Long noncoding RNA Pvt1 promotes the proliferation and migration of Schwann cells by sponging microRNA-214 and targeting c-Jun following peripheral nerve injury
Bin Pan , Di Guo , Li Jing, Ke Li, Xin Li Gen Li Xiao Gao Zhi-Wen Li, Wei Zhao, Hu Feng , Meng-Han Cao
Abstract Research has shown that long-chain noncoding RNAs (lncRNAs) are involved in the regulation of a variety of biological processes, including peripheral nerve regeneration, in part by acting as competing endogenous RNAs.c-Jun plays a key role in the repair of peripheral nerve injury.However, the precise underlying mechanism of c-Jun remains unclear.In this study, we performed microarray and bioinformatics analysis of mouse crush-injured sciatic nerves and found that the lncRNA Pvt1 was overexpressed in Schwann cells after peripheral nerve injury.Mechanistic studies revealed that Pvt1 increased c-Jun expression through sponging miRNA-214.We overexpressed Pvt1 in Schwann cells cultured in vitro and found that the proliferation and migration of Schwann cells were enhanced,and overexpression of miRNA-214 counteracted the effects of Pvt1 overexpression on Schwann cell proliferation and migration.We conducted in vivo analyses and injected Schwann cells overexpressing Pvt1 into injured sciatic nerves of mice.Schwann cells overexpressing Pvt1 enhanced the regeneration of injured sciatic nerves following peripheral nerve injury and the locomotor function of mice was improved.Our findings reveal the role of lncRNAs in the repair of peripheral nerve injury and highlight lncRNA Pvt1 as a novel potential treatment target for peripheral nerve injury.
Key Words: cell migration; ceRNA; c-Jun; lncRNA; microarray; miR-214; nerve regeneration; peripheral nerve injury; Pvt1; Schwann cells
Introduction
Peripheral nerve injury (PNI) is a critical problem that affects the patient’s quality of life and brings high economic burdens to the family and society.Approximately 1.5–2.8% of trauma patients are affected by PNI, which often results in disability (Noble et al., 1998).The peripheral nervous system,unlike the central nervous system, exhibits an intrinsic regenerative power(He et al., 2016; Kubiak et al., 2020).Schwann cells, the main glial cells of the peripheral nervous system, play a key role in PNI repair.After PNI, the distal end of the injured axon degrades and produces debris.Schwann cells remove the degenerating axon and myelin debris either directly or by recruiting macrophages (Li et al., 2022; Walker, 2022).Schwann cells and the recruited macrophages then produce nerve growth factors that promote peripheral nerve regeneration (Lee et al., 2016).However, for severe PNI, the regeneration ability is limited (Qian et al., 2018).A previous study reported that c-Jun is essential for the restoration of damaged nerves.c-Jun is rapidly upregulated in Schwann cells after PNI, and in the absence of c-Jun, the biological activity of Schwann cells is markedly decreased (Jessen and Mirsky,2016).
In recent years, increasing research has focused on the relationship between long noncoding RNAs (lncRNAs) and Schwann cells after PNI (Zhou et al.,2021).lncRNAs are a class of RNA molecules over 200 nucleotides in length that lack protein-coding capacity (Li et al., 2013).Studies have shown that lncRNAs play epigenetic, transcriptional, and post-transcriptional regulatory roles in various biological processes.lncRNAs can also function as competing endogenous RNAs (ceRNAs) through microRNA (miRNA) response element sequences and competitively bind and ‘sponge’ miRNAs, thereby upregulating the expression of miRNA target genes (Thomson and Dinger,2016).lncRNAs play a regulatory role in the occurrence and development of various human diseases (Xue et al., 2017).Several studies have shown that lncRNAs play key roles after PNI.Pan et al.(2017a) reported that the lncRNA NONMMUG014387 promotes the proliferation and migration of Schwann cells after PNI.Yao et al.(2015) found that lncRNA uc.217 controls the outgrowth of dorsal root ganglion neurons after PNI.Some reports revealed that lncRNAs participate in peripheral nerve regeneration through their functions as ceRNAs.After diabetic peripheral neuropathy, lncRNAs competitively bind miR-212-5p to control the expression of target genes and affect the proliferation and migration of Schwann cells (Wang et al.,2020a).Better understanding of lncRNAs and ceRNA networks may provide new insights into the underlying molecular mechanism of peripheral nerve regeneration.
