Aptasensing biosynthesized phosphatidylserine with a AuNPs nanozyme-based colorimetric aptasensor
Sai Wang,Rui Ma,Chengqiang Li,Ling Zhang,Haiyang Zhang,Xuehan Li,Xiangzhao Mao,c,
a Qingdao Key Laboratory of Food Biotechnology, College of Food Science and Engineering, Ocean University of China, Qingdao 266404, China
b Key Laboratory of Biological Processing of Aquatic Products, China National Light Industry, Qingdao 266404, China
c Laboratory for Marine Drugs and Bioproducts of Qingdao National Laboratory for Marine Science and Technology, Qingdao 266237, China
Keywords: Phosphatidylserine Chimeric aptamer AuNPs nanozyme Colorimetric aptasensor
ABSTRACT Sensitive monitoring of the target products during the biosynthesis process is crucial,and facile analytical approaches are urgently needed.Herein,phosphatidylserine (PS) was chosen as the model target,a colorimetric aptasensor was developed for the rapid quantitation in biosynthesis samples.A chimeric aptamer was constructed with two homogeneous original PS aptamers.Specific recognition between the chimeric aptamer and PS results in the desorption of aptamer from the surface of the AuNPs nanozyme,and the peroxidase-like enzymatic activity of the AuNPs nanozyme was weakened in a relationship with the different concentrations.The developed aptasensor performed well when applied for analyzing PS in biosynthesis samples.The aptasensor offers good sensitivity and selectivity,under optimal conditions,achieving monitoring and quantitation of PS in the range of 2.5-80.0 μmol/L,with a limit of detection at 536.2 nmol/L.Moreover,the aptasensor provides good accuracy,with comparison rates of 98.17%-106.40%,when compared with the HPLC-ELSD.This study provides a good reference for monitoring other biosynthesized products and promoting the development of aptamers and aptasensors in real-world applications.
1.Introduction
Currently,marine bioactive substances such as phosphatidylserine,polyunsaturated fatty acids,laminarin,and holothurian glycosides are drawing more and more attention[1-3].The biosynthesis method can realize the green,pollution-free,and efficient preparation of marine substances.Thus,the application of biosynthesis technologies to prepare marine bioactive substances has gradually become a research hotspot in recent years.Phosphatidylserine (PS) is a phospholipid that can regulate the functional status of essential proteins in cell membranes[4].It has functions of relieving stress,treating Alzheimer’s disease,repairing brain damage,improving brain function,treating depression,activating enzyme cofactors[5-8],etc.PS can be obtained by extracting separation from animal and plant cells[9],while it is rare in nature,and the complex separation processes and complicated impurities make it much more diff icult.At present,biosynthesis technology based on enzymatic bioconversion is used to prepare PS[10].During biosynthesis,it is critical to recognize and quantitate PS so as to attain precise monitoring of the target product.
Traditionally,high performance liquid chromatography (HPLC)is mostly adopted for the quantitation of the PS and other bioactive substances[11].Precise quantitation can be achieved.However,the extensive use of organic solvents,complicated operation,high pretreatment requirements,and time-consuming protocols makes it more suitable for certif ied conf irmation.To achieve rapid monitoring during the biosynthesis process,it is necessary and significant to develop facile and rapid detection methods for the quantitation of PS with high sensitivity and specif icity.
Aptamers,initially proposed in 1990[12-14],are RNAs or singlestranded DNAs (ssDNAs) containing 15-100 nucleotides.Specific aptamers can be obtained throughin vitroselection technology termed Systematic Evolution of Ligands by EXponential enrichment(SELEX),binding with targets in variety such as proteins[15],small molecules[16],antibiotics[17],ions[18],cells[19],nucleic acids[20],pathogens[21],toxins[22],and even viruses[23].Aptamers have distinct merits including rapid preparation with low-cost and no batch differences,ready availability,high chemical stability,and wide acceptability of chemical modification[24].In recent years,a variety of aptamers have been reported to be successfully screened and applied for biosensing detection[25],as well as drug analysis[26]and targeted therapy[27].Among them,biosensing detection with aptamer-based biosensors (aptasensors) is developed and reported for the detection of substances including antibiotics[28],bacteria[29],banned drugs[30],proteins[31],etc.A binding between the aptamers and targets is transduced into measurable signals,such as colorimetric signals,electrochemical signals,fluorescent signals,optical signals are being constructed[32],and thus achieving rapid and accurate detection.
