Antibacterial mechanism of kojic acid and tea polyphenols against Escherichia coli O157:H7 through transcriptomic analysis
Yilin Lin,Ruifei Wng,Xioqing Li,Keren Agyekumw Addo,Meimei Fng,Yehui Zhng,Yigng Yu,c,
a School of Food Sciences and Engineering, South China University of Technology, Guangzhou 510640, China
b Sericulture &Agri-food Research Institute Guangdong Academy of Agricultural Sciences, Key Laboratory of Functional Foods,Ministry of Agriculture and Rural Affairs, Guangdong Key Laboratory of Agricultural Products Processing, Guangzhou 510610, China
c Research Center of Food Safety and Detection, College of Food Science and Engineering, South China University of Technology, Guangzhou 510640, China
Keywords: Kojic acid Tea polyphenols Antibacterial mechanism Escherichia coli O157:H7 RNA-seq
ABSTRACT Escherichia coli O157:H7 is one of the major foodborne pathogenic bacterial that cause infectious diseases in humans.The previous found that a combination of kojic acid and tea polyphenols exhibited better activity against E.coli O157:H7 than using either alone.This study aimed to explore responses underlying the antibacterial mechanisms of kojic acid and tea polyphenols from the gene level.The functional enrichment analysis by comparing kojic acid and tea polyphenols individually or synergistically against E.coli O157:H7 found that acid resistance systems in kojic acid were activated,and the cell membrane and genomic DNA were destructed in the cells,resulting in “oxygen starvation”.The oxidative stress response triggered by tea polyphenols inhibited both sulfur uptake and the synthesis of ATP,which affected the bacteria's life metabolic process.Interestingly,we found that kojic acid combined with tea polyphenols hindered the uptake of iron that played an essential role in the synthesis of DNA,respiration,tricarboxylic acid cycle.The results suggested that the iron uptake pathways may represent a novel approach for kojic acid and tea polyphenols synergistically against E.coli O157:H7 and provided a theoretical basis for bacterial pathogen control in the food industry.
1.Introduction
Escherichia coliO157:H7,one of the most common foodborne bacterial pathogens,can infect the human body through contaminated food at low infective doses to cause hemorrhagic diarrhea,enteritis,and even hemolytic uremic syndrome[1-2].According to the center for disease control and prevention,this bacterium annually infected 20 000 people causing life-threatening conditions[3].E.coliO157:H7 strongly tolerates many food-related environmental stresses,such as acid stress,low temperatures,osmotic stress,and poor nutrition[3].Any food and beverage that may contact animal feces is a potential source ofE.coliO157:H7 outbreaks that signif icantly contaminate beef[4-5].Thus,many researchers are currently trying to f ind antimicrobials to controlE.coliO157:H7 and other pathogens.
Recently,many natural antimicrobial agents from plant,animal,or microbial fermentation have been developed with the insecurity of synthetic antibacterial agents and the increasing demand of consumers for green antibacterial agents[6].Kojic acid (5-hydroxy-2-hydroxymethyl-4H-pyran-4-one,KA),a natural product byAspergillusandPenicilliumand existing in many fermented foods such as soy sauce,exhibits inhibitory effects on a wide range of bacteria,especially gram-negative bacteria[7].Previous studies already showed KA combined with grafted chitosan oligosaccharides could be employed to treatStreptococcus pyogenes,Staphylococcusaureus,andSalmonella typhimurium[8].In addition,KA has been recognized as a natural food additive in the food industry for preventing enzymatic due to its exciting bioactivities[9-11].Similarly,tea polyphenols (TP),as natural polyphenols with many beneficial physiological activities such as antitumor and antioxidation,have also been proved to have broad-spectrum antibacterial activities,serving as a promising candidate to replace synthetic preservatives[12].A range of studies has demonstrated that TP has inhibited biofilm formation of Campylobacter Jejuni[13],drug-resistant ofS.aureus[14]and preservation of fish and meats in the food industry fields[15-16].Then,the growth of gram-positive bacteria was more easily inhibited than gram-negative bacteria by TP treatment[17].
The KA and TP againstE.coliO157:H7 have caused varying degrees of cell membrane integrity destruction,genomic DNA degradation and affected biofilm formation[18].Some studies have expounded the antibacterial mechanism of TP or its main components(epigallocatechin gallate,etc.) against various bacteria by phenotypic experiments,including membrane damage,inhibition of critical enzymes,and induction of oxidative stress[19-20].Moreover,the responses ofPseudomonas aeruginosa[21]andS.aureus[22]to TP stress have also been revealed by transcriptomics.However,the response ofE.coliO157:H7 to TP at the transcriptional level has not been elucidated.On the other hand,the antibacterial mechanisms of KA and its derivatives have only been reported in limited literature,and no transcriptomics study on bacterial response to KA has been reported until now.Besides,our previous study has stated that a combination of kojic acid and tea polyphenols exhibited better againstE.coliO157:H7 than using either alone.KA and TP exhibited synergistic inhibitory effects on gram-negative pathogens and spoilage bacteria based on their different subcellular targets and mechanisms,showing a good application prospect in aquatic products[18].Therefore,a deeper understanding of about the antibacterial mechanisms and molecular targets of KA combined with TP on bacteria is required for their practical applications as disinfectants or preservatives in the food industry.
Thus,the objectives of this study were to reveal the responses ofE.coliO157:H7 to the stresses of KA and TP individually or synergistically by transcriptomic analysis and to indirectly elucidate the antibacterial mechanisms of KA and TP againstE.coliO157:H7 and the reason behind their synergistic inhibitory effect,which will promote their application in the food industry.
2.Materials and methods
2.1 Bacterial strain and chemicals
E.coliO157:H7 ATCC 35150 was obtained from the American Type Culture Collection (Manassas,USA),and it was stored in tryptic soy broth (TSB) with 25% glycerol at -80 °C before use.The bacterium was activated by incubation at 37 °C for 24 h after streaking the stock culture onto tryptone soy agar (TSA).Then a single colony from TSA was inoculated into TSB to reach a stationary phase after incubation at 37 °C for 18 h.Stock solutions of kojic acid (KA,99%;Aladdin Reagent Co.,Ltd.,Shanghai,China),tea polyphenols (TP,polyphenol content ≥ 98%,EGCG content ≥ 50%;Hongxing Pharmaceutical Co.,Ltd.,Anhui,China) were prepared just before use and briefly stored at 4 °C.
