Transcriptomics integrated with metabolomics reveals the mechanism of CaCl2-HCl electrolyzed water-induced glucosinolate biosynthesis in broccoli sprouts
Cui Li,Shuhui Song,Ynn He,Hijie Liu,
a College of Food Science and Nutritional Engineering, China Agricultural University, Beijing 100083, China
b Institute of Agri-food Processing and Nutrition, Beijing Academy of Agriculture and Forestry Sciences, Beijing 100097, China
Keywords: Broccoli sprouts CaCl2-HCl electrolyzed water Glucosinolates Transcriptomics Metabolomics
ABSTRACT Glucosinolates are important phytochemicals in Brassicaceae.We investigated the effect of CaCl2-HCl electrolyzed water (CHEW) on glucosinolates biosynthesis in broccoli sprouts.The results showed that CHEW treatment significantly decreased reactive oxygen species (ROS) and malondialdeh yde (MDA) contents in broccoli sprouts.On the the 8th day,compared to tap water treatment,the the total glucosinolate content of broccoli sprouts with CHEW treatment increased by 10.6% and calcium content was dramatically enhanced from 14.4 mg/g DW to 22.7 mg/g DW.Comparative transcriptome and metabolome analyses revealed that CHEW treatment activated ROS and calcium signaling transduction pathways in broccoli sprouts and they interacted through MAPK cascades.Besides,CHEW treatment not only promoted the biosynthesis of amino acids,but also enhanced the expression of structural genes in glucosinolate synthesis through transcription factors (MYBs,bHLHs,WRKYs,etc.).The results of this study provided new insights into the regulatory network of glucosinolates biosynthesis in broccoli sprouts under CHEW treatment.
1.Introduction
Broccoli (Brassicaoleracea L.var.italica),as a vegetable,is consumed worldwide due to its diverse bioactive components such as phenols,f lavonoids,carotenoids and glucosinolates.Epidemiological evidence suggests that the the consumption of broccoli has many beneficial effects on human health,such as anti-inflammatory[1],antioxidant[2],hypoglycemic[3],fight against obesity[4],etc.,which effects are primarily attributed to glucosinolates and their hydrolyzate isothiocyanates.
Glucosinolates are a class of plant secondary metabolites containing nitrogen and sulfur,which mainly exist in Brassicaceae and play an important role in plant resistance to pests and diseases[5].To date,more than 130 glucosinolates have been identified and Brassicaceae contains more than 40 kinds of glucosinolates[6-7].According to the side chain R (derived from 8 amino acids),glucosinolates can be classif ied into aliphatic glucosinolates,indole glucosinolates and aromatic glucosinolates[8-9].Glucoraphanin (GRA),one of aliphatic glucosinolates,is the most abundant glucosinolate in broccoli,accounting for more than 50% of the total glucosinolate,and its content in broccoli sprouts is dozens of times that of mature broccoli[10].Therefore,researches on glucosinolates in broccoli sprouts are gradually increasing.
The biosynthesis of glucosinolate in broccoli sprouts has been clarif ied,which mainly includes three stages: (a) side-chain elongation of amino acid;(b) formation of glucosinolate core structure;(c) secondary modification of glucosinolate side chain[11].The formation of the glucosinolate core structure is not only a common part of all glucosinolates,but also a key step in glucosinolate synthesis.The genes involved in this process includeCYP79F1,CYP79B1,GGP1,UGT74B1,SOT18,etc.[12].Numerous researches have been done on the accumulation of glucosinolates in broccoli sprouts with diverse treatments.Previous studies found that the contents of GRA and isothiocyanates in broccoli sprouts were enhanced after 10 mmol/L CaCl2treatment,and the mechanism was mainly explained from the perspectives of electrolyte leakage,changes in antioxidant enzyme activities and gene expression[2,13-14].Slightly Acidic Electrolyzed Water (SAEW) was used for the cultivation of broccoli sprouts,and the contents of glucosinolates and GRA in the SAEW treatment were higher than those in the tap water control group[15].Besides,ZnSO4plus CaCl2treatment enhanced the content of glucosinolates in broccoli sprouts as well as the expressions ofMYB28,Elong,CYP79F1,CYP83A1,FMOGS-OX1andAOP2[16].Current research mainly explains the regulation mechanism of exogenous substances treatment on broccoli sprouts from the aspects of enzyme activity and glucosinolate synthesis-related structural gene expression.However,the regulatory network of glucosinolate synthesis is relatively complex.The study of transcription factors that regulate glucosinolate synthesis can further enrich the regulatory mechanism of glucosinolate synthesis.Current studies have shown thatMYB28,MYB29,MYB76,MYB34,MYB51andMYB122of MYB transcription factor family play a major role in regulating the synthesis of glucosinolates in cruciferous plants;the bHLH (Basic Helix-Loop-Helix) transcription factorsMYC2,MYC3andMYC4regulate the synthesis of glucosinolates by acting on the MYB transcription factors,while the WRKY transcription factorsWRKY18andWRKY40cooperate withCYP81F2to negatively regulate the synthesis of indole glucosinolates[17].
The development of transcriptome and metabolome technology has increased our our understanding of the network between genes and key metabolites in plants.A previous study found that integrative analysis of the metabolome and transcriptome profiles provided some novel insights into anthocyanin accumulation and the molecular mechanism of anthocyanin biosynthesis in asparagus[18].Aliphatic glucosinolates and indolic glucosinolates were found to be increased significantly in the CaCl2treated broccoli microgreens using metabolomic approaches,but the molecular mechanism was not explained[14].Thus,transcriptome and metabolome analysis could be considered to elucidate regulatory networks between genes and glucosinolates biosynthesis in broccoli sprouts.
A complete understanding of the the regulatory network of glucosinolates biosynthesis in broccoli sprouts is crucial to develop functional foods to meet the increasing health demand in our diet.Our previous study found that CaCl2-HCl electrolyzed water(CHEW) treatment promoted glucosinolate synthesis and generated more bioactive isothiocyanates in broccoli sprouts[19].Therefore,this study combined metabolomics and transcriptomics to further explore the molecular mechanism of CHEW treatment-induced glucosinolate biosynthesis in broccoli sprouts.The data from this study enhanced our understanding of the regulatory mechanism of glucosinolate synthesis,and provided a theoretical basis for variety improvement and high-quality cultivation of broccoli sprouts with high glucosinolate content.