The colorimetric technology has advantages of intuitive judgment and simple operation,thus showing a high application promise for realizing visual and rapid detection.Gold nanoparticles(AuNPs) are a kind of precious metal nanomaterials with great application prospects.Due to its excellent size dependence,surface plasmon resonance characteristics,and large specific surface area,it has been widely used in the development of visual and high-throughput colorimetric aptasensors.What’s more,the AuNPs showed peroxidase-like enzymatic activity and can act as peroxidase-like nanozyme.Developing aptasensors with AuNPs nanozyme helps to achieve simple,visualized,and sensitive detection[33].Further,ssDNAs like aptamers can adsorb onto the surface of AuNPs and greatly increase their peroxidase-like catalytic activity[34].Sensitive AuNPs-based colorimetric aptasensor can be developed according such features of the AuNPs and aptamers.
Herein,the biosynthesized PS was taken as a representative of biosynthesized products,a chimeric aptamer showing high affinity towards PS was generated,and a sensitive colorimetric aptasensor was developed with the chimeric aptamer and the AuNPs nanozyme.Biolayer interferometry (BLI) assay and computational assays were applied to assist in the generation of the chimeric aptamer.Based on AuNPs’ peroxidase-like activity,a colorimetric aptasensor was then fabricated for facile,rapid,and sensitive quantification of PS in biosynthesized samples.Moreover,the study helped to promote the development of aptamers and aptasensors in real-world applications.For a long time,although aptamers show great advantages,however,their application was limited when they are used in complex systems.Biosynthesis samples are simple,and the constructed aptasensor can provide a facile method to meet the urgent need for rapid quantification in this field,which means this research is extremely meaningful.
2.Material and methods
2.1 Reagents and instruments
All DNAs were commercially provided by Sangon Biotech Co.,Ltd.The sequence of the original PS aptamer was as follows:5’-AAAGAC-3’[35].The binding buffer offering working condition for the binding of aptamers and PS was prepared with 150 mmol/L NaCl and 50 mmol/L K2HPO4(pH 7.6).PS standards were obtained from Sigma-Aldrich Co.Avanti Polar-Lipids,Inc.provides phosphatidylcholine (PC).Beijing Solarbio Science &Technology Co.,Ltd.provided the 3,3’,5,5’-tetramethylbenzidine(TMB).Shanghai Yuanye Bio-Technology Co.,Ltd.provided phosphatidylethanolamine (PE),phosphatidic acid (PA),and phosphatidylinositol (PI) standards.Corning Incorporated Co.,Ltd.provided the microplates.Octet RED96e provided the streptavidinprecoated BLI chips.Other reagents were all analytical-grade domestic reagents.
Octet RED96e (Fortebio,USA) was applied for the characterization and measurment of the binding affinity of the aptamers.Centrifuge 5425 (Eppendorf,Germany) was used to obtain concentrated AuNPs.The JEOL-2100 was used to characterize the AuNPs with TEM imaging.Multiskan Sky (Thermo scientific,USA)were used to obtain the absorption spectra when aptasensing PS.UV-2550 Spectrophotometer (Shimadzu,Japan) were used to obtain the absorption spectra of the prepared AuNPs.
