A benchtop brain injury model using resected donortissue from patients with Chiari malformation
Jacqueline A.Tickle, Jon Sen, Christopher Adams, David N.Furness, Rupert Price, Viswapathi Kandula, Nikolaos Tzerakis,Divya M.Chari,
Abstract The use of live animal models for testing new therapies for brain and spinal cord repair is a controversial area.Live animal models have associated ethical issues and scientific concerns regarding the predictability of human responses.Alternative models that replicate the 3D architecture of the central nervous system have prompted the development of organotypic neural injury models.However, the lack of reliable means to access normal human neural tissue has driven reliance on pathological or post-mortem tissue which limits their biological utility.We have established a protocol to use donor cerebellar tonsillar tissue surgically resected from patients with Chiari malformation (cerebellar herniation towards the foramen magnum, with ectopic rather than diseased tissue) to develop an in vitro organotypic model of traumatic brain injury.Viable tissue was maintained for approximately 2 weeks with all the major neural cell types detected.Traumatic injuries could be introduced into the slices with some cardinal features of post-injury pathology evident.Biomaterial placement was also feasible within the in vitro lesions.Accordingly, this ‘proof-of-concept’ study demonstrates that the model offers potential as an alternative to the use of animal tissue for preclinical testing in neural tissue engineering.To our knowledge, this is the first demonstration that donor tissue from patients with Chiari malformation can be used to develop a benchtop model of traumatic brain injury.However, significant challenges in relation to the clinical availability of tissue were encountered, and we discuss logistical issues that must be considered for model scale-up.
Key Words: biomaterial; Chiari malformation; cerebellar slice; human tissue; injury model; neuroregeneration; organotypic; traumatic brain injury
Introduction
Approximately 69 million people globally suffer a traumatic brain injury (TBI)each year (Dewan et al., 2018); penetrating TBI has the worst outcomes in head injury cases (Duckworth et al., 2013).Foreign body penetration can cause tissue cavitation and compression of nerve fibers/blood vessels, with extensive injury to, and loss of, neural cells (Vakil and Singh, 2017).Limited post-injury regeneration can result in a poor clinical prognosis with severe paralysis, functional impairment, and mental health issues (Peterson et al., 2020).Current clinical treatment is largely supportive and there is a need to develop pro-regenerative therapies.Preclinical research shows the significant promise of implantable biomaterials (including surgical grade ones) functioning as reparative bridges that facilitate regenerative processes including delivery of stem/precursor cells, immunomodulation, and reduced glial scarring (Purvis et al., 2020).
Therapeutic advances to mitigate disability have traditionally been tested in live animal models of neurological injury/disease.This is a controversial area,with a steady decline in public approval for animal testing [75% (2010) – 66%(2012) – 51% (2017) (Van Norman, 2020)].Major ethical concerns are shared by public/scientific communities, in relation to the potential for substantial animal suffering (Bracken, 2009).Rodents are used extensively in preclinical studies, but the data are not always reliable in predicting the outcome in human studies (Vandamme, 2014).For example, inter-species differences in brain structure and function result in different pathological and behavioral response following induced TBI (Xiong et al., 2013).Sex differences also show inter-species variability with human trials showing better outcomes following TBI for males.By contrast, animal trials can show more favorable outcomes for females, with progesterone suggested as having a neuroprotective role in animals (Gupte et al., 2019).A recent study trialing antibodies to combat neurodegenerative disorders found an increased neuroinflammatory response in human neural cells, a response not observed in mouse microglia(Trudler et al., 2021).The reasons underpinning such differences are likely to be complex and out of the scope of this introduction.However, approximately 79% of phase II trials fail to progress due to safety/efficacy issues (Dowden and Munro, 2019), based on the inherent difficulties in extrapolating drug data from animal to human (Basketter et al., 2012).Accordingly, there is a requirement for experimental models that offer the best predictability for regenerative therapy in humans.