2.Materials and methods
2.1 Preparation of CHEW
CaCl2-HCl electrolyzed water (CHEW) with a near-neutral pH and containing available chlorine,can be generated by electrolyzing CaCl2-HCl solution using a nonmembrane electrolytic cell.Available chlorine refers to the oxidized chlorine contained in chlorinecontaining compounds,and its concentration is expressed in mg/L or % (g/100 mL).In CHEW,the available chlorine mainly refers to HClO and ClO-.
CHEW was produced according to the method described previously with minor modifications[19].5 mmol/L CaCl2acidic solution (pH was adjusted to 4.40 ± 0.05 by concentrated hydrochloric acid) was electrolyzed with a 3 A electrical current for 40 s,then concentrated hydrochloric acid was used again to adjusted the pH to 5.5 ± 0.05,and CHEW with available chlorine concentration of 10 mg/L was obtained finally.The control treatment used tap water without electrolyzed.Analytical-grade reagents: hydrochloric acid(36%-38%) and anhydrous calcium chloride (96%) were purchased from Sinopharm Chemical Reagent Co.,Ltd.
2.2 Plant materials,cultivation and treatment
Broccoli seeds (BrassicaoleraceaL.[Italica group] cv.‘zhongqing’) were used in this study.The cultivation conditions of broccoli seeds were conducted according to previous method[19].After soaking with CHEW solution and tap water respectively,the seeds were evenly distributed in a polypropylene box and kept in a growth chamber (PRx-450C,Ningbo Saifu Instrument Corporation,Ningbo,China) at 25 °C,80% relative humidity under light.On the 4thand 8thdays during the growth,broccoli sprouts were carefully harvested,immediately frozen with liquid nitrogen,and stored in a freezer at -80 °C before the phytochemicals were analyzed.
2.3 ROS and MDA content determinations
H2O2and MDA contents were determined according to the methods described previously[19].AnDetection Kit (BC1290,Solarbio,Beijing,China) was used to assay thecontent.Approximately 100 mg broccoli sprouts were homogenized with 1 mL extraction solution in an ice bath and then centrifuged at 4 °C,12 000 r/min for 20 min.Afterwards,200 µL supernatant and reaction solutions were mixed according to the manual,and the absorbance of the solution was measured at 530 nm.content was expressed as µmol/g FW.For each treatment,measurements were made in triplicate.
2.4 Determination of glucosinolate and calcium content
Glucosinolates and calcium content were determined according to the method described previously[19].For each treatment,measurements were made in triplicate.
2.5 Metabolite extraction and analysis
Broccoli sprouts were freeze-dried (Scientz-100F) and then pulverized (30 Hz,1.5 min) to powder using a grinder (MM 400,Retsch).Broccoli sprout’s dry powder (100 mg) was homogenized with 1.2 mL of 70% methanol,and placed in a 4 °C refrigerator overnight.After centrifugation for 10 min at 12 000 r/min,the supernatant was filtered through a 0.22 µm membrane filter before it was injected into a UPLC-MS/MS analysis.
Ultra Performance Liquid Chromatography (UPLC) (SHIMADZU Nexera X2) and Tandem mass spectrometry (MS/MS) (Applied Biosystems 4500 QTRA) were used in this study.Liquid phase conditions: column (Agilent SB-C181.8 µm,2.1 mm × 100 mm);phase A is ultrapure water (add 0.1% formic acid),phase B is acetonitrile(add 0.1% formic acid).The ratio of phase B is 5% at 0.00 min,then increases linearly to 95% within 9.00 min,and maintained at 95% for 1 min.10-11 min: the ratio of phase B decreases to 5%,and maintained 5% equilibrate to 14 min.The flow rate was 0.35 mL/min,the column temperature was 40 °C and the injection volume was 4 µL.The ESI source operation parameters: ion source,turbo spray;source temperature 550 °C;ion spray voltage (IS) 5 500 V(positive ion mode)/-4 500 V (negative ion mode);ion source gas I(GSI),gas II (GSII) and curtain air (CUR) were set to 50,60,and 25.0 psi,respectively,and the collision-induced ionization parameter was set to high.Instrument tuning and mass calibration were performed with 10 and 100 µmol/L polypropylene glycol solutions in triple quadrupole (QQQ) and LIT modes,respectively.QQQ scans use MRM mode and set the collision gas (nitrogen) to medium.
Kyoto Encyclopedia of Genes and Genomes (KEGG) compound database (http://www.kegg.jp/kegg/compound/) was used to identify identify metabolites.Significantly changed metabolites between groups were determined by VIP ≥ 1 and absolute log2FC (fold change) ≥ 1.Pathways with significantly changed metabolites were were then fed into MSEA (metabolite sets enrichment analysis),and hypergeometric test’sP-values determined their significance.
2.6 Transcriptome analysis
2.6.1 RNAextraction
Total RNA was extracted using RNA prep Pure Plant Plus Kit(DP441,Tiangen Biotech,Beijing,China) according to the operation specifications.RNA purity was checked using the Nano Photometer®spectrophotometer (IMPLEN,CA,USA).RNA concentration was measured using Qubit® RNA Assay Kit in Qubit®2.0 Fluro meter(Life Technologies,CA,USA).RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies,CA,USA).
2.6.2 Libraryconstructionandsequencing
The starting RNA for library construction was 1 µg total RNA.NEB Next® Ultra RNA Library Prep Kit for Illumina® (NEB,USA)was used in library construction.PolyA-tailed mRNAs were enriched by Oligo(dT) magnetic beads,and then interrupted the mRNAs with divalent cations randomly in NEB Fragmentation Buffer.Using fragmented mRNA as a template and random oligonucleotides as primers,the first strand of cDNA was synthesized in the M-MuLV reverse transcriptase system,and then the RNA strand was degraded by RNase.In DNA polymerase I system,dNTPs were used to synthesize the second strand of cDNA.The purified double-stranded cDNA was end-repaired,A-tailed,and connected to a sequencing adapter.AM Pure XP beads were used to screen 250-300 bp cDNA,and PCR amplification was performed,AM Pure XP beads were used to purify the PCR product again.Finally,we get the library.
After the library was constructed,Qubit 2.0 Fluorometer was used for preliminary quantification.Dilute the library to 1.5 ng/µL,and then used Agilent 2100 bioanalyzer to detect the insert size of the library.After the insert size was as expected,qRT-PCR accurately determined the effective concentration (above 2 nmol/L) of the library.Entrust Wuhan Metware Biotechnology Co.,Ltd.to conduct on-machine sequencing.