2.2 Computational analysis
With the assistance of AutoDock Tools,the molecular binding between PS and its aptamer was studied using molecular docking simulation which was performed.Before that,the pdb format file of aptamer was obtained from the Rnacompose website (http://rnacomposer.cs.put.poznan.pl/),and the sdf format file of PS was provided by the Zinc website (http://zinc.docking.org/).The molecular docking was then performed.First,the three-dimensional structure of PS was made flexible by adjusting the AutoDock Tools torsion parameters.Then,the aptamer was hydrotreated,and the charges were added before the atom type was determined.Next,the PS file was further converted to get the pdbqt format,along with which,the aptamer file was exploited to generate the corresponding Grid Box to obtain Grid energy files.After that,these files were processed with the Auto-Grid procedure.Molecular docking was then carried out after specifying relevant parameters[36].Binding mechanism was analyzed based on the output results.
2.3 BLI analysis
First,the 500 nmol/L biotin-labelled aptamer was heated at 95 °C and cooled,and 200 µL of pretreated aptamer was added to the 96-well plate.Then the aptamer was loaded onto the chip surface to form immobilized functional probes,which would bind selectively with PS in 200 µL sample solution and dissociate subsequently in the buffer solution.The program parameters were set according to our previous study[33].As a control,buffer solution without PS was employed during the measuring procedure.The variations on the thickness of the biolayer on the chip surface were tracked in real time and transformed into measurable and obvious concentration-dependent signals.After that,the Octet Data Analysis Software was used to get the concentration-signal response curves and the dissociation constant(Kd) with specific values indicating the aptamers’ binding affinity to PS was calculated.
2.4 Preparation and characterization of AuNPs
The traditional method based on trisodium citrate reduction was applied to prepare AuNPs,and then the AuNPs were concentrated before use.To begin with,100 mL of 0.01% chloroauric acid was steadily heated until boiling.Then,4 mL of 1% trisodium citrate solution was added quickly,and the solution was heated for another 10 min until the solution became wine red.With centrifugation at 12 000 r/min for 20 min at room temperature and subsequent removal of a certain amount of the supernatant,concentrated AuNPs solution was obtained with an enriched concentration of 10 nmol/L.The prepared AuNPs were characterized by UV-vis measurement and TEM observation.And the prepared AuNPs were kept at 4 °C in the dark.
2.5 Quantitation of PS with the developed aptasensor
Aptamers were pretreated by rapid heating (95 °C,5 min) and cooling (95 °C to room temperature).After that,the pretreated aptamers and PS were mixed and incubated for 20 min.Next,the solution was diluted with ultrapure water by 10 times,and an equal volume of the solution containing 10 nmol/L AuNPs was mixed and incubated for 15 min.To transduce the molecular binding into colorimetric signals,TMB,H2O2,and acetic acid solution were added under optimal conditions to allow the enzymatic transduction.The absorption spectra of 500-750 nm were recorded,and the absorbance value at 650 nm (A650nm) was collected for further analysis.TheA650nmvalues decreased gradually along with the increment of PS concentration.Thus,the change rate of the absorbance values was calculated and termed as signal change rate ((A0-Ai)/A0× 100%,whereAirepresented theA650nmwhen PS was present at a particular concentration andA0represented that in the absence of PS).
2.6 Optimization of the aptasensor fabricated with AuNPs nanozyme and chimeric aptamer
To get the optimal concentration of chimeric aptamers in the reaction system,the aptamer at concentrations of 0.312 5,0.625,1.25,2.5,5,10,and 20 µmol/L were mixed and incubated with the AuNPs nanozyme,respectively.After that,the mixture was then incubated with a mixture of TMB,H2O2,and acetic acid at a ratio of 3:7.After that,the absorption spectra at 500-750 nm were measured,and theA650nmwas collected.
To obtain superior sensing performance,components of the substance system were optimized with gradient concentrations of TMB (0.125,0.25,0.5,1,2,and 4 mmol/L),gradient concentrations of H2O2(1%,5%,10%,15%,20%,and 25%),and different pH of acetic acid (2.0,3.0,4.0,5.0,6.0,and 7.0).
For each optimization,the absorption spectrum obtained from each sample and the absorbance values recorded at 650 nm were collected and compared.
2.7 Preparation and pretreatment of biosynthesis samples
Biosynthesis products were prepared by the transphosphatidylation catalyzed by phospholipase D (PLD) towards PC andL-serine[37],then quantified with the HPLC-ELSD.Then,the prepared biosynthesis samples were diluted with cyclopentyl methyl ether and stored at 4 °C for analysis with the developed aptasensor.