However, modeling central nervous system (CNS) pathology presents a unique challenge, due to its intricate cytoarchitecture containing multiple, specialist cell types.Models which include human cells such asin vitro
co-culture systems, microfluidic bionetworks, ‘organ-on-a-chip’, and 3D organoid systems have gained substantial popularity recently.While extremely useful in their reductionist and high throughput element, these currently have limited ability to mimic the complexities of neural cytoarchitecture, extracellular matrix, and injury pathology.Specifically, we refer to the molecular microenvironment of lesion sites and the spatial/temporal pattern of neuropathological sequelae post-injury, such as glial scarring and immune responses.‘Brain-on-a-chip’systems deploying 3D cell cultures and microfluidic systems can simulate diffuse axonal injury in TBI after a strain injury (Dolle et al., 2014), but the capacity to model the complexity of penetrating TBI on a chip is currently limited.Similarly, human brain organoids are a revolutionary technologyusing pluripotent stem cell sources and do replicate features of neural architecture and development to a significant extent.It has recently been proven feasible to study TBI in these systems using controlled cortical impact with an evaluation of neuronal and astroglial pathology; additional immune components are needed, however, for adequate patho-mimicry (Jgamadze et al., 2020; Ramirez et al., 2021).Human iPSCs are a valuable platform forin vitro
, patient/pathology-specific models to test drug and therapy efficacy(Alia et al., 2019), with the use of support media and substrates that mimic the extracellular matrix and signaling environment (Logan et al., 2019).This is a transformative technology but there are difficulties in correlating organoid neurodevelopment to the human brain (Lancaster et al., 2013).Further, iPSCs have a poor conversion rate for genetic modification and require a substantial time commitment to establish cell lines.Importantly, any system based on the use of iPSCs lacks immune components, critical for studies in regenerative neurology (Doss and Sachinidis, 2019).The subtype and maturation of both neurons and astrocytes are difficult to correlate within vivo
findings, with regional variation within the CNS resulting in variability between cell-based organoids (Logan et al., 2019).Lack of vascularisation (Auerbach et al.,2003) and necrosis are concerns (Logan et al., 2019), and to our knowledge,traumatic injury models have not been described in such systems.Organ-like or ‘organotypic’ models, including those incorporating neurological injuries, can provide an alternative approach in this regard.These are highly versatile for the investigation of neural development, pathophysiological mechanisms, and pharmaceutical interventions.The models maintain neural cytoarchitecture, vascular networks, and structural relationships of cells, allowing for parameters of neural regeneration and cell survival to be examined.A range of species can be used as donor tissue sources and some human neural tissue organotypic models have been attempted previously(Hendriks et al., 2021).However, these have usually relied on post-mortem donor tissue, epileptiform tissue, or tumor biopsies (Waldvogel et al., 2008;Schwarz et al., 2019).There are critical limitations associated with the use of tissue derived from such sources.Tissue quality relies on ante and postmortem factors such as respiratory stress, cause of death, post-mortem interval, tissue preservation method, and length of storage (Gomez-Nicola and Boche, 2015).A further limitation is variability between samples as the neural origin is dependent on the focus of the pathology (Jones et al., 2016)and the severity of the neurological insult to the surrounding areas.Further,most resected tissue is retained for diagnostic purposes leaving a scarcity of available tissue for investigatory research purposes (Jones et al., 2016).There are also limitations in relation to the pathology and exposure to patient medications (Jones et al., 2016) – points of contention when extrapolating the findings of such studies.
Here, we have evaluated the feasibility of developing anin vitro
, organotypic TBI model using human cerebellar tissue excised from patients with Chiari malformation (Type I) during decompressive surgery.This is a congenital abnormality characterized by downward displacement of the cerebellar tonsils beneath the skull base foramen magnum into the cervical spinal canal.The condition is typically diagnosed in adolescence or adulthood and may be associated with severe symptoms (e.g., headaches, dizziness, altered sensation, episodes of collapse, visual disturbance).In a proportion of cases,surgical treatment to decompress the foramen magnum and amputate the cerebellar tonsils is utilized.Thus, the tonsillar tissue is resected due to being ectopically positioned, as opposed to the majority of resective neurosurgery that is performed due to pathophysiological processes that give rise to tumor tissue or epileptogenic foci, for example.While there is a paucity of literature evaluating the cellular features of resected cerebellar tonsil tissue,it is considered to be within a range of cellular normality compared with e.g.,tumor or epileptogenic tissue.The study aims were to: (i) identify optimal culture conditions to maintain slices from resected tissue, with reliable detection of major neural cell populations; (ii) develop a protocol to introduce a focal traumatic injury in slices; (iii) establish if cardinal neuropathological responses are observed postinjury; and (iv) evaluate the feasibility of implanting a surgical grade scaffold into the injury site.
Methods
Patient recruitment
The study population was any patient (≥ 18 years) that had been diagnosed with Chiari malformation and subsequently required surgery to decompress the foramen magnum and remove the cerebellar tonsils.Exclusion criteria were unscreened tissue; patients with a history of blood-borne disease(e.g., human immunodeficiency virus), and patients that presented with neurological comorbidity e.g., epilepsy; neurodegenerative disorder.Eligible patients were identified in neurosurgical outpatient clinics and approached at the stage where surgery had been identified as the appropriate management strategy.Here, patients were provided with an information sheet and given at least 48 hours to read this, with informed consent taken from those who agreed to participate in the study.Patients were also informed of their right to withdraw consent at any point in the study.Where consent was given, the laboratory research team was advised 24 hours prior to surgery.All tissue samples were anonymized after collection.All study procedures for patient recruitment and tissue use were approved by the National Health Service(NHS) Research Ethics Committee (Yorkshire and the Humber; Reference: 17/YH/0010) on January 24, 2017.Tissue was handled in accordance with the Human Tissue Act 2004 and patient consent was also obtained for publication of any data arising from the study.The intended duration of the study was two years following which, in discussion with the clinical and research team we opted to terminate the study, with application to the NHS REC committee to conclude the permissions at this time.Accordingly, no further patients were recruited after 2019.