2.6.3 Bioinformaticsanalysis
Use fastp v 0.19.3 to filter the raw data,mainly to remove reads with adapters;when the N content in any sequencing read exceeds 10% of the bases of the reads,the paired reads are removed;when any sequencing reads When the number of low-quality (Q ≤ 20)bases contained in the reads exceeds 50% of the bases of the read,the paired reads are removed.All subsequent analyses were based on clean reads.Download the reference genome and its annotation files from the http://brassicadb.cn/#/ website,use HISAT v2.1.0 to build the index,and align the clean reads to the reference genome.Gene alignments were calculated using feature Counts v1.6.2,and then the FPKM for each gene was calculated based on gene length.
Differential expression analysis between the two groups was performed using DESeq2 v1.22.1,withP-values corrected using the Benjamini &Hochberg method.AdjustedP-value and |log2FC|as threshold for significant differential expression.The enrichment analysis is performed based on the hypergeometric test.For KEGG,the hypergeometric distribution test is performed with the unit of pathway;for GO,it is performed based on the GO term.
2.7 Quantitative real-time PCR (qRT-PCR) analysis
The qRT-PCR was used to verify the reliability of RNA-seq results.The DEGs were randomly selected based on RNA-seq data.Total RNA was extracted from broccoli sprouts using a plant RNA kit(ZP405K,ZOMANBIO).First-strand cDNA was synthesized from 1 ng to 5 µg RNA using Hifair II 1st Strand cDNA Synthesis SuperMix for qPCR (11123ES60,YEASEN).qRT-PCR were performed using Hieff qPCR SYBR Green Master Mix (11202ES08,YEASEN).The PCR amplification was performed using 3-step cycling conditions:95 °C for 2 min,followed by 45 cycles of 95 °C for 30 s,58 °C for 30 s and 72 °C for 60 s.The primers listed in Table S1 andActinwas used as internal reference gene.Relative expressions were calculated using the 2-△△CTmethod.Three biological replications were used and four technical repetitions were performed on each replicate.
2.8 Statistical analysis
Statistical analysis was conducted using IBM SPSS Statistics 19(SPSS Inc.,NY,USA).All data were expressed as means ± standard error of triplicate independent cultures.Data were statistically analyzed by one-way ANOVA and Duncan’s multiple-range tests.Differences atP< 0.05 were considered significant.
3.Results and discussion
3.1 Effect of CHEW on ROS,MDA,calcium and total glucosinolate conents of broccoli sprouts
To investigate the effects of CHEW on the physiological changes in broccoli sprouts,we found that the contents of H2O2and total glucosinolates were significantly decreased,while calcium content increased significantly during the growth of broccoli sprouts(Fig.1).Compared with the tap water treatment,CHEW treatment significantly decreased ROS and MDA contents in broccoli sprouts,which indicated that CHEW could relieve lipid peroxidation of the cell membrane of broccoli sprouts and reduce the level of ROS(Figs.1A-C).Compared to the tap water treatment,the calcium content in broccoli sprouts treated with CHEW on day 8 was dramatically enhanced from 14.4 mg/g DW to 22.7 mg/g DW(Fig.1D).In addition,compared with tap water,the total glucosinolate content of broccoli sprouts treated with CHEW on day 4 and day 8 increased by 10.8% and 10.6%,respectively (Fig.1E).The total glucosinolate content in the CHEW treatment was consistently higher than that in the tap water treatment during the growth of broccoli sprouts.
Oxidative components of CHEW such as HClO and ClO-,will produce oxidative stress to plants.Under oxidative stress,plants are supposed to produce a large amount of ROS which is an important second messenger[20].Previous studies have shown that the glucosinolates content of broccoli sprouts was significantly increased with slightly acidic electrolyzed water treatment[15].Same as ROS,Ca2+is also a second messenger and plays an important role in the growth and development of plants.Previous study found that exogenous and endogenous calcium can promote glucosinolates biosynthesis in broccoli sprouts under ZnSO4stress[16].These results show that Ca2+and ROS of CHEW play important roles in glucosinolate synthesis in broccoli sprouts.
3.2 Metabolome analysis
To explore the effect of CHEW treatment on the growth and glucosinolate biosynthesis of broccoli sprouts,metabolome analysis was performed on the 4thand 8thdays under tap water and CHEW treatments.Principal component analysis (PCA) was conducted and showed the isolation trend of the detected metabolites among the samples in “TW4”,“CHEW4”,“TW8” and “CHEW8” groups (Fig.S1a).Correlation analysis was performed on the three biological replicates in each sample,andR2> 0.8,indicating the samples had good repeatability (Fig.S1b).The results implied that the metabolites of broccoli sprouts were remarkably changed under CHEW treatment.In this study,a total of 994 differentially accumulated metabolites(DAMs) were detected which were divided into 15 categories and sorted by quantity as follows: phenolic acids,lipids,flavonoids,amino acids and derivatives,alkaloids,organic acids,lignans and coumarins,saccharides and alcohols,nucleotides and derivatives,glucosinolates,vitamin,terpenoids,tannins,stilbene and other metabolites (Fig.2A).Compared with tap water treatment,159 DAMs (84 up-regulated and 75 down-regulated) on the 4thday and 218 DAMs (94 up-regulated and 124 down-regulated) on the 8thday in broccoli sprouts with CHEW treatment were filtered from the annotated metabolites by setting a threshold of VIP ≥ 1 and absolute log2FC ≥ 1 (Fig.2B).The results implied that CHEW treatment induced more metabolite changes during growth of broccoli sprouts.
KEGG enrichment analysis of these DAMs revealed that compared to tap water treatment,the DAMs in broccoli sprouts with CHEW treatment were significantly enriched in metabolic pathways,biosynthesis of secondary metabolites,amino acids and cofactors(Figs.2C,D).This observation meant that CHEW treatment might affect the biosynthesis of secondary metabolites during the growth of broccoli sprouts by promoting the biosynthesis of cofactors and amino acids,the precursors of secondary metabolites.This result was in agreement with the the previous report,which showed that slightly acidic electrolyzed water promotes the biosynthesis of glucosinolates and anthocyanin in broccoli sprouts,which were associated with an increase in their precursor,methionine and phenylalanine respectively[21].