2.2 Sample preparation
According to the previously determined sublethal concentrations of KA and TP againstE.coliO157:H7,four sets of samples were selected for transcriptomic analyses to explore the antibacterial mechanisms of KA,TP,and their combination[18].The bacterial suspensions were treated with: 1) CK: sterile TSB without antibacterial substances;2) T1: sterile TSB with 1/2 MIC KA;3) T2: sterile TSB with 1/2 MIC TP and 4) T3: sterile TSB with 1/5 MIC KA+1/5 MIC TP,and then incubated at 37 °C for 8 h.After incubation,the cell suspensions were centrifuged at 8 000 ×gfor 3 min to remove the culture medium.The obtained cell pellets were immediately inactivated with liquid nitrogen and temporarily stored at-80 °C before the following RNA extraction.
2.3 RNA extraction and cDNA library construction
Firstly,the appropriate bacterial pellets were fully ground in liquid nitrogen to ensure that bacterial cells could be rapidly and sufficiently lysed in the subsequent process.According to its operation manual,total RNA was extracted using the TRIzol reagent (Invitrogen,Carlsbad,USA).Briefly,the intracellular substances were released from bacterial cells under the treatment of TRIzol reagent.Total RNA was separated from the DNA and proteins after the addition of adding RNA was precipitated by isopropanol and ethanol (75%,V/V) and then dried under vacuum.The quality and integrity of total RNA were assessed using an Agilent 2100 Bio-analyzer (Agilent Technologies,Palo Alto,USA) and RNase-free agarose gel electrophoresis.After removing rRNA from total RNA by Ribo-ZeroTM Magnetic Kit(Epicentre,Madison,USA),the total mRNA was fragmented into short fragments with 200-700 nt using fragmentation buffer.The mRNA fragments were reverse transcripted into cDNA with random hexamer primers,and the second strand of cDNA was synthesized with dNTP to promote DNA polymerase I.The synthesized cDNA fragments were purified using a QiaQuick PCR extraction kit(Qiagen,Venlo,the Netherlands) and then subjected to end repair,the ligation of poly(A) and sequencing adapters.The ligated cDNA fragments were size screened by the agarose gel electrophoresis.Finally,the selected cDNA fragments were amplified and sequenced using Illumina HiSeq2500 by Gene Denovo Biotechnology Co.(Guangzhou,China).
2.4 Read alignment and normalization of gene expression levels
The quality trimming and adaptor removal of raw reads were conducted using in-house Perl scripts to obtain clean reads with high quality according to the principle (i.e.,removing reads containing adapters,more than 10% of unknown nucleotides (N) and more than 50% of low quality (Q-value ≤ 20) bases).Short reads alignment tool Bowtie2 was used to exclude the reads mapped to the rRNA database[23].The remaining clean reads were then mapped to theE.coliO157:H7 str.Sakai genome (NCBI accession: NC_002695.2,NC_002128.1 and NC_002127.1) using TopHat2[24].Based on our previous method,the gene expression levels were normalized using StringTie software[25].In brief,the gene expression abundance was quantified using fragments per kilobase of transcript per million(FPKM) method.
2.5 Differentially expressed genes (DEGs) and function enrichment
The edgeR package identified differentially expressed genes between the samples[26].The transcripts' false discovery rate (FDR)and fold change (FC) were calculated.The criteria of significant differential expression were FDR ≤ 0.01 and the absolute value of log2FC ≥ 2.DEGs functional enrichment was performed using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases.Significantly enriched GO terms and KEGG pathways were obtained with the criteria of an adjustedP-value ≤ 0.05.
2.6 Validation of RNA-seq data by qRT-PCR
The quantitative real-time reverse transcription-PCR (qRTPCR,Applied Biosystem,Foster,USA) was used to identify and validate the RNA-Seq data.Forward and reverse PCR primers for nine genes were designed using Primer3 software and are listed in Table 1.We regarded the E-16S rRNA gene ofE.coliO157:H7 as the internal control gene.The total RNA was extracted according to the manufacturer's instructions,and cDNA was synthesized by NEB 7530 kit (New England Biolabs,USA).All qRT-PCR reactions were performed at 45 °C for 10 min,then 95 °C for 2 min,and 45 consecutive cycles consisting of 95 °C for 5 s,and 60 °C for 10 s,and 72 °C for 5 s.Each target gene was performed in triplicate using total RNA as the starting material.
3.Results
3.1 Data processing and analysis
After RNA-seq using the Illumina sequencing platform,3 236 715 600,3 150 001 800,2 318 529 900 and 3 115 076 100 raw reads were obtained from the CK,T1,T2 and T3 sample,respectively.The clean reads were harvested by strict quality control and data filtration,and CK,T1,T2 and T3 groups yielded 3 134 015 299(96.83%),3 022 657 995 (95.96%),2 227 762 714 (96.09%) and 2 938 529 294 (94.33%) clean reads correspondingly.
The Q20 quality scores of the 4 transcriptomes were all above 98%,and the GC contents were all above 52%.After further removing the rRNA sequence reads,the remaining reads were mapped to the reference genome,which contained 4 802,5 015,4 754 and 4 884 unigenes,respectively.Accordingly,the gene detection ratio of all genes was 90.76%,94.78%,89.85% and 92.31% for CK,T1,T2 and T3,demonstrating that the sequencing data quality was good enough for further subsequent analysis.
3.2 Identification of differentially expressed genes (DEGs)
DEGs were identified from the collection of mapped genes with the criteria of |log2FC| ≥ 2 and FDR ≤ 0.01.The results demonstrated that in the comparison to CK,978 DEGs were identified in the T1 group,among which 837 were up-regulated and 141 were down-regulated (Fig.1A);135 DEGs were identified in the T2 group,among which 83 were up-regulated and 52 were down-regulated(Fig.1B);232 DEGs were identified in T3 group,among which 167 were up-regulated and 65 were down-regulated (Fig.1C).The Venn diagram shows the number of DEGs of different comparisons(Fig.1D).Strikingly,36 genes were generally altered in all three comparisons (CK-vs-T1,CK-vs-T2 and CK-vs-T3).Besides,75 genes were specifically differentially expressed in the T3 group but not altered in the T1 and T2 groups.Thus,these 111 genes might be the key to understanding the synergistic antibacterial mechanism of KA and TP againstE.coliO157:H7.