3.Results and discussion
3.1 Scheme of the colorimetric aptasensor fabricated with a chimeric aptamer and AuNPs nanozyme
Fig.1 illustrates the scheme of development of the chimeric aptamer-based colorimetric aptasensor and the analysis of biosynthesized PS using the aptasensor.Firstly,a chimeric aptamer with enhanced affinity was generated with two homogeneous PS original aptamers.With the cooperative folding of the two sets of functional bases,the aptamer would exhibit a higher affinity towards PS.Then,the generated aptamer was designed to be combined with the AuNPs nanozyme to fabricate the proposed colorimetric aptasensor.The free aptamer would adsorb easily on the AuNPs,improving the peroxidase-like enzymatic capability of the AuNPs nanozyme towards TMB.The specific molecular binding between the chimeric aptamer and PS would lead to the desorption of aptamer from the surface of the AuNPs nanozyme,leading to a weaker enzymatic capability of the nanozyme towards TMB.As a result,with the increment of PS concentration,the aptamers tended to dissociate from the AuNPs,and the peroxidase-like catalytic ability of AuNPs nanozyme decreased,making the absorbance values at 650 nm weaken gradually,thus achieving highly sensitive detection of PS.With the developed colorimetric aptasensor fabricated with the chimeric aptamer and AuNPs nanozyme,rapid and sensitive quantitation of PS could be realized thereof.
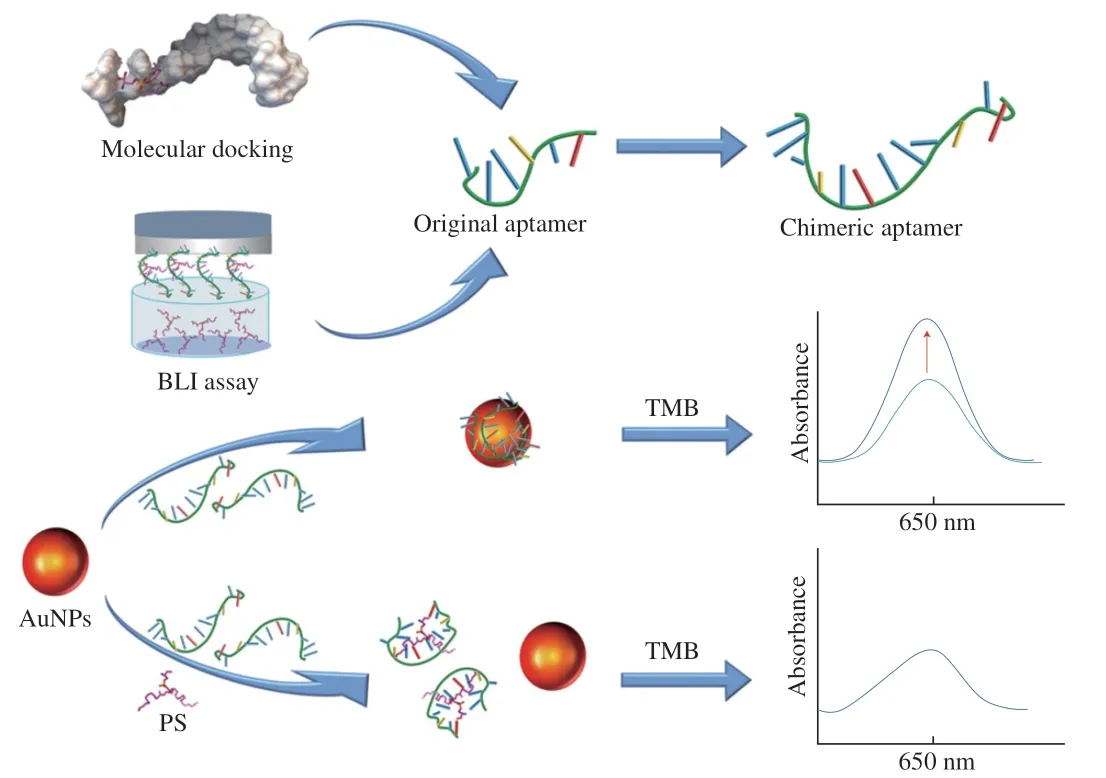
Fig.1 Schematic illustration of the design of the facile colorimetric aptasensor fabricated with the chimeric aptamer and AuNPs nanozyme.