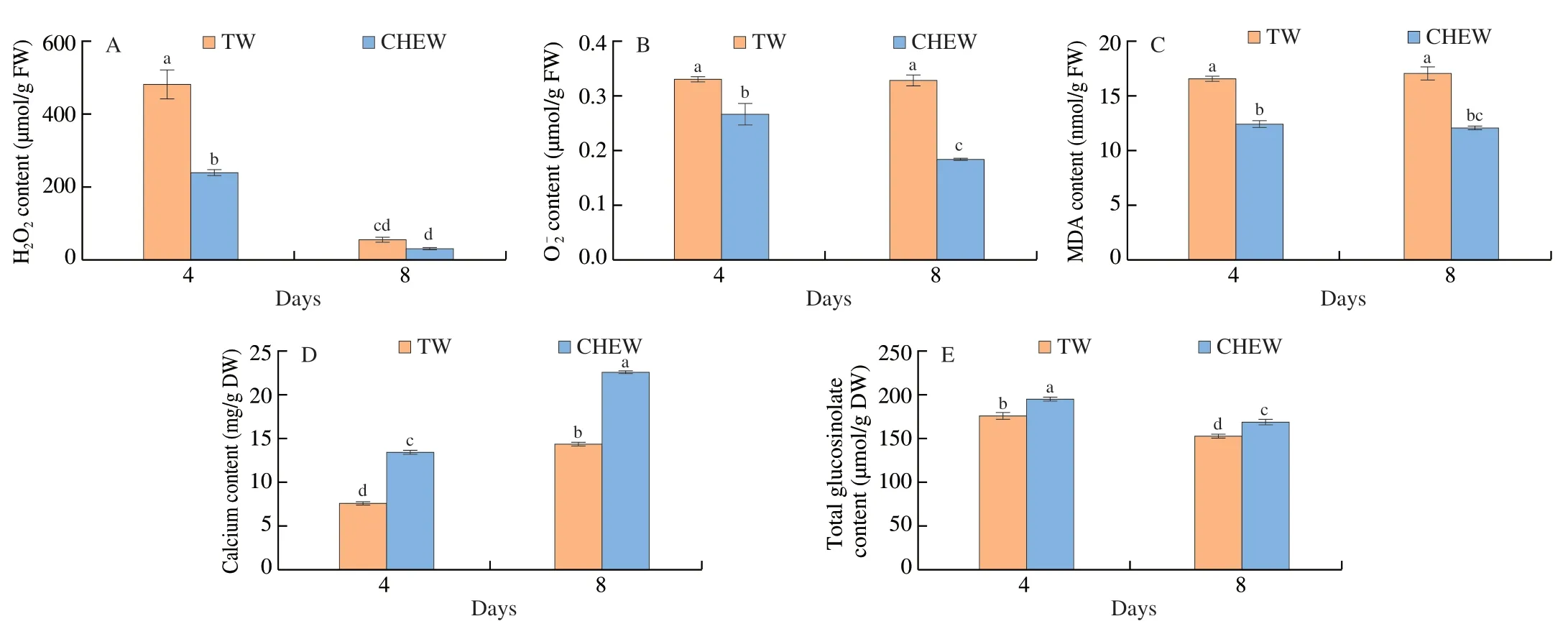
Fig.1 Contents of H2O2 (A),(B),MDA (C),Calcium (D) and total glucosinolate (E) in broccoli sprouts under different treatments during growth.TW,tap water,CHEW,CaCl2-HCl electrolyzed water.Each value is the mean of 3 replicates per treatment (mean ± SE).Values not sharing a common letter are significantly different for each time point at P < 0.05.
3.3 Transcriptome analysis
To explore the molecular events of glucosinolate biosynthesis in broccoli sprouts under CHEW treatment,transcriptome analysis was performed.We investigated the changes in gene expression levels among the samples (“TW4”,“CHEW4”,“TW8” and “CHEW8”),each of which included 3 replicates.After removing the low-quality reads,we obtained clean reads with an average number of 46 180 865.The percentages of Q30 and GC were 92.73%-93.99% and 46.71%-47.66%,respectively,indicating that the quality of transcriptome sequencing data is high (Table S2).PCA indicated that all biological replicates were grouped together (Fig.S2a),and correlation analysis revealed obviously good repeatability of the samples (Fig.S2b).According to the results of transcriptomic analysis,there were 1 014 differentially expressed genes (DEGs) between TW4 and CHEW4,of which 223 were upregulated and 791 were downregulated.There were 1 372 DEGs between TW8 and CHEW8,including 1 096 upregulated genes and 276 downregulated genes (Fig.S2c).The volcano plots were drawn to intuitively represent the differential gene distribution between each tap water treatment and CHEW treatment during the growth of broccoli sprouts.The results indicated obviously that the CHEW affected the expression of numerous genes of broccoli sprouts during growth,and the number of upregulated expressed genes increased significantly on the 8th day (Figs.3A and B).
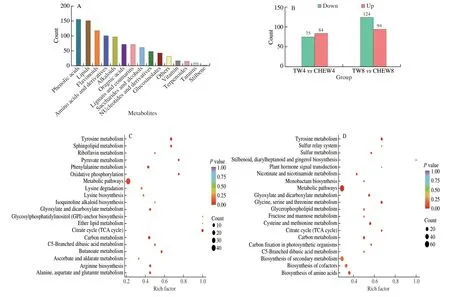
Fig.2 Preliminary analysis of metabolomics data.(A) Classification and statistical analysis of all metabolites detected.(B) Statistical data of DAMs in broccoli sprouts.(C) KEGG enrichment analysis of DRMs between TW4 and CHEW4.(D) KEGG enrichment analysis of DRMs between TW8 and CHEW8.
3.4 GO and KEGG enrichment analysis of DEGs
The DEGs were classified into 3 groups,biological process,cellular component and molecular function by GO classification and plotted as shown in Figs.3C and D.Compared with tap water treatment,the processes of cellular response to external stimulus,metal ion transport,antioxidant and enzyme activities,and cell wall formation were significantly activatated in broccoli sprouts under CHEW treatment on the 4thday.On the 8thday,regulation of response to the the external stimulus was the largest group in the biological process of broccoli sprouts under CHEW treatment,followed by a response to reactive oxygen species and the the metal ion transport process.The genes related to “membrane” was predominant in the group of cellular components.In the molecular function group,the genes were mostly involved in calcium ion binding.
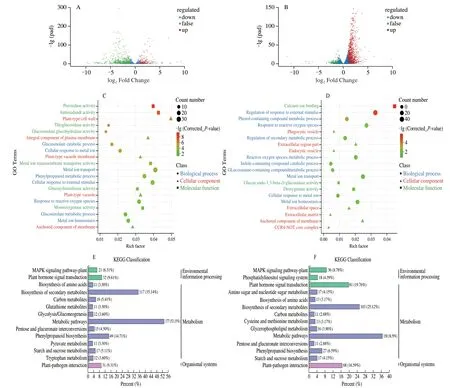
Fig.3 Preliminary analysis of transcriptomics data.(A) Volcano plots of DEGs between TW4 and CHEW4.The green dots indicate the down-regulated expressed genes (fold change,log2FC < -1),the red dots indicate the up-regulated expressed genes (log2FC > 1),and the blue dots indicate the non-differentially expressed genes (-1 ≤ log2FC ≤ 1).The abscissa indicates the differential expression multiple,and the ordinate indicates the level of difference in the gene significance.(B) Volcano plots of DEGs between TW8 and CHEW8.(C) GO enrichment results of DEGs between TW4 and CHEW4.(D) GO enrichment results of DEGs between TW8 and CHEW8.(E) KEGG enrichment analysis of DEGs between TW4 and CHEW4.(F) KEGG enrichment analysis of DEGs between TW8 and CHEW8.