In this study, we explored the mechanisms of lncRNAs in nerve injury repair after PNI.We established a mouse model of sciatic nerve injury and detected the expression changes of lncRNA and mRNA in the injured segment after sciatic nerve injury through microarray technology.We performed bioinformatics analysis and prediction to analyze potential ceRNA regulatory networks during peripheral nerve regeneration and verified our findings usingin vitro
andin vivo
experiments.Methods
Animals and PNI model
To eliminate the influence of sex and hormone levels of experimental animals, only male mice were used in this study.In total, 117 C57BL/6 male mice (8-week-old, 22–27 g, Lab Animal Research Center of Xuzhou Medical University; license No.SCXK (Su) 2020-0011) were used in this study.Animals were raised under specific-pathogen-free conditions at 22–25°C with 12 hours of sufficient light.
In initial experiments, 72 mice were randomized into four groups (days 0, 3,7, and 14;n
= 18/group) and underwent sciatic nerve crush surgery (Liu et al.,2019).Mice were anesthetized by intraperitoneal administration of 1 mg/10 g ketamine (Nhwa Pharmaceutical, Xuzhou, China) and 0.1 mg/10 g xylazine(Sigma, Darmstadt, Germany)).The disappearance of voluntary movement indicated that anesthesia was successful.The sciatic nerve was exposed layer by layer and clipped 1 cm above the bifurcation of the tibia and fibula nerves with a vascular clamp for 20 seconds.The vascular clamp was opened after 20 seconds, and the incision was sutured layer by layer.The sciatic nerve tissue distal to injury was collected on days 0, 3, 7, and 14 after surgery.The sciatic nerve tissue was frozen in liquid nitrogen, and the operation was repeated three times.The other 45 mice were used forin vivo
experiments described in a later section.The experimental processes are shown in Figure 1.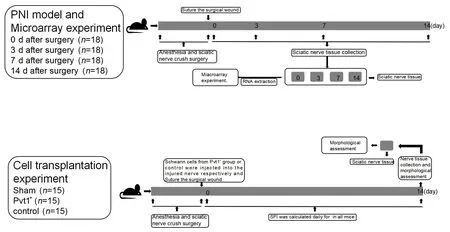
Figure 1|Schematic of the animal experiments.
All the experiments were approved by the Animal Ethics Committee of Xuzhou Medical University of China (approval No.L20210226440) on February 28,2021.All experiments were designed and reported according to the Animal Research: Reporting ofIn Vivo
Experiments (ARRIVE) guidelines (Percie du Sert et al., 2020).Microarray analysis
Total RNA was extracted from sciatic nerve tissue from each group using TRIzol (Invitrogen, Carlsbad, CA, USA) and purified with RNeasy spin columns(Qiagen, Hilden, Germany).The NanoDrop ND-2000 and Agilent Bioanalyzer 2100 instruments (Agilent Technologies, Santa Clara, CA, USA) were used to verify the purified RNA.Microarray was used to detect the changes of mRNA and lncRNA in the distal nerve at 0, 3, 7, and 14 days after PNI through the Array platform (Agilent Technologies).Results were normalized using GeneSpring v11.0 software (Agilent Technologies).The experiments were performed following the manufacturer’s protocols (Yu et al., 2013).
Bioinformatics analysis
Bioinformatics was used to identify differentially expressed mRNAs and lncRNAs.In hierarchical clustering, the Z-scores were determined based on expression levels for mRNAs and lncRNAs, and Euclidean distance measurements were employed when calculating lncRNA time and distance.For expression pattern clustering, expression patterns for differentially expressed mRNAs and lncRNAs were assessed.We defined some unique profiles using an approach to short-time-series gene expression data clustering.Expression model profiles were related to the number of genes associated with a given profile.Multiple comparisons tests were performed on the expression model using Fisher’s exact test (Liao et al., 2011).
The target miRNAs of differentially expressed lncRNAs were predicted using DIANA (http://diana.imis.athena-innovation.gr), miRWalk (http://mirwalk.umm.uni-heidelberg.de) (Sticht et al., 2018), miRDB (http://www.mirdb.org),and RNA22 (https://cm.jefferson.edu/rna22/) databases.These databases were also used to predict miRNAs that interact with c-Jun mRNA.Integration and analysis of the results were performed using an online Venn diagram tool(http://bioinformatics.psb.ugent.be/webtools/Venn).
