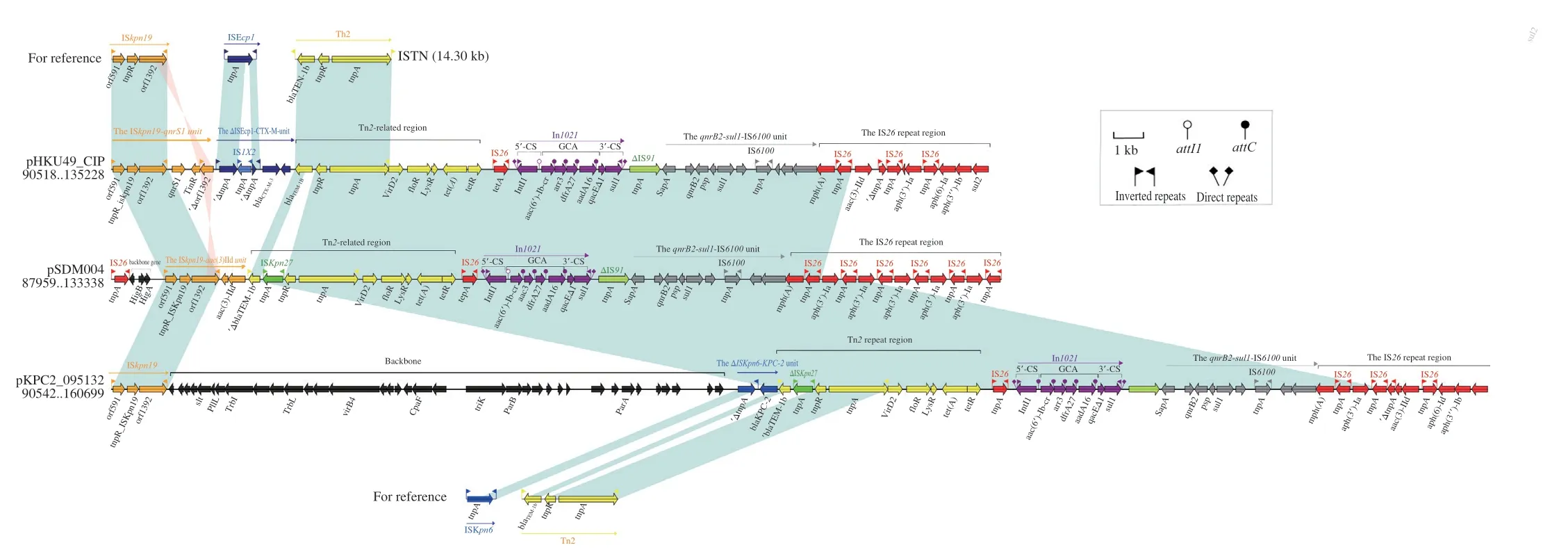
Fig.5 Accessory resistance regions from pSDM004,pHKU49_CIP,and pKPC2_095132.Arrows denote genes.Based on function classification,genes,mobile elements,and other features are colored.Shading denotes regions of homology (> 95% nucleotide identity).
As demonstrated in Fig.5,different structures were observed in ISKpn19and downstream.In pSDM004,ISKpn19carriedaac(3)-IIdconstituting a single unit followed by the Tn2 region.In pHKU49_CIP,an inversed ISKpn19remnant,carryingqnrS1and ISEcp1containingblaCTX-M-3,existed in the adjacent area.pKPC2_095132 includes another new complex region containing truncated ISKpn19,Slt,PilL,TrbL,Vir84,CpaF,triK,parB,andparAin the length of nearly 31 kb.The IS26repeat region,associated with genes responsible for resistance to aminoglycosides,macrolides,and sulfonamides,was at the end of all plasmids.In pSDM004,multiple resistance genes were bracketed by IS26,includingaph(3’)-Ia,tnpA,andmph(A).However,ARGs in the IS26repeat region of pHKU49_CIP and pKPC2_095132 showed some diversities inaph(3’)-Ia,aac(3)-IId,aph(6)-Id,aph(3’’)-Ib,andsul2.
4.Discussion
Enterobacteriaceae originated from livestock and humans,and they are a significant reservoir for ARGs.These resistance genes might be transferred from livestock to humans through the food chain.Due to the prevalence of waterfowl bacterial infections and the extensive use of antibiotics,the spreading of MDR Enterobacteriaceae containing MGEs has accelerated in duck farms and industry[40].
The phenotypes,genotypes,plasmids,and related genes of antibiotic-resistant Enterobacteriaceae isolated from 6 Zhejiang duck farms were studied.Most tested Enterobacteriaceae strains (186/215,86.51%) were MDR.The resistance rates to tetracycline,oxacillin,amoxicillin,trimethoprim,sulfamethoxazole,and chloramphenicol were higher than 70%.Most of these 6 antibiotics were categorized as “high important antimicrobials” for humans by the World Health Organization (WHO)[41].In a similar study,Salmonellastrains were isolated from duck farms in Qingdao,Jinan,and Zibo regions,China.The strains showed strong resistance to tetracycline and ciprofloxacin with drug resistance rates of 92.9% and 80.4%,respectively.MostSalmonellaisolates (89.3%) exhibited resistance to multiple drugs[42].E.colistrains isolated from duck farms in Guizhou Province,China showed a high amoxicillin resistance rate (100%)[43].Among 165E.colistrains isolated from duck farms in Sichuan Province,China,163 (98.8%) exhibited MDR.Most isolates revealed high resistance to ampicillin (95.8%),chloramphenicol (89.7%),and trimethoprimsulfamethoxazole (84.2%)[40].These results indicate that controlling multi-drug resistance in Enterobacteriaceae in duck husbandry requires particular vigilance.