Table 1 Primer pairs used for validation of RNA-Seq data by qRT-PCR.

Fig.1 DEGs of E.coli O157:H7 between different treatments Volcano plots of DEGs of CK-vs-T1 (A),CK-vs-T2 (B) and CK-vs-T3 (C).Venn diagram of DEGs (D).CK: control sample,T1: 1/2 MIC KA,T2: 1/2 MIC TP,T3: 1/5 MIC KA+1/5 MIC TP (The criteria of DEGs: |log2FC| ≥ 2,FDR ≤ 0.01).nodiff: no difference.
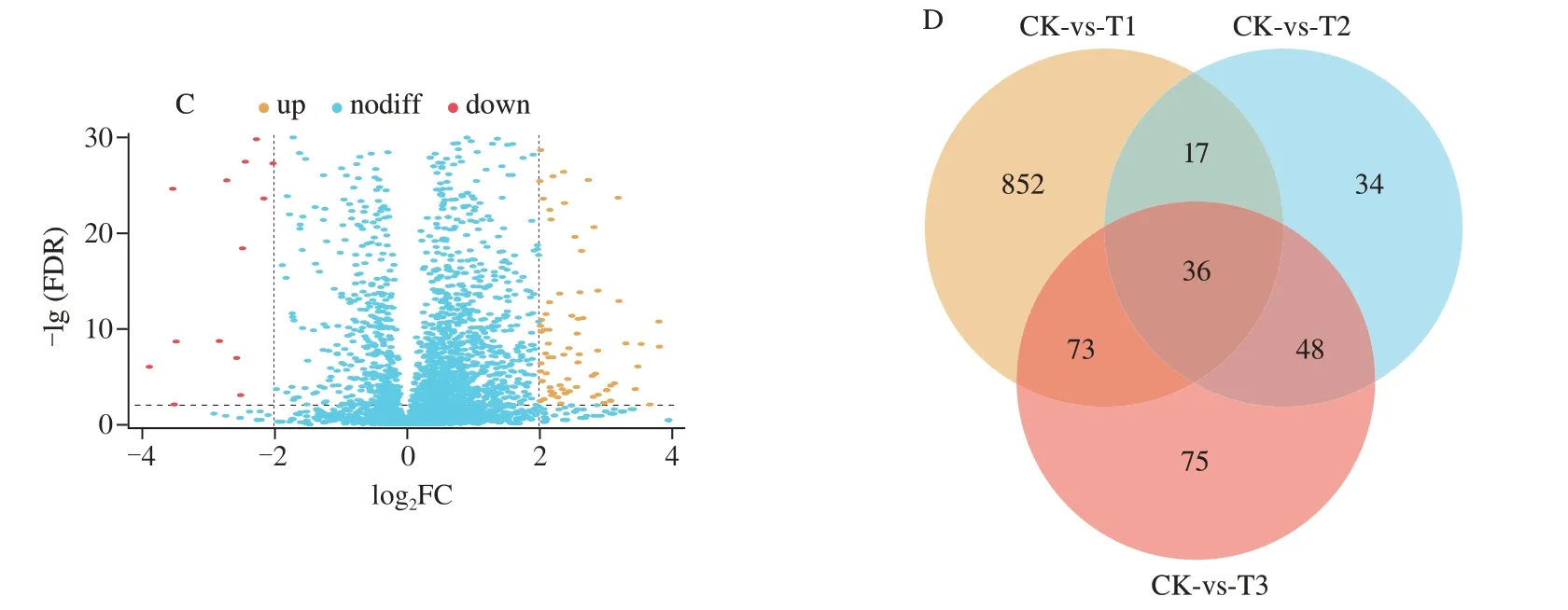
Fig.1 (Continued)
3.3 GO analyses of DEGs
GO and KEGG were applied to investigate DEGs functions to understand the transcriptional responses ofE.coliO157:H7.GO was applied to define and standardize the function of genes and proteins,including biological process (BP),molecular function (MF)and cellular component (CC).GO provided the top 5 significantly enriched between CK-vs-T1 and CK-vs-T2 were different,indicating the KA and TP responded differently (Fig.2).BP of CK-vs-T3 were the same as CK-vs-T2,while the 4 top terms in MF were the same as CK-vs-T1.Moreover,protein complex,periplasmic space and integral component of the membrane of CC terms in CK-vs-T3 were shared with CK-vs-T2.In addition,membrane and intrinsic components of the membrane of CC terms in CK-vs-T3 were shared with CK-vs-T1.The results demonstrated that KA and TP exhibited a synergistic effect on the transcription ofE.coliO157:H7 when used in combination,among which KA mainly altered its BP and TP mainly altered MF.
3.4 KEGG analyses of DEGs
KEGG is a giant database to help further analyze and understand transcriptional responses.The results show the top 10 significantly enriched KEGG pathways of the 3 comparisons in Fig.3.The enriched pathways between CK-vs-T1 and CK-vs-T2 were almost entirely different,which was similar to the corresponding results of GO.However,7 significantly enriched pathways in CK-vs-T3 were the same as those in CK-vs-T2,while CK-vs-T3 shared only one KEGG pathway with CK-vs-T1.In addition,biosynthesis of siderophore group nonribosomal peptides,other glycan degradation and galactose metabolism occurred in CK-vs-T3,suggesting thatE.coliO157:H7 would suffer some new stress when a combination of KA and TP.
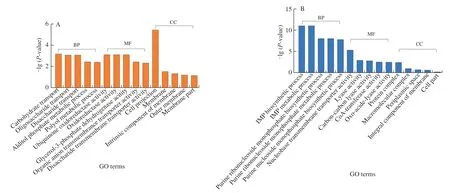
Fig.2 The top 5 significantly enriched GO annotations in the GO terms of BP,MF and CC.(A) CK-vs-T1,(B) CK-vs-T2,(C) CK-vs-T3.CK: control sample,T1: 1/2 MIC KA,T2: 1/2 MIC TP,T3: 1/5 MIC KA+1/5 MIC TP.

Fig.3 The top 10 significantly enriched KEGG pathways of (A) CK-vs-T1,(B) CK-vs-T2 and (C) CK-vs-T3.CK: control sample,T1: 1/2 MIC KA,T2: 1/2 MIC TP,T3: 1/5 MIC KA+1/5 MIC TP.