3.2 Generation of the chimeric aptamer
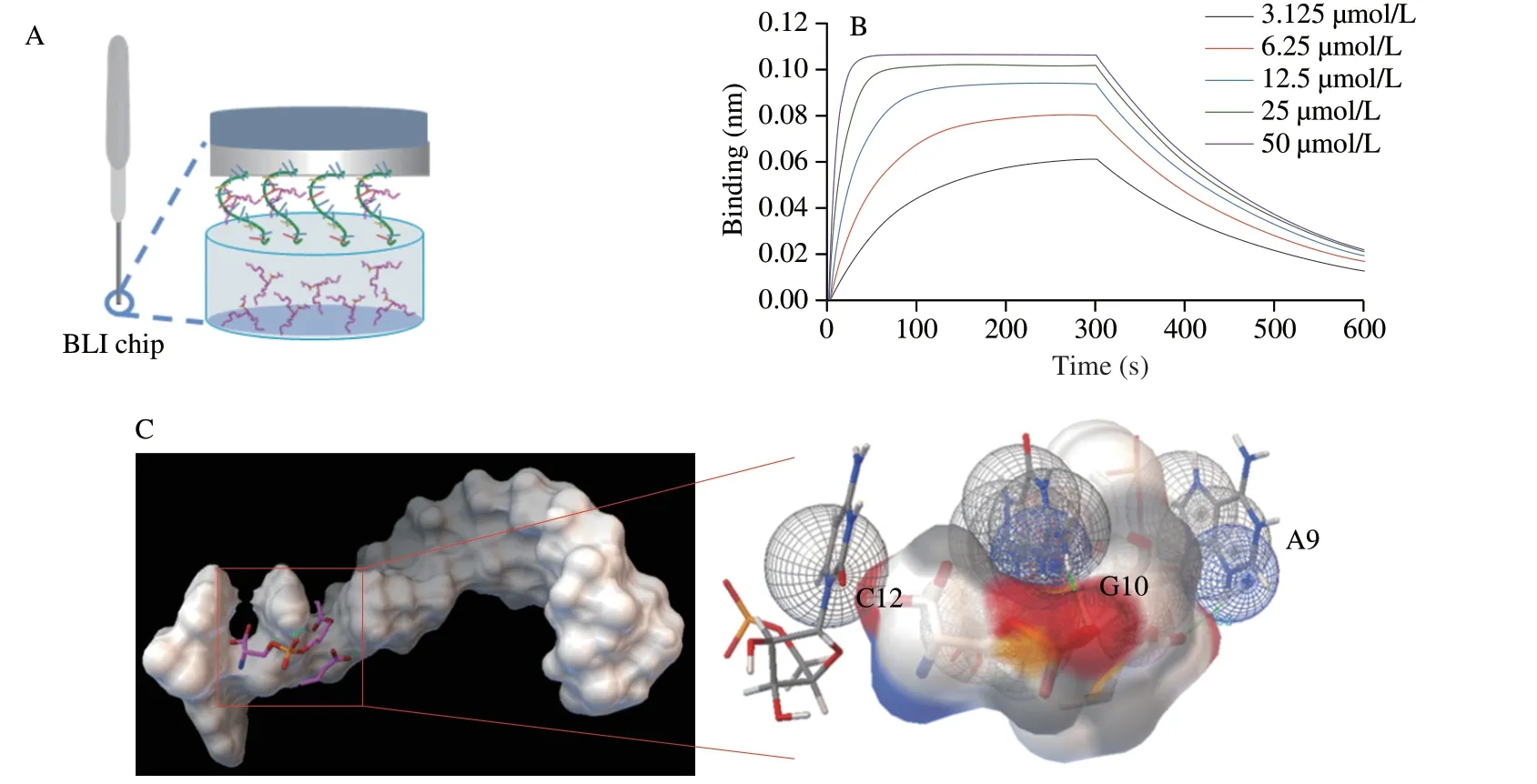
Fig.2 (A) Scheme of BLI assay for characterizing the binding affinity of the chimeric aptamer.(B) BLI assay results of the characterization of the chimeric aptamer and PS.(C) Molecular docking of the chimeric aptamer and PS.