Materials (including test media details)
All media, supplements, and reagents, unless specified otherwise, were sourced from Fisher Scientific, Loughborough, UK.All plastic consumables were sourced from Greiner Bio One Ltd., Stonehouse, UK.For detailed information, see Additional file 1.
Human cerebellar tonsil tissue collection and slice derivation
Twenty-four hours before surgery, culture plates were prepared for organotypic slice maintenance using the liquid: air interface protocol –a proven method in line with previously published protocols from our laboratory (Weightman et al., 2014).Briefly, 1.1 mL medium was added to a 35 mm petri dish containing a membrane insert.Membrane inserts (30 mm;0.4 μm) and confetti (0.45 μm pore size) were both sourced from Millipore,Watford, UK.Three media with different chemical compositions were trialed for slice maintenance, each based on published protocols – Medium 1(Merz et al., 2013); Medium 2 (Brewer et al., 1993; Eugene et al., 2014), and Medium 3 (Andersson et al., 2016).Medium 1 comprised 50% minimum essential medium +GlutaMAX (MEM); 25% normal horse serum; 25% Hank’s Balanced Salt Solution (no phenol red, Ca, Mg); 1% L-glutamine; 1%penicillin/streptomycin (both SLS, Nottingham, UK); 1% glucose (Sigma-Aldrich,Dorset, UK).Medium 2 comprised 98% Neurobasal-A; 2% B27 Supplement;0.05 mM L-Glutamine (SLS).Medium 3 comprised 50% MEM; 25% normal horse serum; 18% Hank’s Balanced Salt Solution (no phenol red, Ca, Mg);2% B27 Supplement; 0.5% penicillin/streptomycin; 2 mM L-Glutamine; 11.8 mM glucose; 20 mM sucrose (Sigma-Aldrich).Three pieces (approximately 5 ×5 mm) of pre-cut confetti were placed onto the membrane, with one corner abutting the membrane edge, which could be held with tweezers to prevent disruption when lesioning tissue or for removing confetti.Prepared Petri dishes were placed in a large square, lidded petri dish and incubated at 37°C in 5% CO/95% humidified air.
Hibernate-A was used for the collection, transport, and initial processing of cerebellar tissue samples.Cerebellar tonsil tissue was resected by the neurosurgical team and placed into 50 mL falcon tubes, containing Hibernate-A, on ice.During the surgery, the neurosurgeon may deem tissue resection unnecessary or judge the resected tissue not viable for the study,for example, if the tissue was necrotic.Where tissue was taken, samples were immediately transferred to the laboratory research staff for transport to the laboratory (20–30 minutes).
For processing, tissue samples were transferred to a 90 mm petri dish containing chilled Hibernate-A, and the dimensions of each fragment were measured to estimate tissue volume (Figure 1).Organotypic slices were then derived from the tissue fragments according to published protocols(Weightman et al., 2014) modified for the slicing of human cerebellar tissue.Here, tissue samples were transferred using a wide-bore Pasteur pipette to a dampened cutting disc on a McIlwain tissue chopper.The blade was also dampened with Hibernate-A to prevent tissue sticking.Three slice thicknesses(200, 250, and 350 μm) were trialed for slice maintenance.Slicing was initiated within the tissue sample (~1 mm internal to the tissue edge) as it was observed that tissue fragmented when the blade was programmed to approach the tissue.Following 10–15 downward strokes of the blade,the chopper was halted, and slices were collected in a 35 mm petri dish by washing chilled Hibernate-A over the disc.This petri dish was then placed on ice for 90 minutes.The timeframe from excision to the slicing of alltissue samples was within 1 hour.Following the 90 minutes on ice, all slices were examined for damage (e.g., ‘raggedness’; tissue maceration; bruising/ruptured blood vessels), with wholly intact slices individually placed on confetti within a membrane insert and maintained at 37°C in 5% CO/95%humidified air.The overall processing time from tissue excision to culture on confetti was ~4 hours.