4.Discussion
4.1 Acid stimulation under the KA stress
Organic acids can pass the cell membrane directly in the undissociated lipophilic and subsequently dissociate in the cytoplasm,resulting in the acidification of the intracellular environment and disorder of cell metabolism[27-28].In addition,the organic acid is at least four acid resistance (AR) systems inE.coliand it would be activated under sub-lethal acid exposures[29].As a natural organic acid produced by microbial fermentation,the antibacterial mechanism of KA may be similar to other organic acids.
The genes expression of acid resistance under KA stress was shown in Table 2,which showed that the arginine decarboxylase system (AR-3) and the cadBA system (AR-4) were up-regulated,while more effective the oxidative systems (AR-1) and the glutamatedependent system (AR-2),and sigma factor RpoS were significantly down-regulated (Fig.4).The AR-3 was composed of an arginine decarboxylase AdiA,a transcriptional activator AdiY and an arginine/agmatine antiporter AdiC,controlled explicitly by regulator CysB[30].This system functions mainly through AdiA and AdiC,which upregulated 1.9 and 2.0 log2FC.The arginine decarboxylase AdiA restored appropriate intracellular pH by replacing the carboxyl groups of arginine with a proton recruited from the cytoplasm,while AdiC was used to deliver the arginine to,and remove the agmatine from,the cytoplasm.Similarly,AR-4 is generally used to resist mild acid stress inE.coliO157:H7.This system possesses a similar set of genes working analogously to the arginine decarboxylase system,and the difference is that the AR-4 employs lysine as the substrate[31].Therefore,KA could stimulate the acid stress response ofE.coliO157:H7 to cause varying degrees of cell membrane integrity destruction,but the acid stress caused by KA is relatively mild,which only partially contributed to its antibacterial effect.

Table 2 The expression of some essential genes related to kojic acid stress in E.coli O157:H7.
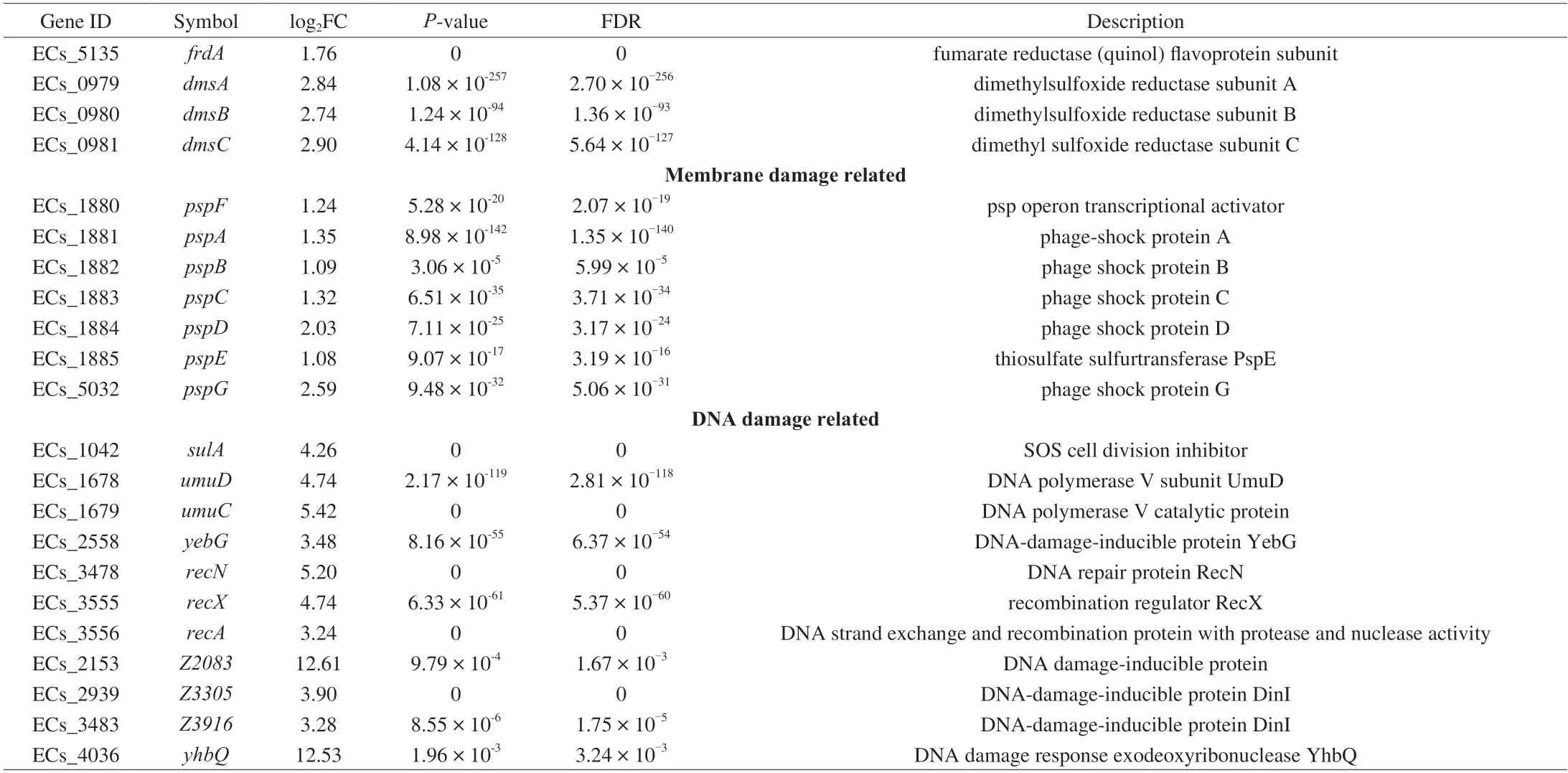
Table 2 (Continued)
4.2 Oxygen starvation under the KA stress
Two-component systems are a series of critical signal transduction systems which can reflect the stress state and metabolic changes of bacteria[32].The results suggested that significantly altered 44 genes were mainly related to the two-component system OmpR family,NarL family,NtrC family and CitB family,and most of them were involved in anaerobic respiration (Table 2).Under environmental stress,some bacteria tend to switch their metabolism from aerobic respiration to anaerobic respiration,during which some alternative electron acceptors such as nitrate,nitrite,dimethylsulfoxide (DMSO),trimethylamine-N-oxide (TMAO) and fumarate were employed to release energy[33].ThefdnGHIoperon encoding the formate dehydrogenases-N (Fdh-N) was significantly up-regulated by around 3 log2FC.Fdh-N is produced maximally under anaerobic conditions,reducing nitrate to nitrite in the formate-nitrate respiratory chain and Nar and generating the proton motive force[34-35].However,the Nar protein is encoded by thenarGHJIoperon,which was significantly suppressed in the present study.This phenomenon might be due to the lack of nitrate in the medium or the inhibition of nitrate transport by KA because the expression ofnarGHJIis positively correlated with the availability of nitrate[36].