Two original PS aptamers (5’-AAAGAC-3’) withKdof 6 528 nmol/L[35,38]were connected to generate a chimeric aptamer.With the cooperative folding the two aptamers,the chimeric aptamer would show a higher affinity.As is known,the BLI assay has the advantage of real-time optical analysis.To verify the design,BLI assay was carried out to characterize and measure aptamers’ binding affinity.Fig.2A illustrates the scheme of BLI assay,which was performed based on the fiber-optic biosensors for measuring the interactions between the immobilized chimeric aptamer and the PS[39].As shown in Fig.2B,the response changed in an increasing tendency along with the increment of the PS concentration varying from 3.125 µmol/L to 50 µmol/L.With assistance of the Octet Data Analysis Software,the affinity curve was fitted,and theKdof the original and the chimeric aptamer were determined to be 6 528 nmol/L[38]and 2 405 nmol/L,respectively,indicating an improvement on the high affinity of the chimeric aptamer towards PS.Moreover,the affinity between the chimeric aptamer and PS was investigated by molecular docking simulation.The information in Fig.2C showed that the A9,G10,C12 were the crucial bases facilitating the binding between the chimeric aptamer and PS.
3.3 Preparation of the AuNPs nanozyme
AuNPs nanozyme with a diameter around 13 nm were prepared and further condensed to 10 nmol/L.The sharp absorption around 520 nm (Fig.3A) and TEM imaging results (Fig.3B) indicated the successful preparation of AuNPs.
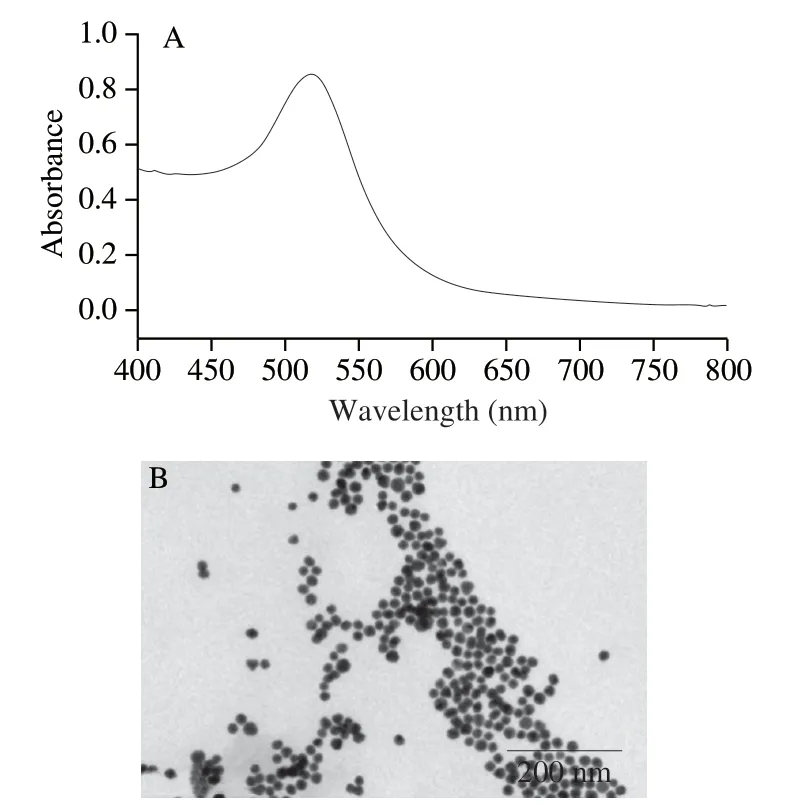
Fig.3 (A) The absorption characterization and (B) TEM characterization of the AuNPs.