Organotypic slice culture maintenance
After 24 hours in culture, all the medium was removed, and fresh medium added to each petri dish.Subsequently, slices were maintained through 50%medium changes every 2–3 days for up to 40 days, dependent on experimental condition (detailed in results).In initial trials, Medium 3 (Andersson et al., 2016)proved the most effective in maintaining slice viability and, therefore, was used throughout the remainder of the study.At 7 days of slice maintenance,B27 Supplement was withdrawn from Medium 3 and replaced with MEM.All media were stored at 4°C and warmed to 37°C before use.At the indicated time points, slices were fixed (4% paraformaldehyde; 30 minutes; room temperature)for further processing or subject to live/dead staining.
Organotypic slice lesioning and implantation of DuraGen PlusTM into the lesion site
Following published protocols (Weightman et al., 2014), a penetrating,transecting injury was induced in a subset of slices at 8 days post slice derivation with the tissue from within the lesion removed using an aspirator(Figure 1: inset).Lesioned slices were maintained for a further 21 days.Subsets of slices were fixed at 7-, 14- and 21 days post-lesion.Immediately following injury, under microscopic observation the DuraGen Plus(~2 mm thickness) was cut to the size and depth of the lesioned tissue and implanted into the lesion site of a subset of slices.DuraGen Pluswas a kind gift supplied by Integra LifeSciences (Princeton, NJ, USA).
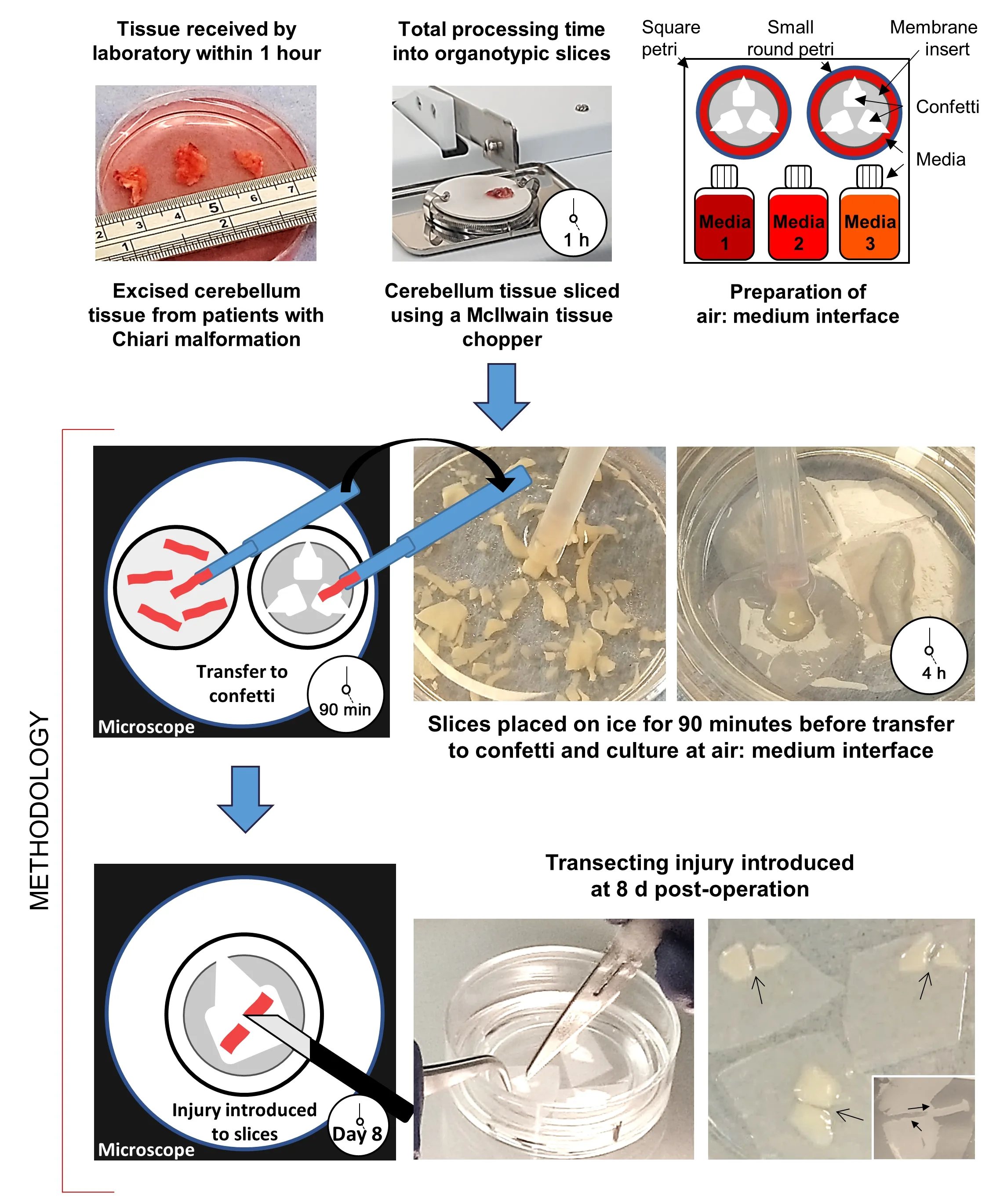
Figure 1 | Schematic diagram showing the production of organotypic neural slices,derived from cerebellar tonsillar tissue resected during decompressive surgery for Chiari malformation.