The up-regulated nitrate transporter NarK also indirectly confirmed a lack of nitrite inE.coliO157:H7.There is a priority hierarchy for the anaerobic respiratory operons whereby nitrate served as the more favorable substrate and is used preferentially before TMAO,DMSO,or fumarate,only when nitrate is unavailable to the cell are the torACD,frdABCD and dmsABC operons expressed optimally[37].Consistent with this conclusion,the other anaerobic respiratory operons (torACD,frdABCDanddmsABC) and the TorS/TorR system were significantly up-regulated in the present study (Fig.4).Besides,except for thetorACDoperon,many genes related to the survival of bacteria at extreme pH were also modulated by the TorS/TorR system,includingtnaAB,gadABC,hdeABDandyhiEM[38].The related genes of the two-component system changed the respiration pattern ofE.coliO157:H7 under the KA stress,driving cells to rely more on anaerobic respiration.
4.3 Other damages under the KA stress
Previous studies confirmed that KA contributes to membrane damage by scanning electron microscope[18].Previous studies about the antibacterial mechanisms stated that cell membranes and genomic DNA might be the critical targets of KA and its derivatives[8,39].Hence,E.coliO157:H7 would repair the cell membrane and eliminate DNA damage by regulating the expression of related genes under sublethal KA stress.
As expected,most phage shock proteins related genes were significantly up-regulated after the KA treatment,among which the expression ofpspDandpspGwas enhanced by more than 2 log2FC(Fig.4).The phage shock protein response was induced by impaired inner membrane integrity inE.coli[40].PspDis a peripherally bound inner membrane (IM) protein used to sense incorrect insertion of membrane protein and to maintain the proton motive force (PMF)of IM[41].As an integral protein to maintain the IM stability,pspGalso directly confront a variety of stress stimulation by switching the cell to anaerobic respiration,thereby ingeniously adjusting and maintaining energy supply[42],which further confirmed the previous discussion about oxygen starvation.On the other hand,therecAwas significantly up-regulation by 3.24 log2FC under KA stress,resulting in the SOS responses genes being up-regulated,which caused a diverse set of physiological changes for DNA damage tolerance and repair[43].
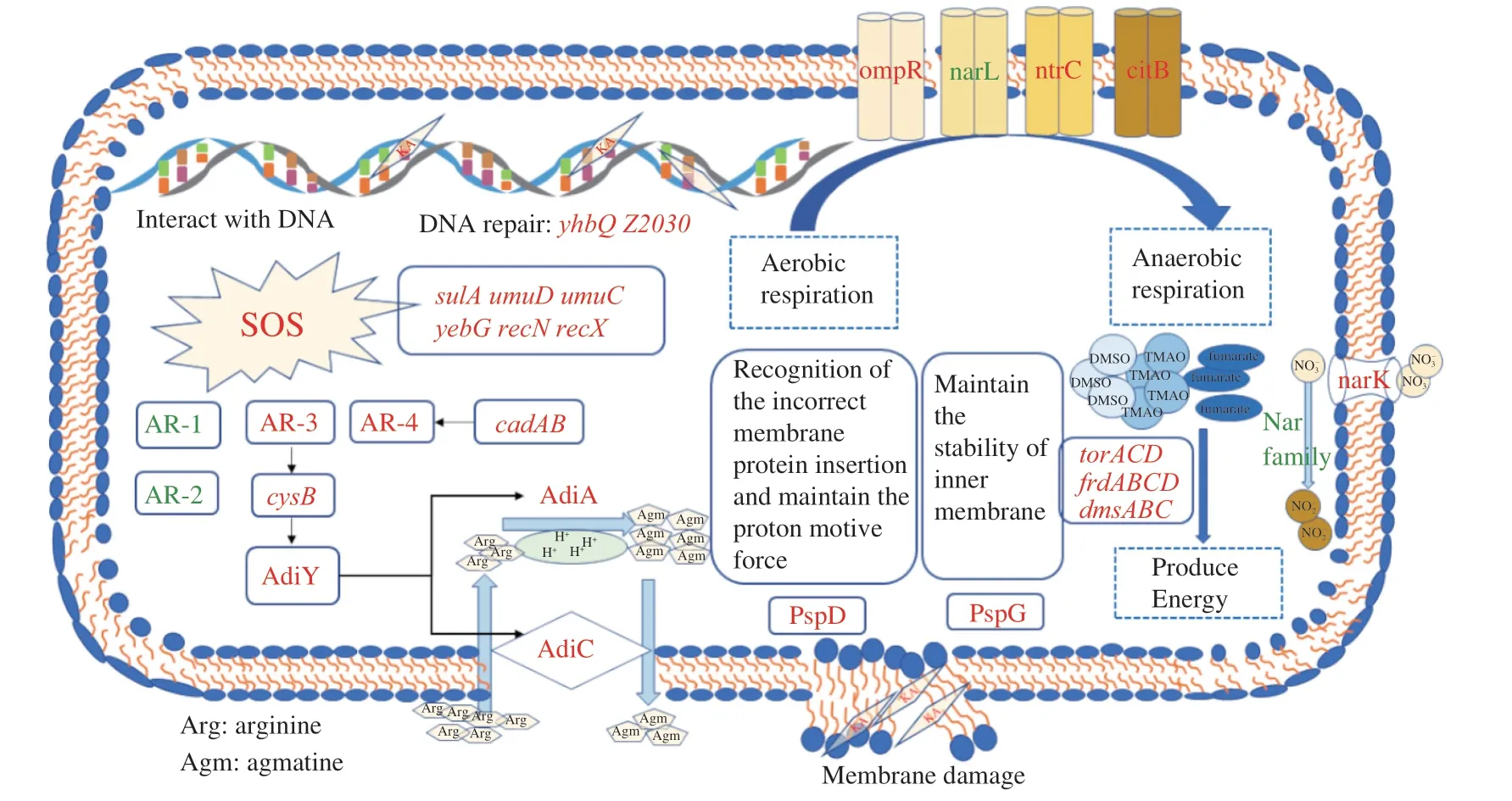
Fig.4 The responses of E.coli O157:H7 against kojic acid in sublethal concentration.Genes colored in red or green represent a higher or lower level than the control.