KEGG enrichment analysis was performed on all DEGs.Compared with the tap water treatment,the DEGs of broccoli sprouts with CHEW treatment were mainly enriched in the the metabolism group on the 4thday,including metabolic pathways and biosynthesis of secondary metabolites (Fig.3E).On the 8thday,the main enrichment pathways changed significantly.The DEGs associated with “plant hormone signal transduction”,“plantpathogen interaction” and “plant MAPK signaling pathway” were significantly enriched (Fig.3F).Analysis of the DEGs involved in the three predominant pathways revealed that there were 81,68 and 36 DEGs in plant hormone signal transduction,plant-pathogen interaction and Mitogen-activated Protein Kinase (MAPK) signaling pathway,respectively (Fig.4A).A total of 77 DEGs were obtained after screening (Supplementary B).As shown in Fig.4B,the DEGs of auxin signaling transduction were significantly down-regulated while others were significantly up-regulated.Previous studies showed that the catabolites of glucosinolates,raphanusamic acid and indole-3-carbinol,inhibited auxin signaling by competing with auxin for binding to the TIR1 auxin receptor[22-23].Therefore,the downregulated of auxin signaling transduction might be associated with an increase in glucosinolates metabolism in broccoli sprouts under CHEW treatment.
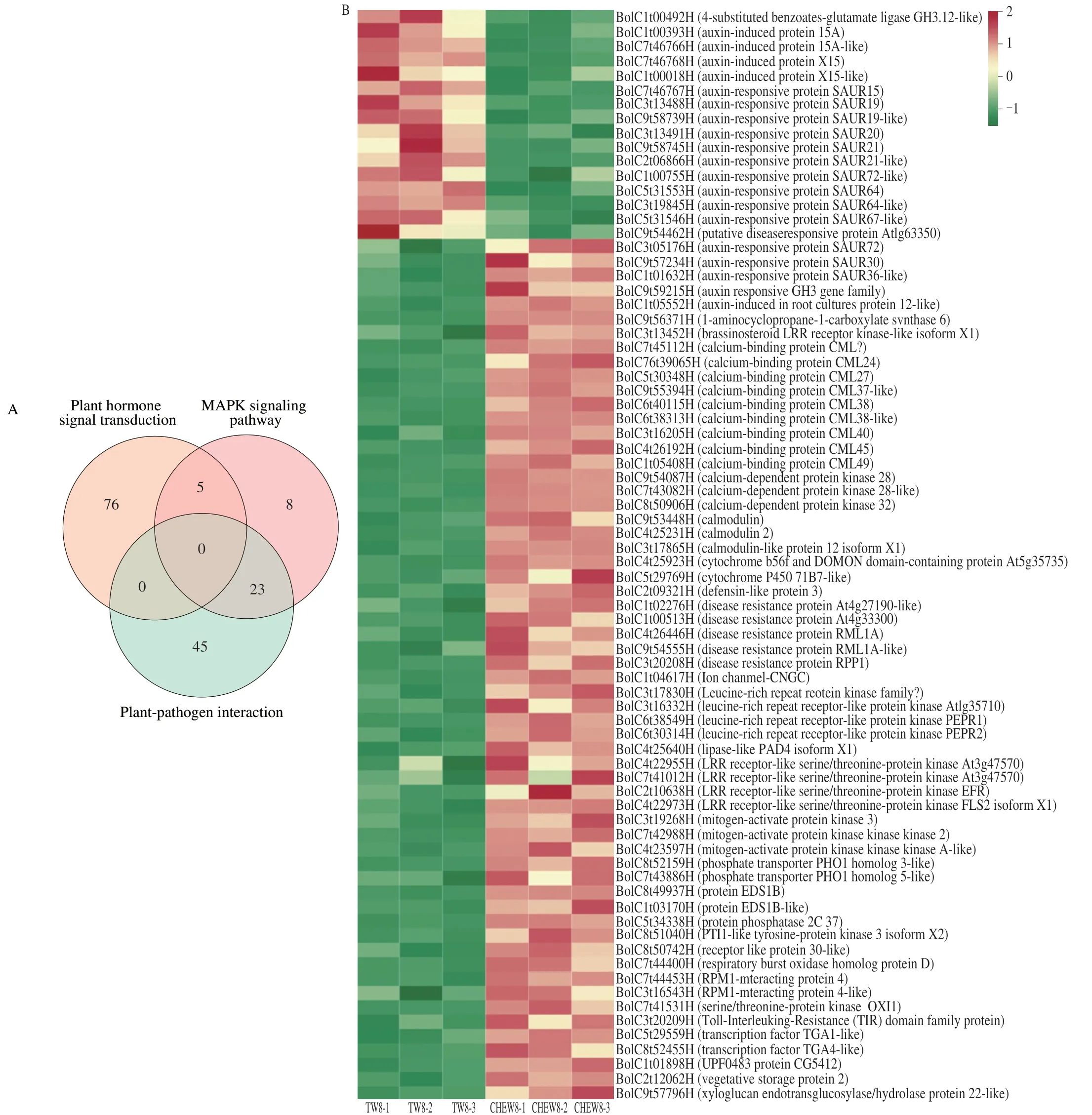
Fig.4 Analysis of DEGs between TW8 and CHEW8 in three predominant pathways (plant hormone signal transduction,MAPK signaling pathway and plantpathogen interaction).(A) Venn diagram of DEGs in three pathways.(B) Cluster heat map of DEGs in three pathways.
Studies showed that the plant MAPK cascades were were widely involved in plant abiotic stress responses by transmitting stress signals,activating downstream signaling pathways and the expression of related genes[24-26].The DEGs that were significantly up-regulated in the MAPK signaling pathway were mainly related to ROS signal and calcium signal transduction.Among them,calciumbinding proteins (CMLs),calcium-dependent protein kinases (CDPKs)and calmodulins (CaMs) were calcium sensors,which could detect calcium signals and trigger the the specific downstream physiological process.RBOHD was a respiratory burst oxidase involved in ROS generation[27].ROS and Ca2+played important roles in the activation of serine/threonine protein kinase (oxidative signal inducible1,OXI1),which could activate the protein kinase (MAPK) cascade (MAPK3/6)to affect plant’s stress resistance[28].The results showed that CHEW treatment first activated ROS signal and calcium signal,and then they interacted through MAPK cascades to affect glucosinolate biosynthesis in broccoli sprouts.