Schwann cell culture and transfection
Mouse Schwann cells (Cat# 60507, RRID:CVCL_4694) were provided by ScienCell (San Diego, CA, USA).The Schwann cells were cultured in Dulbecco’s modified Eagle medium (ScienCell) containing 10% fetal bovine serum and 1%penicillin/streptomycin at 37°C with 5% CO.
Schwann cells were identified with S-100 staining.Cells were fixed with 4%paraformaldehyde and then incubated with anti-S-100 antibody (rabbit, 1:200,Abcam, Cambridge, UK, Cat# ab52642, RRID:AB_882426) overnight at 4°C.Cells were then incubated with Cy3-labeled goat anti-rabbit IgG (1:200, Bioss,Beijing, China, Cat# bs-0295G-Cy3, RRID:AB_10892956) for 1 hour at 37°C.Five non-overlapping regions were selected randomly and observed by a fluorescence microscope (Olympus, Tokyo, Japan).
Schwann cells were divided into four experimental groups.Cells in the control group were transfected with empty pcDNA3.1 plasmids (Synthgene, Nanjing,China).Cells in the Pvt1group were transfected with Pvt1 pcDNA3.1 plasmids(Synthgene) containing Pvt1 complementary DNA (cDNA).Cells in the Pvt1-group were transfected with the pRNAT-U6.1/Neo plasmid (Synthgene)expressing a short hairpin RNA of lncRNA Pvt1.Cells in the Pvt1/miR-214group were transfected with Pvt1 pcDNA3.1 plasmids (Synthgene) and miR-214 mimics (Synthgene) (Wu et al., 2020).Synthgene (Nanjing, China) was used to design and synthesize the plasmids and miR-214 mimics used in this study.Schwann cells were transfected using Lipofectamine 2000 (Invitrogen)at 37°C for 48 hours.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
RNA was isolated from cultured cells by TRIzol (Invitrogen).For analysis of Pvt1 and c-Jun mRNA, cDNA was reverse transcribed from RNA using the PrimeScript RT kit (TaKaRa, Tokyo, Japan).qRT-PCR was performed using SYBR Premix Ex-Taq (TaKaRa) with the ABI Stepone instrument (Applied Biosystems,Santa Clara, CA, USA) in triplicate, following the manufacturers’ protocols.To analyze miR-214 expression, RNAs were reverse-transcribed using primers specific for miR-214 with TIANScript M-ML V (Tiangen, Beijing, China).miR-214 expression was examined by qRT-PCR.The reverse transcription reaction conditions were incubation at 42°C for 30 minutes, denaturation at 85°C for 5 minutes, and heat preservation at 4°C.The qRT-PCR conditions were incubation at 50°C for 2 minutes and incubation at 95°C for 10 minutes,followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute.The results were calculated with the 2method with β-actin mRNA or U6 as an internal reference.Table 1 lists the primer sequences.