Live/dead staining to evaluate slice health
Slice health was assessed by calcein/ethidium homodimer staining at 4, 7,14, and 21 days and, in the case of one sample subset, at 40 days (ethidium homodimer (Sigma-Aldrich, Gilligham, UK) and calcein-AM (VWR, Poole,Leicestershire, UK).Slice cytoarchitecture and LIVE/DEAD proportions were assessed from fluorescence micrographs.Utilizing a previously reported technique (Weightman et al., 2014), the estimation of slice viability was based on comparative integrated density values of calcein (live cells) versus ethidium homodimer (dead cells).For detailed information, see Additional file 1.
Immunocytochemistry
To assess tissue architecture and the relationship with the DuraGen Plus™insert, slices were prepared either for immunocytochemistry or transmission electron microscopy (TEM).Slices were immunostained to detect major neural cell types: astrocytes (GFAP); neurons (Tuj-1); microglia (Iba-1) and oligodendrocytes (MBP).Immunostaining protocols used Tris-buffered saline 10x (Sigma-Aldrich); normal donkey serum (NDS) (Stratech Scientific, Ely,UK); Triton-X 100; gelatin (G-2625; Sigma-Aldrich).Primary and secondary antibodies were goat anti-Iba-1 (ionising calcium-binding adaptor protein-1)(Abcam, Cambridge, UK, Cat# ab5076, RRID: AB_2224402), mouse anti-Tuj-1(beta tubulin III) (Biolegend, London, UK, Cat# 801201, RRID: AB_2313773),rabbit anti-GFAP (glial fibrillary acidic protein) (DakoCytomation, Ely, UK,Agilent Cat# Z0334, RRID: AB_10013382); rat anti-MBP (myelin basic protein)(Bio-Rad, Watford, UK, Cat# MCA409S, RRID: AB_325004) and AffiniPure Cy3- and FITC-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA, Cat# 111-166-003, RRID: AB_2338007;Jackson ImmunoResearch Labs, Cat# 111-095-003, RRID: AB_2337972).Vectashield mounting medium with DAPI (4′,6-diamidino-phenylindole)was sourced from Vector Laboratories, Peterborough, UK.For detailed information, see Additional file 1.
Transmission electron microscopy
For electron microscopy, a subset of slices at 21 days post-lesion were glutaraldehyde fixed and post-fixed with 1% OsOprior to embedding in Spurr (Spurr, 1969) resin.Digital images were acquired from ultrathin (100 nm) sections examined on a JEOL 1230 TEM.For detailed information, see Additional file 1.
Results
Patient recruitment and tissue processing
Of the ten patients presenting with the condition and referred to the research team during the study period, nine patients consented to donate their tissue to the study.Of these, widespread tissue necrosis in the herniated cerebellar tonsils was observed in one case and three cases required decompression only.Resected tissue from five surgeries was therefore available for the research study.The volume of tissue received, and the number of slices generated was highly variable.The total volume (mm) of each tissue sample,calculated at the time of processing, was between 198–3510 mm.‘Intact’slices derived per sample and utilized within the study varied from 35–50(Table 1).