3.4 Optimization of the developed aptasensor fabricated with the AuNPs nanozyme and the chimeric aptamer
The concentration of aptamer,TMB,pH value of acetic acid buffer,and concentration of H2O2in the catalytic reaction were crucial to the limit of detection,detection range,and sensitivity of the constructed aptasensor.To realize the excellent performance of the aptasensor,the concentration of chimeric aptamer,concentration of TMB,concentration of H2O2,and pH value of acetic acid buffer were optimized in detail.
Firstly,it is critical to utilize a rational concentration of the chimeric aptamer to achieve a higher sensitivity of the fabricated aptasensor.The insufficient concentration of the chimeric aptamers would result in very limited enhancement of the peroxidase catalytic performance of the AuNPs nanozyme,thus leading to a narrow detection range.On the contrary,with a high concentration of chimeric aptamers,the excessive aptamers not bound with PS would always be adsorbed on the surface of AuNPs,making the sensitivity of detection very limited.As shown in Figs.4A-B,the collectedA650nmfrom the sample solutions increased with a positive correlation with the concentration of the chimeric aptamer.No obvious improvement was observed when the concentration was higher than 10 µmol/L.Therefore,the concentration of the chimeric aptamer at 10 µmol/L was chosen as the optimal.Meanwhile,the parameters concentration of TMB,pH value of acetate buffer,and concentration of H2O2in the catalytic reaction were optimized.
The absorbance values reflected the catalytic capacity of the system with the change of independent variables.The higher theA650nmvalue,the stronger the catalytic ability.According to the absorbances obtained under different conditions,the results of significant analysis and the principle of saving reagent,the following experimental conditions were screened as optimal: The optimal concentration of TMB,1 mmol/L (Figs.4C-D);optimal pH value of acetic acid,4.0 (Figs.4E-F);the optimal concentration of H2O2,10%(Figs.4G-H).
3.5 Analytical curves and LOD of the aptasensor fabricated with the AuNPs nanozyme and the chimeric aptamer
Under the optimal conditions and with the high-affinity aptamer and the peroxidase-like catalytic ability of AuNPs nanozyme,the proposed colorimetric aptasensor for aptasensing PS was developed.The response curves of the aptasensor fabricated with the AuNPs nanozyme and the chimeric aptamer were attained by adjusting the concentration of PS.After analysis,the resulting spectra and signal diagrams are presented in Figs.5A-B.The binding between the chimeric aptamer and PS resulted in the desorption of chimeric aptamers from the surface of AuNPs nanozyme,leading to the decrement of the peroxidase-like enzymatic capability of AuNPs nanozyme,as well as theA650nm.As shown in Fig.5B,with the PS in gradient concentrations added,the value ofA650nmvaried with a decreasing tendency.And thus,the signal change rate presents a progressive increment.As shown in Fig.5C,within the concentration range of 2.5-80.0 µmol/L,the signal change rate caused by the PS had a good linear correlation with the logarithm of the concentration of PS.Limit of detection (LOD) of the colorimetric aptasensor fabricated with the AuNPs nanozyme and the chimeric aptamer was calculated to be 536.2 nmol/L,estimated as the concentration that corresponded to 3 standard deviations above the blank.The LOD was much lower than those of the HPLC-based methods,which were reportedca.1.93 × 105[40],6.42 × 104[41],or 1.05 × 105nmol/L[43],indicating a higher sensitivity of the present study.

Fig.5 (A) Spectra of the solution with different concentration of PS added in the solution.(B-C) The response and the calibration curves of the colorimetric aptasensor fabricated with the AuNPs nanozyme and the chimeric aptamer.The linear equation was y=15.30 lg(concentration of PS)+12.40 (R2=0.995 7).(D) Selectivity assay of the aptasensor.