The damage genes ofyhbQandZ2083were drastically upregulation by 12.53 and 12.61 log2FC,respectively (Fig.4).YhbQis a metal-dependent endonuclease (Mg2+,Mn2+and Co2+) with high activity against nucleotide excision repair and DNA recombination intermediates[43].Consistently,our former research showed that KA is bound with DNA similarly to ethidium bromidein vitro,which might interfere with DNA replication,transcription and other processes[18].In conclusion,these phenomena suggested that KA could inactivateE.coliO157:H7 by causing acid stress and oxygen starvation and destroying cell membranes and genomic DNA,and DNA seemed to be the most important target to the relatively higher up-regulation of related genes.
4.4 Oxidative stress under the TP stress
The studies have reported that TP is a pro-oxidant to inhibit Pseudomonas spp.and could induce the expression of oxidative stress-related genes[21,44-45].There are three major regulators related to oxidative stress inE.coliO157:H7,includingoxyR,soxR,andrpoS[46].In our study,only thesoxRgene was significantly upregulation by 1.57 log2FC under the TP stress (Table 3).SoxRis activated by superoxide radicals and regulates several genes affiliated with oxidative stress responses,includingsodA(MnSOD)for superoxide dismutation,nfo(endonuclease IV) for DNA repair,andyggXfor iron-sulfur protein protection[47].The up-regulated expression of yggX indicated that TP could lead to the accumulation of intracellular superoxide radicals,which might oxidize Fe-S cluster proteins or interfere with their synthesis (Fig.5),whileE.coliO157:H7 confronted these problems by initiating a soxR-dependent oxidative stress response.
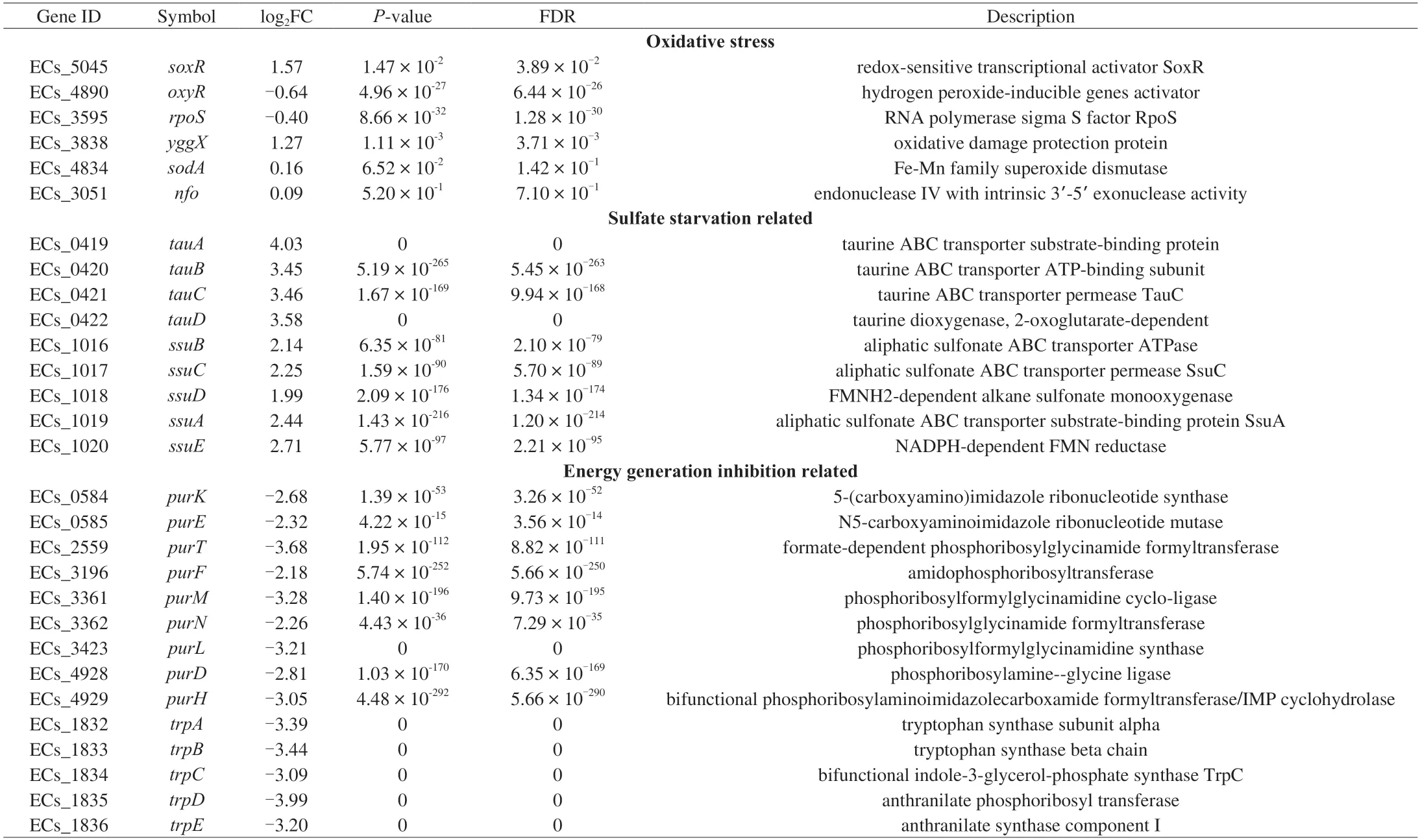
Table 3 The expression of some important genes related to tea polyphenols stress in E.coli O157:H7.