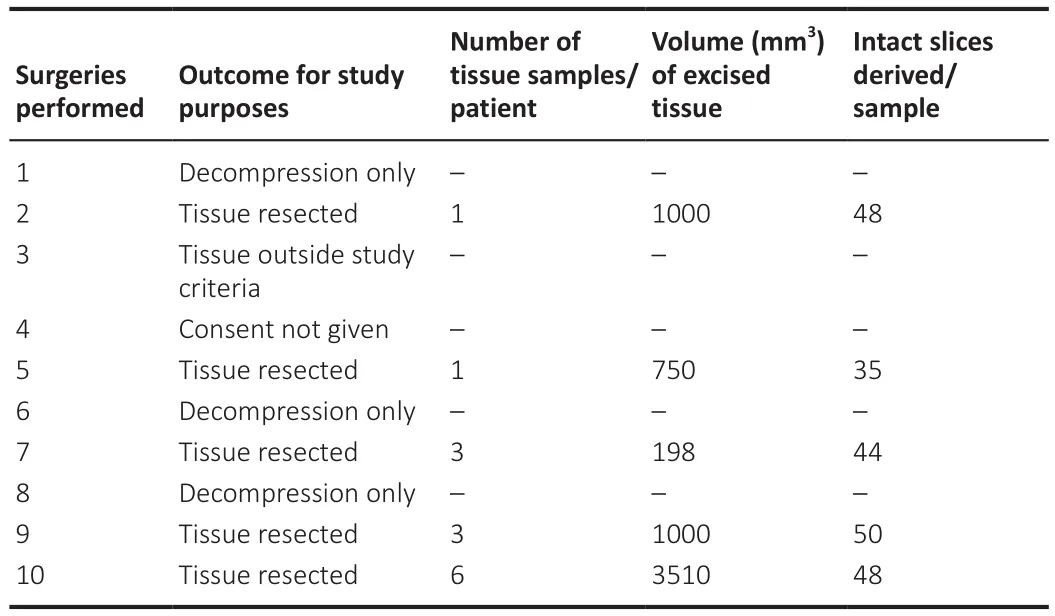
Table 1 |The number of patients requiring decompressive surgery for Chiari malformation referred to the research team, and the outcome for study purposes
Chiari tissue slice viability
During culture, microscopic observations of tissue provided an indication of overall slice health.Low viability/non-viable slices showed evidence of tissue shrinkage with an opaque, darkened appearance versus the translucent ‘intact’appearance of healthy tissue slices.Using this method, it was estimated that at 24 hours post-slice derivation, the average number of viable organotypic slices derived from each tissue sample was 45 (approximately 88% of initially derived slices; data not shown).After performing calcein/ethidium homodimer staining, it was observed that tissue viability at each experimental time point was high overall [> ~80% (Figure 2A)].Slices were observed to be viable for 21 days and in three cases, beyond this time point (Figure 2B–E).In one case, tissue was maintained for 40 days, and viable cells could be observed at this late time point [~72% viable (Figure 2F)].
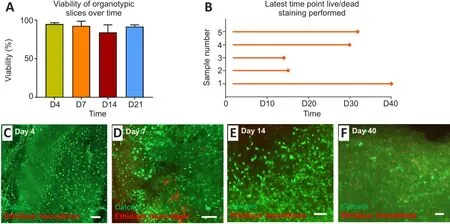
Figure 2 | Study outcomes and viability assessment of cerebellar tissue slices.
Major neural cell types in Chiari slices
All the major cell types of the CNS could be detected in the slices at 7-and 14-days post tissue derivation (Figure 3).Specifically, neurons (Tuj-1-immunoreactive cells), astrocytes (GFAP-immunoreactive cells), and microglia(Iba-1-immunoreactive cells) (Figure 3A–D) could all be reliably stained and detected, with oligodendrocytes (MBP-immunoreactive cells) also observed on day 14 (Figure 3E–H).Occasionally, cells showing features consistent with reactive astrogliosis [GFAP upregulation and hypertrophy (Figure 3B)]could be observed in non-experimental lesion areas, potentially due to local insult during surgical tissue resection.Morphological changes characterizing microglial activation could be seen at the slice margins on days 7 and 14, with Iba-1-immunoreactive cells showing evidence of rounding with shortened,thicker processes (Figure 3D and G), potentially reflecting immune cell activation during tissue excision from the brain or due to slicing procedures.
Slice transecting injury and pathological responses
A penetrating (transecting) injury could be reliably introduced into the slices under microscopic guidance, at 8 days post slice derivation (Figure 4A and B).The average lesion width was 342.7 ± 6.7 μm.At 7 days post-lesion,pathological features consistent with reactive gliosis could be observed at the lesion margins as a dense band of GFAP upregulation (Figure 4C).There was also evidence of short-range axonal sprouting, with cellular processes immunostained for Tuj-1 seen extending beyond the lesion edge (Figure 4D).

Figure 3|Major neural cell types contributing to pathology are detectable in cerebellar tissue slices.
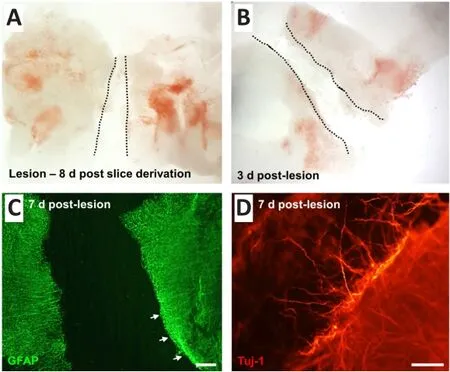
Figure 4 | A reproducible, penetrating (transecting) lesion can be introduced into the cerebellar tissue slice at 8 days post-slice derivation.