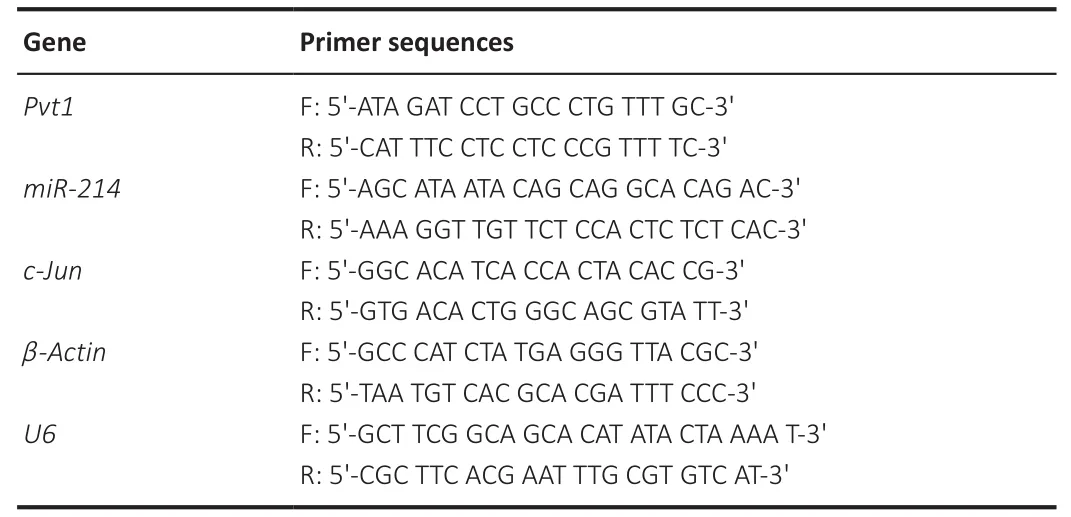
Table 1 |Primer sequences for polymerase chain reaction
Western blot analysis
Cultured Schwann cells were lysed by radioimmunoprecipitation assay lysis buffer (Sigma) and the protein concentrations of lysates were quantified using the BCA kit (Sangon Biotech, Shanghai, China).The protein samples were separated via polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Merck Millipore, Darmstadt, Germany).The membrane was blocked using 5% milk and then incubated with anti-c-Jun antibody (1:1000, Abcam, Cat# ab40766, RRID: AB_731602) and antiβ-actin antibody (1:1000, Sigma, Cat# A5441, RRID: AB_476744) overnight at 4°C.Goat anti-rabbit IgG (1:5000, Sigma, Cat# AP132P, RRID: AB_90264)was added and incubated for 1 hour at 4°C.Band detection was performed using an enhanced chemiluminescence kit (NCM Biotech, Suzhou, China).The results were analyzed by ImageJ software v1.8.0 (National Institutes of Health,Bethesda, MD, USA) as previously described (Schneider et al., 2012).
Isolation of cytoplasmic and nuclear RNA in Schwann cells
RNA was isolated from Schwann cell nuclear and cytoplasmic samples with the PARIS Kit (Invitrogen) as previously described (Wu et al., 2020).Expression of the lncRNA Pvt1 was detected from RNA by qRT-PCR.
Fluorescence in situ hybridization
Schwann cells were incubated with 10 μL probes labeled with digoxigenin(Invitrogen) in a humectant box at 37°C for 12–16 hours and then washed.Nuclei were stained with 4′,6-diamidino-2-phenylindole (Qiagen).Cells were observed with a fluorescence microscope.
Luciferase reporter assay
The sequences of c-Jun mRNA and Pvt1 were obtained from the NCBI database (https://archive-dtd.ncbi.nlm.nih.gov/).The c-Jun- and Pvt1-binding sequences in miR-214 were obtained from the BiBiServ2-RNAhybrid website(https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/submission.html).The wild-type (WT) and mutant (MUT) c-Jun-binding sequences in miR-214 and Pvt1-binding sequences in miR-214 were inserted into the psiCheck2 plasmids to construct psiChech2-c-Jun-WT, psiChech2-c-Jun-MUT, psiChech2-Pvt1-WT and psiChech2-Pvt1-MUT (Wu et al., 2020).
The plasmids were co-transfected with miR-214 or NC mimics into Schwann cells using Lipofectamine 2000 (Invitrogen) for 48 hours at 37°C.A dual luciferase reporter assay system (Promega, Corporation, WI, USA) was used to evaluate luciferase activity as described in a previous study (Wu et al., 2020).
Cell proliferation assays
The 5-ethynyl-2′-deoxyuridine (EdU) uptake assay was performed following a previously published method (Pan et al., 2017b).Schwann cells were divided into three groups: control group, Pvt1group (cells with Pvt1 overexpression), and Pvt1/miR-214group (cells with both Pvt1 and miR-214 overexpression).Cells were incubated for 2 hours with 50 mM EdU (Beyotime,Shanghai, China), followed by fixation at 25°C with 4% paraformaldehyde and permeabilization with 5% Triton X-100.Five non-overlapping regions were selected randomly from each group and observed by a fluorescence microscope.The proportion of EdU-positive cells/4′,6-diamidino-2-phenylindole-positive cells was calculated.