3.5 The metabolome and transcriptome combined analysis of glucosinolate synthesis pathways in broccoli sprouts
Among DAMs,a total of 36 glucosinolates were identified,including 26 aliphatic glucosinolates,5 indole glucosinolates(neoglucobrassicin,4-hydroxyglucobrassicin,4-methoxyglucobrassicin,1,4-dimethoxyindol-3-ylmethyl glucosinolate,sulfoglucobrassicin)and 5 aromatic glucosinolates (gluconasturtiin,clucosinalbin,3-phenylpropyl glucosinolate,4-benzoyloxybutylglucosinolate,5-(benzoyloxy) pentyl glucosinolate) (Supplementary C).The aliphatic glucosinolate-glucoraphanin had the highest glucosinolate content in broccoli sprouts,accounting for more than 50% of the total glucosinolate content[29].Our study also confirmed this (Table S3).As shown in Fig.5A,the contents of most glucosinolates in broccoli sprouts with tap water treatment showed a decreasing trend from day 4 to day 8.Compared with the tap water treatment,CHEW treatment significantly increased the contents of aliphatic glucosinolates on the 8thday.In addition,the glucoraphanin content in the CHEW treatment was consistently higher than that in the tap water treatment during the growth of broccoli sprouts.The results indicated that CHEW can be a useful tool for enhancing the content of secondary metabolites(glucosinolates,flavonoids,phenolic acids,etc.) and calcium in broccoli sprouts intended for fresh consumption as a functional food.
To explore the difference in glucosinolate biosynthesis between tap water treatment and CHEW treatment during the growth of broccoli sprouts,DEGs in the glucosinolate synthesis pathway were identified.A total of 19 structural genes related to glucosinolate biosynthesis were screened from all DEGs and the expression of them were represented in the cluster heat map (Fig.5B).
The process of glucosinolate biosynthesis was shown in Fig.5C.Methionine (Met) was extended by a branched-chain amino acid aminotransferaseBCAT4to generate the 2-oxo acid.Then the 2-oxo acid undergoed a continuous multi-step reaction including condensation with acetyl-CoA by a methylthioalkylmalate synthaseMAM1and isomerization by an isopropylmalate isomeraseIPMI2.The product was transaminated by aBCAT3to chain-elongated Met and entered the core glucosinolate structure pathway[30].The DEGs involved in the formation of the glucosinolate core structure mainly included cytochromes P450 of the CYP79 family (CYP79F1,CYP79B1-3),γ-glutamyl peptidaseGGP1,glucosyltransferasesUGT74B1and the sulfotransferasesSOT16-18.The methylthioalkyl glucosinolates were obtained after the formation of aliphatic glucosinolates’ core structure,and then were oxidized by the flavin monooxygenaseFMOGS-OXto generate methylsulfinylalkyl glucosinolates.Besides,the side chains of indole glucosinolates were derived from tryptophan (Trp),and CYP81F (1-4) could catalyze glucobrassicin (GBS) to form 4-hydroxyglucobrassicin(4OHGBS) and 1-hydroxy-glucobrassicin (1-hydroxy-I3M).Then they were catalyzed byO-methyltransferasesIGMT1andIGMT2to form methoxyglucobrassicin (4MOGBS) and 4-neoglucobrassicin(nGBS),respectively[31-32].Phenylalanine (Phe) is the precursors of aromatic glucosinolates biosynthesis[33].During the growth of broccoli sprouts,the DEGs related to methionine side chain elongation were significantly up-regulated while other structural DEGs were significantly down-regulated.Besides,compared with the tap water treatment,CHEW treatment could significantly increase the expression of most structural DEGs,which is consistent with the metabolome results.The results indicated that CHEW treatment promoted glucosinolate biosynthesis mainly by enhancing the expression of DEGs related to the formation of glucosinolate’ core structure and secondary modification of glucosinolate side chain.
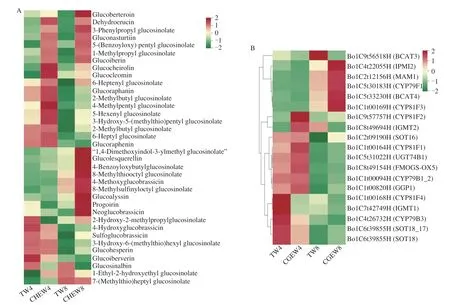
Fig.5 Combined metabolome and transcriptome analysis of glucosinolate synthesis pathways.(A) Cluster heat map of glucosinolates of different treatments.(B) The heatmap of structural DEGs in glucosinolate biosynthesis pathway according to the FPKM value.(C) The identification of structural DEGs in glucosinolate biosynthesis pathway.(D) RT-qPCR validation of 6 genes (BCAT4,MAM1,UGT74B1,SOT18,SOT16,CYP81F1,respectively) related to glucosinolate synthesis of broccoli sprouts.

Fig.5 (Continued)
RT-qPCR was used to analyze the expression levels of 6 structural genes related to glucosinolate biosynthesis in broccoli sprouts (Fig.5D).The results showed that the expression patterns of these genes were consistent with the RNA-seq results,indicating that the RNA data are valid and reliable.
3.6 The analysis of glucosinolates biosynthesis regulation network in broccoli sprouts under CHEW treatment
Transcription factors are important regulators of glucosinolate accumulation and typically act by regulating the expression of structural genes in the glucosinolate biosynthesis pathway.By searching the transcriptome annotation results,a total of 663 transcription factors in all DEGs were identified in this study.As shown in Fig.6A,most of transcription factors belonged to AP2/ERF-ERF,bHLH,MYB,NAC,WRKY and C2H2 families.Previous studies showed that MYB,bHLH and WRKY transcriptionfactors were important factors in the biosynthesis of glucosinolates in cruciferous plants,and were also involved in plant secondary metabolism,signal transduction,and stress response[34-36].To understand the regulation of transcription factors,the correlation tests between the expression levels of these transcription factors and the total glucosinolate content were carried out.Based on the results (Table 1),a total of 14 key transcription factors were highly associated with the glucosinolate content (|r| ≥ 0.95,P< 0.05).There were 7 negatively correlated transcription factors (WRKY9,bHLH118,MYB38,MYB51and 3 NAC family genes) and 7 positively correlated transcription factors (MYB44,AZF3,2 bHLH family genes and 3 AP2/ERF-ERF family genes),indicating that they played important roles in glucosinolate biosynthesis of broccoli sprouts under CHEW treatment.