Implantation of a surgical grade matrix into lesions: light and electron microscopic analyses
DuraGen Pluswas implanted into the lesion site immediately following injury.At 21 days post-lesion, in slices without biomaterial implantation,elevated GFAP expression was apparent at the lesion margin (Figure 5A).Cellular integration could be observed at the lesion-DuraGen Plusinterface(Figure 5B), with infiltration of astrocyte processes immunostained for GFAP(Figure 5C) and short-range sprouting of axons labeled with Tuj-1 (Figure 5D).
Ultrastructural observations of the tissue edge of the lesion adjacent to the DuraGen Plussupport observations with fluorescence microscopy, that cellular interactions occur at the interface between the slice and biomaterial insert.Individual cell types were difficult to identify at the edge of the lesion due to cellular degeneration.Some intact cells with dense cytoplasm could be observed (Figure 5E), reminiscent of neuroglia, although not definitively so.The DuraGen Plusitself was generally amorphous and inhomogeneous,with denser and lighter areas within, but little substructure (Figure 5F).Where the two interacted, cell membranes of adjacent cells were closely opposed to the DuraGen Plus, and potentially bound to it by some form of adhesion molecules, for example, cell surface receptors, suggesting the insert had become integrated to a degree with the cells in the slice (Figure 5G).The materials also appeared to interact with cell processes from within the slice and occur underneath the surface, likely meningeal cells in places (Figure 5G).

Figure 5 | Integration of cellular tissue and biomaterial occurs at the lesion-DuraGen® Plus interface.
Discussion
This ‘proof-of-concept’ study demonstrates that it is feasible to grow 3D neural tissue arrays using brain tissue derived from patients undergoing decompressive surgery for the treatment of cerebellar herniation (Chiari malformation type I tissue) to develop a viable organotypic model of TBI.Such injuries prevail in areas of high incidence of terrorism/violence presenting the worst clinical outcome in head injury cases.Several live animal penetrating TBI models have been developed, where large animal models have been used to create brain lesions through gunshot/stab wounds or penetrating ballistic-like brain injury -rifle pellet injury (Plantman, 2015).The ethical and technical considerations associated with such approaches are considerable.To our knowledge, this is the first report of successful organotypic slice derivation from Chiari tissue.Our model adds to the growing body of literature that supports the use of human tissue-based organotypic models for experimental research.There is a clear argument for the development of such a reductionist approach for experimental research and it has been suggested that “best predictability is achieved with human organotypic models that mimic the 3D microenvironment of human tissues” (Heinonen,2015).We also demonstrate the contact alignment of an implanted surgical grade scaffold that can potentially integrate into the injured tissue to modify the regenerative response.Such integration, in our view, likely occurs due to a combination of cellular processes including microglial infiltration of the biomaterial, along with ingrowth of astrocytic processes and neural fibers into the pores of the material.Further studies could examine such processes in detail, along with biomaterial degradation and the receptors/adhesion molecules mediating cell-material interactions.
There were several technical/logistical challenges to consider in the development of the current model.HTA-approved protocols for the collection and transfer of tissue samples from the operating theatre to the laboratory necessitated an extensive collaborative effort between the neurosurgical and experimental teams.The laboratory team received 24 hours’ notice prior to surgery.Delivery of transport medium to the clinical site for storage in refrigerated conditions close to the specified operating theatre then had to be arranged, with tissue collection immediately post-resection to reduce processing time.The proximity between hospital and laboratory was likely key to achieving high tissue viability.In all cases, transfer to the laboratory and slice derivation was conducted within one hour of the tissue being excised.
Any tissue displaying gross pathological insult noted at the time of excision(not evident on pre-operative imaging thereby invalidating its use), was disposed of by the neurosurgical team in accordance with their treatment of clinical waste.The absence of overt pathology means the entire section of resected tissue is available for organotypic slice culture.Crucially, there was no variability in tissue origin between specimens, establishing consistency in the study findings from sample to sample.Of the five cases in this study,there was variation in size and number of tissue samples, although viable slices were derived from every sample.In our hands, it was not feasible to orient the tissue in a specified plane for cutting.This, together with some cellular degeneration and post-slicing changes, made clear identification of anatomical areas and tracts within the slice difficult to achieve at the light and electron microscopic level, although specific neural subtypes could be reliably detected.To preserve tissue integrity the tissue was sliced from within thetissue, as slicing directly from the edge resulted in minced, rather than sliced,tissue.The air: medium interface protocol used here to maintain the sliceshas previously proven successful within our laboratory for rodent spinal cord(Weightman et al., 2014).Assessment of slice health was comparable with other studies (see Jones et al.(2016) for review) with slices showing viability for up to 21 days, and up to 40 days (in one sample).