In Cell Counting Kit-8 (CCK8) experiments, 100 μL of Schwann cells (3 × 10cells/mL) were added to each well of 96-well plates and incubated for 0, 1, 2, 3,and 4 days.After treatment with 10 nM CCK8 reagent (Beyotime), the optical density (OD) value was observed at 450 nm using a microplate reader (Agilent Technologies).
Cell migration
A Transwell chamber (Life Technologies, Shanghai, China) was placed in the culture plate.Schwann cells were added to the upper chamber, and 800 μL of medium with 10% fetal bovine serum was added to the lower chamber.After incubation at 37°C for 24 hours, cells were fixed with 4% paraformaldehyde for 30 minutes at 25°C and stained with crystal violet (Beyotime) for 20 minutes at 25°C.Cells in the upper chamber were wiped off using a cotton swab.Images were obtained from three random fields and stained cells were counted under a phase-contrast microscope (Olympus).
In vivo cell injection experiments
Forty-five mice were randomized into three groups (n
= 15/group): a sham group and two crush groups.The mice of the two crush groups underwent sciatic nerve crush surgery as described above.Schwann cells from Pvt1+group (Schwann cells with Pvt1 overexpression) or control group (Schwann cells without any treatment) mixed with Matrigel (BD Biosciences, San Jose,CA, USA) were injected into the injured nerves, as previously described (Zhou et al., 2014), and the incision was sutured layer by layer.All mice were raised under free conditions after wound healing.Nerve tissue was collected on day 14 after surgery and immunostained with anti-β-tubulin III antibody (mouse, 1:2000, Santa Cruz Biotechnology, Dallas,TX, USA, Cat# sc-80005, RRID: AB_2210816) overnight at 4°C and then stained with Alexa Fluor 488-labeled goat anti-mouse IgG (1:5000, Beyotime, Cat#A0428, RRID: AB_2893435) at 37°C for 40 minutes, as previously described(Li et al., 2022).Nuclei in nerve tissue were stained by 4′,6-diamidino-2-phenylindole.Finally, the nerve tissue was observed under a fluorescence microscope, and the number of axons and Schwann cells was calculated.
Sciatic functional index
Sciatic functional index (SFI) was daily calculated for 14 days in all mice from the three treatment groups.The hind paws of all the mice were stained.All mice whose hind paws were stained were allowed to move independently on white paper to leave traces.Print length (PL), toe spread (TS) and intermediate toe spread (ITS) were determined according to the footprints of the healthy side (normal side, N) or surgical side (experimental side, E).SPI was calculated as 109.5(ETS – NTS)/NTS-38.3(EPL – NPL)/NPL+13.3(EIT – NIT)/NIT – 8.8.
Statistical analysis
No statistical methods were used to predetermine sample sizes; however,our sample sizes were similar to those reported in a previous publication (Pan et al., 2017b).No animals or data points were excluded from the analysis.Evaluators were blinded to the assignment.GraphPad Prism 8.0.2 (GraphPad Software, San Diego, CA, USA, www.graphpad.com) was used to analyze the data.Two-group statistical significance was determined by the Student’st
-test.P
< 0.05 was considered statistically significant.All data are expressed as the mean ± standard deviation (SD).Results
Expression profile analysis of lncRNAs and mRNAs after PNI
The mouse model of PNI was prepared to detect the differential expression of RNA in the injured nerve.Distal segments of the injured site were collected on days 0, 3, 7 and 14 after PNI.Differentially expressed mRNAs and lncRNAs were identified using microarray analysis.Hierarchical clustering showed systematic variations in the expression of lncRNAs at various time points (P
< 0.05).In total, 5844 lncRNAs were differentially expressed on day 3 after PNI, 4986 lncRNAs were differentially expressed on day 7 after PNI, and 4195 lncRNAs were differentially expressed on day 14 after PNI (Figure 2A).The expressions of 5042 mRNAs were altered on day 3 after PNI, the expressions of 4274 mRNAs were altered on day 7 after PNI, and the expressions of 3229 mRNAs were altered on day 14 after PNI (Figure 2B).For expression pattern clustering, we summarized mRNA expression with 26 model profiles.Of these profiles, nine exhibited significantP
values (P
< 0.001; Figure 2C).We then summarized lncRNA expression with 26 model profiles.Among these profiles,11 exhibited significant P values (P
< 0.001; Figure 3A).The expression of c-Jun,a key factor in the repair of PNI, was elevated after injury, with the highest level detected on day 7 (Chen, 2019).The expression trend of c-Jun was consistent with that of profile 23.In the microarray results, the expression trend of the 318 lncRNAs from profile 23 was consistent with that of c-Jun(Figure 3B).These results suggest that 318 lncRNAs may be related to c-Jun expression following PNI.Therefore, we performed further analyses of the 318 lncRNAs.The expression trend of lncRNA Pvt1, c-Jun and miR-214 after PNI
Research has demonstrated that lncRNAs play key regulatory roles by acting as ceRNAs.We therefore hypothesized that c-Jun may be regulated by one of the differentially expressed lncRNAs through a ceRNA mechanism.We first used RNA22, miRDB, miRWalk and DIANA databases to predict candidate miRNA targets of the 318 lncRNAs and identified 1204 overlapping miRNAs in the four databases.We used the same four databases to identify miRNAs that may bind c-Jun mRNA, and seven miRNAs were obtained.Only one miRNA,miR214, overlapped between the 1204 miRNAs that potentially interact with differentially expressed lncRNAs and the 7 candidate miRNAs binding c-Jun mRNA (Figure 4A).