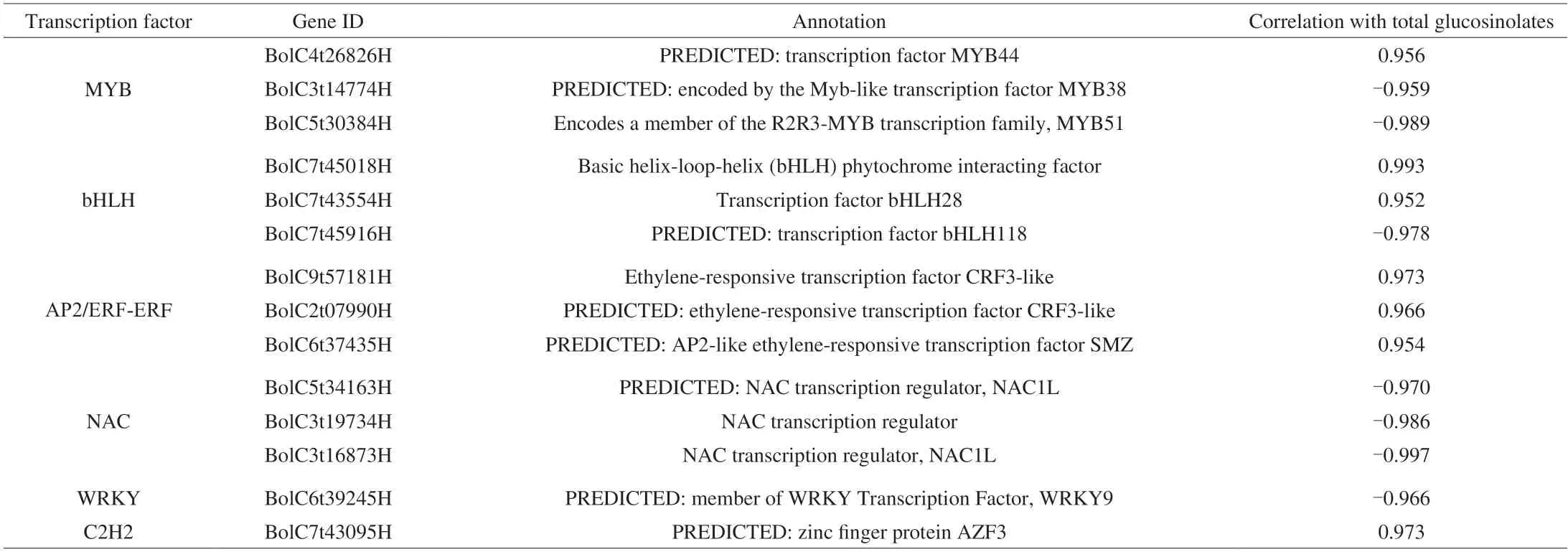
Table 1 Differentially expressed transcription factors involved in glucosinolates biosynthesis.
To further understand the regulatory network of glucosinolates biosynthesis in broccoli sprouts with CHEW treatment,we carried out a correlation analysis between the changes in glucosinolates and transcripts of genes.16 glucosinolates (12 aliphatic glucosinolate:glucoraphanin,2-methylbutyl glucosinolate,4-methylpentyl glucosinolate,3-methylbutyl glucosinolate,6-heptyl glucosinolate,glucoraphenin,8-methylsulfinyloctyl glucosinolate,glucoalyssin,3-hydroxy-6-(methylthio)hexyl glucosinolate,glucohesperin,glucolesquerellin,glucoiberverin;4 indole glucosinolates),10 structural genes (BCAT4,CYP79F1,CYP81F1,CYP81F3,CYP79B1_2,CYP79B3,GGP1,UGT74B1,SOT16,FMOGS-OX5)showing higher correlation coefficient values (|r| ≥ 0.9,P< 0.05)) and 10 transcription factors were obtained to organize an interaction network (Fig.6B).Glucosinolate synthesis in broccoli sprouts under CHEW treatment is mainly regulated by structural genes and transcription factors.Most of transcription factors affect glucosinolate biosynthesis by regulating the expression of structural genes.The results further clarified the regulatory networks of glucosinolate synthesis in broccoli sprouts under CHEW treatment.However,it still needs further molecular biology experiments to confirm.
Studies have found that ROS,Ca2+and key molecules such as MAPK,transcription factors (WRKY,bHLH and MYB family,etc.)have obvious interactions[20,37].The MAPK cascade is an important class of molecules for signal reception,transduction and delivery into the nucleus[38].The mechanism of CHEW treatment-induced glucosinolate biosynthesis in broccoli sprouts was illustrated in Fig.6C.The oxidative components (HClO,ClO-) and calcium of CHEW elicited the stress response in broccoli sprouts,and ROS and calcium signaling transduction pathways were activated.Then,ROS and calcium signals might interact through the MAPK cascade pathway,and promote the biosynthesis of amino acid and expression of transcription factors related to glucosinolate biosynthesis.Finally,the transcription factors promoted the expression of structural genes of glucosinolate synthesis to affect the glucosinolate biosynthesis in broccoli sprouts.