Our proof-of-concept data here suggest that this approach could offer a feasible alternative to the use of pathological tissue for humanin vitro
model development.Tissues excised from Chiari malformation type I patients are surgically reported as displaced/ectopic (as opposed to neuropathological)neural tissue and classified as histologically normal without associated pathology.It may be assumed that a small degree of e.g., gliotic changes may accompany chronic ectopia, but there is very little literature examining this.Therefore, we consider that Chiari malformation type I tissue potentially represents the tissue of highest ‘physiological normalcy’ that is feasible to obtain surgically at the current time.However, there are significant challenges associated with acquiring resected Chiari tissue, as with any surgically resectedtissue.Tissue availability is dependent on several factors.There is no absolute standard of care consensus for the management of Chiari malformation type I; there are different types of decompressive surgery including bony compression alone (without dural opening), decompression with resection of the tonsils, decompression and duraplasty, or simple coagulation of the tonsils depending on the preferred techniques and surgical experience of the neurosurgical team, the age of the patient, and the associated pathology- primarily syrinx formation within a Health Trust (Ferguson et al., 2018;Stewart-Watson and Carroll, 2018), whereby surgical treatment needs to be tailored to individual cases.Patients with the same degree of ectopia and associated symptoms may be offered different management options across centres (e.g., decompression with resection, or decompression and duraplasty).Such factors critically impact the throughput-ness of the model.During the study duration (2 years), of those presenting with the condition,surgery was considered necessary in ten of the presenting patients.Of these, tissue excision was carried out in six cases, of which only five met the study criteria.Accordingly, we have proved that the protocol stated here is technically feasible.However, due to the challenges associated with acquiring resected Chiari tissue, scaling up of this approach as a reproducible, moderate throughputin vitro
system will require wider collaboration across multiple clinical sites.Conclusion
In principle, this study shows promising findings for a long-term benchtop model of human neurological injury for experimental and biological research.The use of histologically normal, adult neural tissue can offer a sophisticated experimental model and useful predictor of pathological responses seen following neurological injury.While we opted to terminate the study at 40 days (the latest time point examined), we consider the tissue can be maintained for longer periods to study regenerative processes occurring over extended time periods.Further, while a penetrating lesion was used to prove the concept that key pathological features can be detected, the model can potentially be adapted to a range of lesion models, including closed TBI.A range of pharmacological and biomaterial-based interventions can potentially be tested in such a system.Our tissue analyses suggest stereotypic injury (scarring) responses in astrocytes, along with a range of observed morphologies in microglia indicative of both resting (ramified)and reactive (amoeboid) phenotypes, supporting the concept that the influence of therapeutic interventions on such injury responses is feasible.Applications can be found in drug and cell therapy and biomaterial testing(for example, immunomodulatory or glial scar attenuation treatment)along with electrophysiological examination, as an alternative to the use of acute brain slices widely used in regenerative neurology investigations.Accordingly, models such as these can have an impact in reducing animal usage and ensuring the rapid progression of effective and safe therapies for CNS disorders, assuming the significant issues in relation to reliable tissue availability can be overcome for scale-up, potentially through multi-center collaboration.
Acknowledgments:
We also gratefully acknowledge help from Holly McGuire, Alda Remogoso, Joanne Hiden, Francis Alipio (UHNM clinical trial research nurse team) and clinical fellow Dr Maria Bushra, for their help with organizational/logistical aspects of the study.
Author contributions:
JAT, CA, JS and DMC conceived, wrote, and edited the manuscript.JAT carried out the experiments, analyzed the experimental results, and generated the table and figures.DMC and NT obtained the funding for this study.NT, RP and VK were the neurosurgical team who provided the surgically resected tissue.DNF supervised the electron microscopy experiments and analyses.All authors approved the final version of this paper.
Conflicts of interest:
The authors declare no conflict of interest.
Availability of data and materials:
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open access statement:
This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional file:
Supplementary Methods.
- 中国神经再生研究(英文版)的其它文章
- Patient-specific monocyte-derived microglia as a screening tool for neurodegenerative diseases
- Molecular hallmarks of long non-coding RNAs in aging and its significant effect on aging-associated diseases
- Inflammation in diabetic retinopathy: possible roles in pathogenesis and potential implications for therapy
- Targeting the nitric oxide/cGMP signaling pathway to treat chronic pain
- Neurosteroids as stress modulators and neurotherapeutics: lessons from the retina
- Myelinosome organelles in pathological retinas:ubiquitous presence and dual role in ocular proteostasis maintenance