In the current study,the most common resistance gene wastet(A),found in 183 (85.12%) strains,followed byblaTEM(78.60%)andsul1(67.44%).The prevalence of antibiotic resistance and ARGs implied the presence of MGEs for the capture,accumulation,and spread of genes.tet(A) gene is mainly associated with nonconjugative transposons.blaTEMgene is the most prevalent resistant gene toβ-lactamase worldwide and is principally known to be spread by plasmids[44].sul1is always associated with class I integrons.Three genes with the highest detection rates,tet(A),sul1,andblaTEM,significantly co-occurred with IS26and integrons.Integrons can capture and mobilize gene cassettes encoding multiple ARGs.They utilize site-specific recombination to transport resistance genes among strains[45].The integrase genesintI1andintI2were mostly co-occurred with 11 resistance genes (aadA1,sul1,sul2,tet(A),tet(B),tet(C),tet(D),tet(E),blaTEM,floR,qnrS),which were detected more than other MGEs.However,we did not detect theintI3gene representing class 3 integrons.Although class 3 integrons are far less common than class 1 and 2 integrons,they have been detected in many clinical and poultry samples[46-47].ISs are MGEs that can move independently and bestow their associated resistance genes randomly to new locations on the same or different DNA molecules.Thus,ISs play a vital role in the rapid evolution of diverse MDR pathogens.As previously reported,these insertion sequences and integrons were often associated with plasmids[48-49].It is noteworthy that a conjugative plasmid pSDM004 was acquired fromP.mirabilisSDM004.This strain was resistant to 18 antibiotics.This isolate also carriedblaNDM-5gene.MostblaNDMare located on plasmids,favoring their spreading[50].However,plasmid sequencing demonstratedblaNDM-5gene was not located on this plasmid,and it was assumed that the gene was mainly located on the chromosome[51-52].Plasmid sequencing also revealed comprehensive genetic contexts of MGEs carried by pSDM004.The other two plasmids pHKU49_CIP and pKPC2_095132 were identical to pSDM004.
Complementary analysis of these three plasmids showed some similarities (such as ΔTn2-related region andqnrB2-sul1-IS6100unit),although several significant differences existed.ISKpn19containedaac(3)-IIdin pSDM004 andqnrS1in pHKU49_CIP.However,it included no resistance genes in pKPC2_095132(Fig.5).Carbapenem-resistantSerratia marcescensfrom hospital resources carriedblaKPC-2-harbouring plasmids,which contained MGEs comprisingqnrS1and ISKpn19[53].The combination of similar genesqnrS1and ISKpn19was also found in other plasmids,such asK.pneumoniaeplasmids pK1HV (HF545434,human,Vietnam) and pNDM-1fa (CP014757,human,USA),E.coliplasmids pPGRT46 (KM023153,human,Nigeria) and pKT58A(JX065631,waterfowl,Slovakia).These findings indicated thatqnrS1gene was related to ISKpn19.In addition to integrons,sul1was carried by IS6100(IS6family).The IS6100has a broad range[54-55].Co-occurrence of IS6100andsul1genes may contribute to the high prevalence ofsul1in the isolates.Interestingly,in pSDM004,another resistance geneaac(3)-IIdwas carried by ISKpn19and linked to the IS26upstream,an outcome of gene recombination mediated by IS26.IS26is the standard component of antibiotic resistance genes.Homologous recombination can occur through IS26,which is more important for creating new MDR regions[56].Besides,the accessory resistance regions were ended with the IS26repeat region in all tested plasmids.The region harbors some genes that encode aminoglycoside resistance.The repeating of IS26in the region is due to the movement mode.A copy of IS26and an adjacent region are inserted next to an existing copy of IS26.Thus,a plasmid containing one copy of IS26is prone to acquire other adjacent IS26units[57].
5.Conclusion
Multi-drug resistance and ARGs were prevalent and diverse in duck farms of Zhejiang,China.Resistance genestet(A) andblaTEM,which had a detection rate above 70%,were significantly associated with integrons and IS26.These MGEs were often carried on plasmids,vital for their spreading.The co-existence of various ARGs on a single plasmid could spread quickly and resist different antibiotic selection pressures.Information on the distribution of MGEs provides a better understanding of the transfer of ARGs to commensal bacteria,reducing the risk to public health and food safety.Monitoring the occurrence and distribution of ARGs and MGEs is essential to shed some light on the complex mechanism of multi-drug resistance.Currently,available information is limited in part by the sample size and the sampling points.More samples and bacterial species are needed for further analysis to obtain a comprehensive antibiotic resistance profile and the prevalence of ARGs and MGEs in the duck production chain.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (32172188) and Science and Technology Cooperation Project of ZheJiang Province (2023SNJF058-3).
Appendix A.Supplementary data
Supplementary data associated with this article can be found,in the online version,at http://doi.org/10.26599/FSHW.2022.9250062.
- 食品科学与人类健康(英文)的其它文章
- Protective effects of oleic acid and polyphenols in extra virgin olive oil on cardiovascular diseases
- Diet and physical activity inf luence the composition of gut microbiota,benef it on Alzheimer’s disease
- Inf luence of nitrogen status on fermentation performances ofnon-Saccharomyces yeasts: a review
- Ganoderma lucidum: a comprehensive review of phytochemistry,eff icacy,safety and clinical study
- Resveratrol combats chronic diseases through enhancing mitochondrial quality
- Demonstration of safety characteristics and effects on gut microbiota of Lactobacillus gasseri HMV18