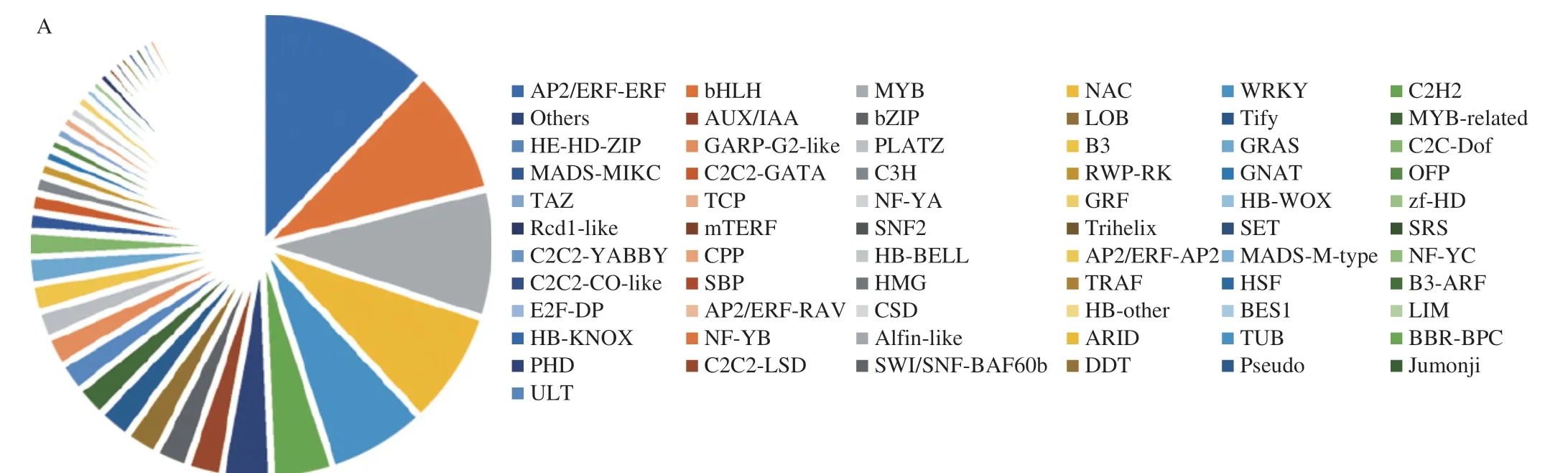
Fig.6 The analysis of glucosinolates biosynthesis regulation network in broccoli sprouts.(A) Classification and statistical analysis of all transcription factors detected.(B) The interaction network between glucosinolates and genes (structural DEGs and transcription factors).(C) Schematic overview of the mechanism of CHEW treatment regulating glucosinolate biosynthesis in broccoli sprouts.
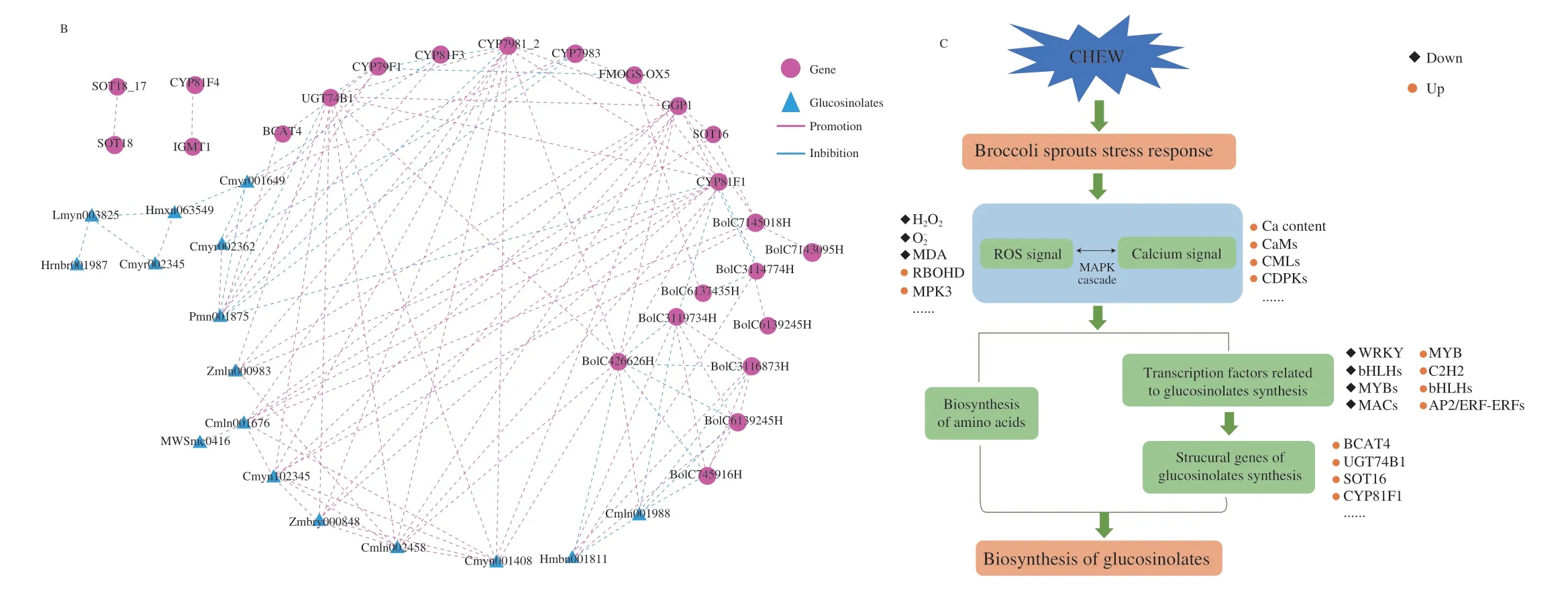
Fig.6 (Continued)
4.Conclusion
Our results showed that CHEW treatment significantly decreased ROS and MDA contents in broccoli sprouts.Compared to the tap water treatment,the calcium content in broccoli sprouts treated with CHEW on day 8 was dramatically enhanced from 14.4 mg/g DW to 22.7 mg/g DW.In addition,the total glucosinolate content of broccoli sprouts with CHEW treatment on day 4 and day 8 increased by 10.8% and 10.6%,respectively.The metabolome analyses implied that CHEW treatment might affect the biosynthesis of glucosinolate in broccoli sprouts by promoting the biosynthesis of cofactors and amino acids,the precursors of secondary metabolites.Transcriptome analyses revealed that CHEW treatment activated the ROS and calcium signaling transduction pathways in broccoli sprouts,and they interacted through MAPK cascades.Transcription factors affected glucosinolate biosynthesis by by promoting the expression of structural genes related to glucosinolate synthesis.The current study provides new insights into the molecular mechanisms of CaCl2-HCl electrolyzed water-induced glucosinolate biosynthesis and provides useful information on the development of effective methods to improve the nutritional quality of broccoli sprouts.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31972091).
Appendix A.Supplementary data
Supplementary data associated with this article can be found,in the online version,at http://doi.org/10.26599/FSHW.2022.9250068.
- 食品科学与人类健康(英文)的其它文章
- Protective effects of oleic acid and polyphenols in extra virgin olive oil on cardiovascular diseases
- Diet and physical activity inf luence the composition of gut microbiota,benef it on Alzheimer’s disease
- Inf luence of nitrogen status on fermentation performances ofnon-Saccharomyces yeasts: a review
- Ganoderma lucidum: a comprehensive review of phytochemistry,eff icacy,safety and clinical study
- Resveratrol combats chronic diseases through enhancing mitochondrial quality
- Demonstration of safety characteristics and effects on gut microbiota of Lactobacillus gasseri HMV18