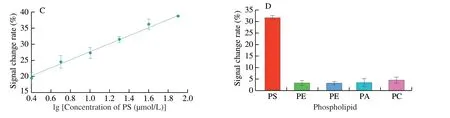
Fig.5 (Continued)
3.6 Selectivity of the aptasensor fabricated with the AuNPs nanozyme and the chimeric aptamer
During the analysis,it might be influenced by the structural analogs of PS,such as PE,PI,PA,and PC.For further evaluation on the selectivity of the aptasensor fabricated with the AuNPs nanozyme and the chimeric aptamer,the aptasensor was tested with PS and these structural analogs.Theoretically,the aptasensor only recognizes PS,and other targets cannot cause the signal changes.As shown in Fig.5D,the signal change rate obtained with the sample containing PS was significantly higher than that from other samples.Other analogs elicited only a low signal change rate of less than 5%.These results indicated that the aptasensor demonstrated remarkable selectivity for PS recognition.
3.7 Analyzing biosynthesized PS with the fabricated colorimetric aptasensor
In order to further evaluate the application potential of the fabricated colorimetric aptasensor,the aptasensor was used to applied to analyze real-world biosynthesized samples.Biosynthesized samples were prepared with conversion rates of 15.20%,36.08%,and 75.9% (Fig.6).The aptasensor was tested with the three samples.The analysis results obtained by the fabricated colorimetric aptasensor were compared with those obtained from HPLC-ELSD with the comparison rates as an indicator.As tabulated in Table 1,the comparison rates ranged from 98.17% to 106.40%,indicating that the fabricated colorimetric aptasensor has good accuracy.Moreover,good repeatability of the developed aptasensor was indicated by the relative standard deviation (RSD) values,which were all lower than 8%.In short,the results suggest that the aptasensor has a good application prospect in the real-world monitoring of biosynthesized PS.

Fig.6 Analysis of PS from bioconversion samples by HPLC-ELSD.Sample 1,2,and 3 were biosynthesized samples prepared with conversion rates of 15.20%,36.08%,and 75.9%,which were equal to 1.95 × 103,4.64 × 103,and 11.75 × 103 µmol/L,respectively.

Table 1 Analysis of biosynthesized PS in bioconversion samples with the aptasensor.
4.Conclusions
In short,PS was chosen and taken as a representative of the biosynthesized product,a chimeric aptamer was generated,and a colorimetric aptasensor based on the chimeric aptamer towards PS and the AuNPs nanozyme was constructed in this study.The chimeric aptamer showed high binding affinity for PS.The difference in the peroxidase-like catalytic performance in the presence and absence of PS was used to develop a facile label-free colorimetric aptasensor for the rapid quantification of PS.The aptasensor showed a LOD at 536.2 nmol/L and a wide linear detection range of 2.5-80.0 µmol/L for PS,which can meet the requirement of sensitivity in practical applications.Furthermore,the aptasensor showed high accuracy and repeatability,with good comparison rates of 98.17%-106.40% and RSD below 8%,indicating high applicability in real world.Overall,this study provides a good reference for convenient,rapid,low-cost,and sensitive quantitation of biosynthesized PS and can be referred to for other target products in the biosynthesis process.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (31922072);the Natural Science Foundation of Shandong Province (ZR2020JQ15);and the Taishan Scholar Project of Shandong Province (tsqn201812020).
- 食品科学与人类健康(英文)的其它文章
- Protective effects of oleic acid and polyphenols in extra virgin olive oil on cardiovascular diseases
- Diet and physical activity inf luence the composition of gut microbiota,benef it on Alzheimer’s disease
- Inf luence of nitrogen status on fermentation performances ofnon-Saccharomyces yeasts: a review
- Ganoderma lucidum: a comprehensive review of phytochemistry,eff icacy,safety and clinical study
- Resveratrol combats chronic diseases through enhancing mitochondrial quality
- Demonstration of safety characteristics and effects on gut microbiota of Lactobacillus gasseri HMV18