4.5 Sulfate starvation under the TP stress
The most significantly enriched KEGG pathway of CK-vs-T2 was sulfur metabolism,implying that the accumulation and utilization inE.coliO157:H7 changed under TP treatment (Fig.3).The DEGs involved in sulfur metabolism were concentrated in two clusters (i.e.,tauABCDandssuEADCB),significantly up-regulation (Table 3).Thetauandssusystems are triggered only in the absence of preferred sulfur sources,such as cysteine and sulfate;these clusters encode an ABC-type transporter system,among whichtauABCandssuABCare required for uptake of taurine and other aliphatic sulfonates,respectively[48].The expression oftauDwas up-regulated by 3.58 log2FC,which encodes a 2-oxoglutarate-dependent dioxygenase to liberate sulfite from taurine[49].Moreover,E.coliO157:H7 utilizes other sulfonates as a sulfite source through the desulfuration ofssuD,an FMNH2-dependent monooxygenase,which was also up-regulated by 1.99 log2FC during TP exposure[50].The sulfite produced or accumulated by the above two pathways is essential for cysteine biosynthesis through the Cys system (Fig.5).Thus,another mechanism of TP againstE.coliO157:H7 might be that the uptake or utilization of cysteine and sulfate were restricted,resulting in the initiation of sulfate starvation responses to supply sulfur sources.

Table 4 The unique DEGs of E.coli O157:H7 responding to the stress of KA and TP in combination.
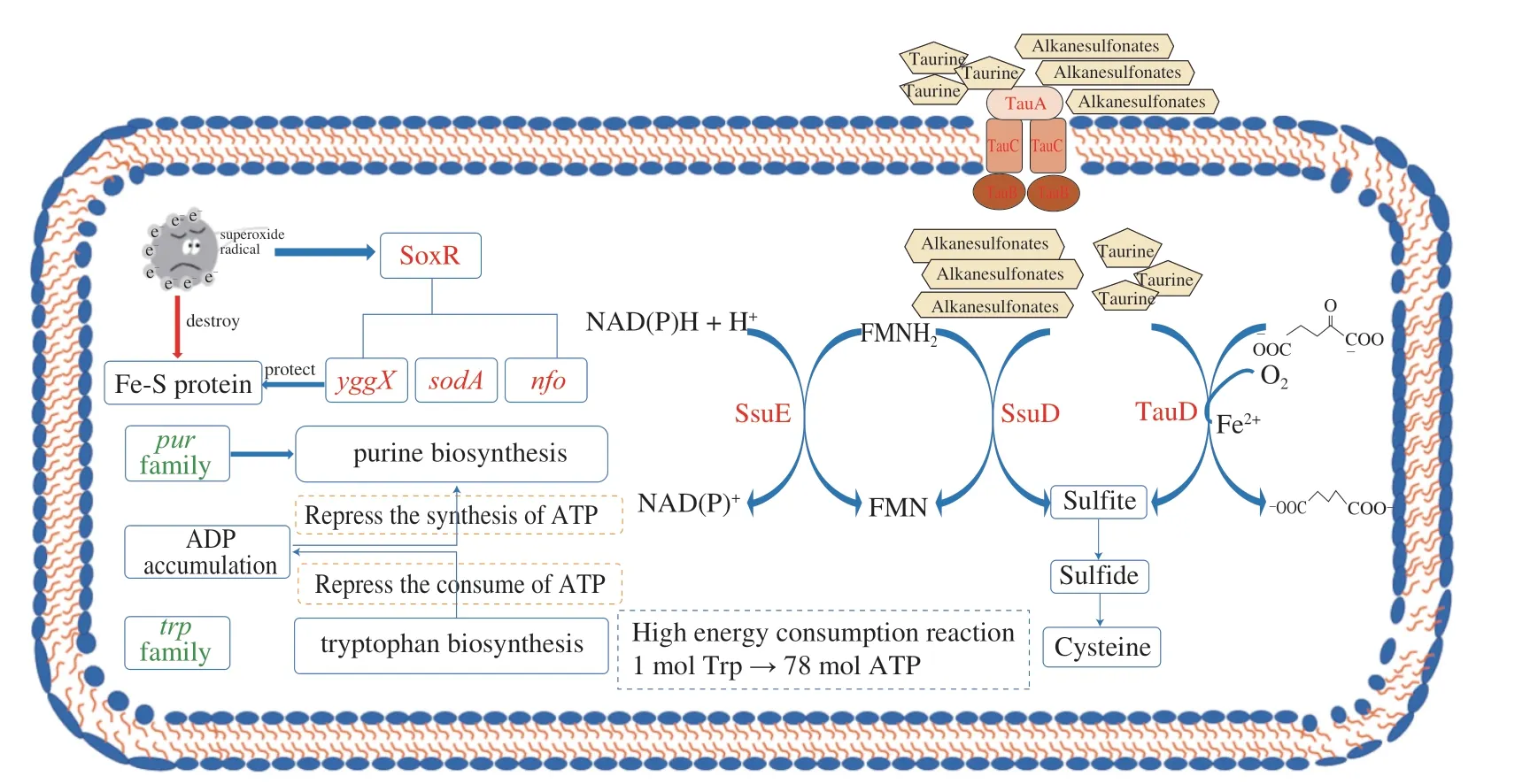
Fig.5 The responses of E.coli O157:H7 against tea polyphenols in sublethal concentration.Genes colored in red or green represent a higher or lower level than the control.
4.6 Inhibiting energy generation under the TP stress
The significantly down-regulated genes were mainly involved in purine nucleotides and tryptophan biosynthesis under TP stress(Fig.5).A total of ninepurgenes were found to be repressed after TP treatment,each encoding an enzyme responsible for a step in thede novopurine biosynthesis pathway (Table 3).Purines and their derivatives,constituting a part of the nucleic acids,cofactors,signal molecules,and the primary carriers of cellular energy,play broad roles in bacterial cellular processes[51].However,several previous studies reported that microbes would turn down the metabolisms related to purine nucleotide biosynthesis to adapt to different stresses,such as phenol and sulfometuron methyl[52].In addition,Jia et al.[52]inferred that the repression of purine nucleotide biosynthesis resulted from increased ADP concentration caused by ATP generation inhibition under external stress.
Five genes participating in tryptophan biosynthesis were significantly repressed by TP stress,includingtrpA,trpB,trpC,trpDandtrpE(Table 3).Recent studies have shown that tryptophan and functional Trp family proteins are critical for pathogenic bacteria to survive in the host and evade the defense of macrophages[53],but the synthesis of 1 mol tryptophan via the Trp pathway requires enormous amounts of ATP (78 mol),which is almost twice the energy required for the synthesis of any other amino acid[54].Hence,we suggest that down-regulation ofpurgenes would block energy generation,causing purine’s high energy consumption metabolisms and tryptophan biosynthesis to be suspended.Besides,the down-regulation of tryptophan and related Trp proteins has decreased the defense to host immunity ofE.coliO157:H7.
4.7 Specific responses under KA binding to TP
KA combined TP has enhanced the damage of cell membrane and genomic DNA inE.coliO157:H7,which was better effects of synergistic inhibitory than using either alone[18].The unique DEGs only appeared in CK-vs-T3 were shown in Table 4,which might be responsible for the synergistic antibacterial effect of KA and TP.Among these unique DEGs,iron uptake genes were significantly down-regulated,mainly including the operonentEBAHFdevoted to the siderophore enterobactin synthesis and two genesfepAandfepCinvolved in the enterobactin transport (Fig.6).Moreover,another iron uptake system utilizing hydroxamate siderophores (fhuAandfhuF)were also repressed by KA-TP treatment.Iron is indispensable for bacterial growth and survival under many external stresses,such as poor nutrition,metal toxicity and oxidative stress[55-57].This conclusion was confirmed by the up-regulation offbpBandfbpC,both of which are involved in the iron-sparing response and devoted to permitting the essential iron-dependent enzymes to access the limited iron pools by inhibiting the transcription of other high iron-utilizing proteins[58].Therefore,one of the reasons behind the synergistic antibacterial effect of KA and TP might be that the iron uptake pathways were hindered,thus making bacteria more sensitive to various stresses,such as oxidative stress.
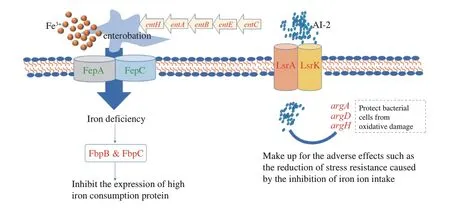
Fig.6 The individual responses of E.coli O157:H7 against kojic acid -tea polyphenols in sublethal concentration.Genes colored in red or green represent a higher or lower level than the control.
Among these unique DEGs,two clusters of genes were upregulation,including three quorum sensing-related genes (lsrA,lsrDandlsrK) and three arginine biosynthesis-related genes (argA,argDandargH).LsrAandlsrDare part of the ABC transporters complex,responsible for the uptake of autoinducer 2 (AI-2).ThelsrKencodes a cytoplasmic kinase LsrK directly regulating the AI-2 uptake system,which co-regulates many other genes associated with biofilm formation and cell motility[59].According to the up-regulated expression oflsrgenes,we speculated thatE.coliO157:H7 coordinate with each other through the AI-2 quorum sensing system to form biofilms,thus enhancing the resistance to KA-TP stress.Interestingly,the up-regulated arginine biosynthesis-related genes also prove the above conjecture.Arginine may directly regulate cell-tocell interactions and an appropriate concentration of extracellular arginine is helpful to bacterial biofilm development[60].In addition,arginine is a compatible solute to improve the resistance of bacteria to osmotic stress and a precursor for the synthesis of putrescine and spermidine,which may reduce oxidative damage in bacterial cells[61-62].It could be concluded thatE.coliO157:H7 promoted biofilm formation and resistance to various stresses by enhancing the quorumsensing signal molecule uptake and arginine biosynthesis so as to neutralize the effect of limited iron uptake caused by KA-TP stress.
4.8 qRT-PCR validation
To provide convincing evidence for the purpose of validating RNA-seq data,nine genes were selected randomly for qRT-PCR analysis.The qRT-PCR data results of this study show well correlation with RNA-seq (R2=0.893 7) (Fig.7).On the whole,this data provided more evidence to understand the positive effects of KA and TP againstE.coliO157:H7.

Fig.7 Gene expression fold changes a resulting from RNA-Seq analysis and qRT-PCR for nice selected genes in difference treatments.The X-axis represents the log2FC of RNA-seq,the Y-axis represents the log2FC of qRT-PCR,and the R2 represents the R-square of the regression line.
5.Conclusion
This study revealed the antibacterial mechanism of sublethal KA and TP againstE.coliO157:H7 using high-throughput transcriptome sequencing technology.Comparing DEGs between CK and T1,KA inactivatedE.coliO157:H7 by causing acid stress and oxygen starvation and destroying cell membranes and genomic DNA,while DNA seemed to be the most crucial antibacterial target.Then,comparing DEGs between CK and T2,E.coliO157:H7 confronted a series of adverse conditions under the TP stress,including oxidative stress,sulfate starvation and energy generation inhibition.Except for the above antibacterial mechanism,the iron uptake pathways were hindered by KA and TP,which might be the reason behind their synergistic antibacterial effect,whileE.coliO157:H7 neutralizes this effect by enhancing the AI-2 uptake and arginine biosynthesis.With this evidence obtained from the current study,exploration of the iron uptake pathways induced by measuring the change and transfer of iron ions inE.coliO157:H7 will significantly explain the antibacterial mechanism of KA combined with TP in the future.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by National Natural Science Foundation of China (31972021),R&D Projects in Key Areas of Guangdong Province (2019B020212003),the Science and Technology Program of Guangzhou,China (202206010177),Guangdong key research and development program (2021B0202060001) Foshan and agricultural academy cooperation project and Guangdong Modern Agriculture project (2022KJ117),Aquatic Products Center Project of GAAS.We thank Guangzhou Gene Denovo Biotechnology Co.,Ltd.,China,for technical assistance.
- 食品科学与人类健康(英文)的其它文章
- Protective effects of oleic acid and polyphenols in extra virgin olive oil on cardiovascular diseases
- Diet and physical activity inf luence the composition of gut microbiota,benef it on Alzheimer’s disease
- Inf luence of nitrogen status on fermentation performances ofnon-Saccharomyces yeasts: a review
- Ganoderma lucidum: a comprehensive review of phytochemistry,eff icacy,safety and clinical study
- Resveratrol combats chronic diseases through enhancing mitochondrial quality
- Demonstration of safety characteristics and effects on gut microbiota of Lactobacillus gasseri HMV18