We performed further analyses to predict the target lncRNAs that interact with miR-214 using the four databases, and the results showed that only one lncRNA, Pvt1, may interact with miR-214.We obtained the sequences of c-Jun mRNA and Pvt1 from the NCBI database and identified the potential Pvt1-binding sequences in miR-214 (Figure 4B) and sequences in miR-214 mediating interactions with c-Jun mRNA (Figure 4C) using the BiBiServ2-RNAhybrid website.
We first examined the expression levels of Pvt1, miR-214, and c-Jun at 0,3, 7, and 14 days after PNI by PCR and western blot.Pvt1 expression was upregulated after PNI and gradually increased until it slightly decreased on day 14 (n
= 3; Figure 4D).The expression trends of c-Jun mRNA and Pvt1 were similar (Figure 4E).miR-214 expression was reduced following PNI and recovered slightly on day 14 (Figure 4F).The western blot results for c-Jun were consistent with the PCR results (n
= 4,P
< 0.05; Figure 4G and H).The results of PCR and western blot preliminarily verify our guess that c-Jun may be regulated by lncRNA Pvt1 through a ceRNA mechanism.lncRNA Pvt1 may regulate c-Jun expression as ceRNA by sponging miR-214 in Schwann cells
Cultured Schwann cells (Figure 5A) were identified by S-100 staining (Figure 5B).In fluorescencein situ
hybridization using a probe for Pvt1 showed fluorescence mostly in the cytoplasm (Figure 5C).Examination of nuclear and cytoplasmic RNAs of Schwann cells by qPCR further confirmed that Pvt1 was primarily present in the cytoplasm.These results indicate that Pvt1 may regulate its target gene at the post-transcriptional level (n
= 3,P
< 0.05;Figure 5D).We next explored the potential regulatory effects of Pvt1 on c-Jun and miR-214 levels by modulating Pvt1 expression by shRNA targeting or plasmid-mediated overexpression.We confirmed that Pvt1 expression was decreased after transfection with Pvt1 shRNA (n
= 5,P
< 0.05; Figure 6A) and upregulated by transfection with pcDNA3.1-Pvt1 (n
= 5,P
< 0.05; Figure 6B)compared with the respective controls.Overexpression of Pvt1 in Schwann cells notably decreased miR-214 expression (n
= 5,P
< 0.05; Figure 6C) and dramatically increased c-Jun mRNA and protein levels (n
= 5,P
< 0.05; Figure 6D–F).In contrast, downregulation of Pvt1 resulted in significantly increased miR-214 expression (n
= 5,P
< 0.05; Figure 6G) and dramatically increased c-Jun mRNA and protein levels (n
= 5,P
< 0.05; Figure 6H–J).These results reveal that there is a potential regulatory relationship among Pvt1, miR-214 and c-Jun in Schwann cells.At the same time, luciferase reporter assays showed that miR-214 mimics reduced luciferase activity driven by the WT-Pvt1 and WT-c-Jun luciferase reporters, with no effects on the MUT-Pvt1 and MUT-c-Jun constructs (n
= 3,P
< 0.05; Figure 5E and F).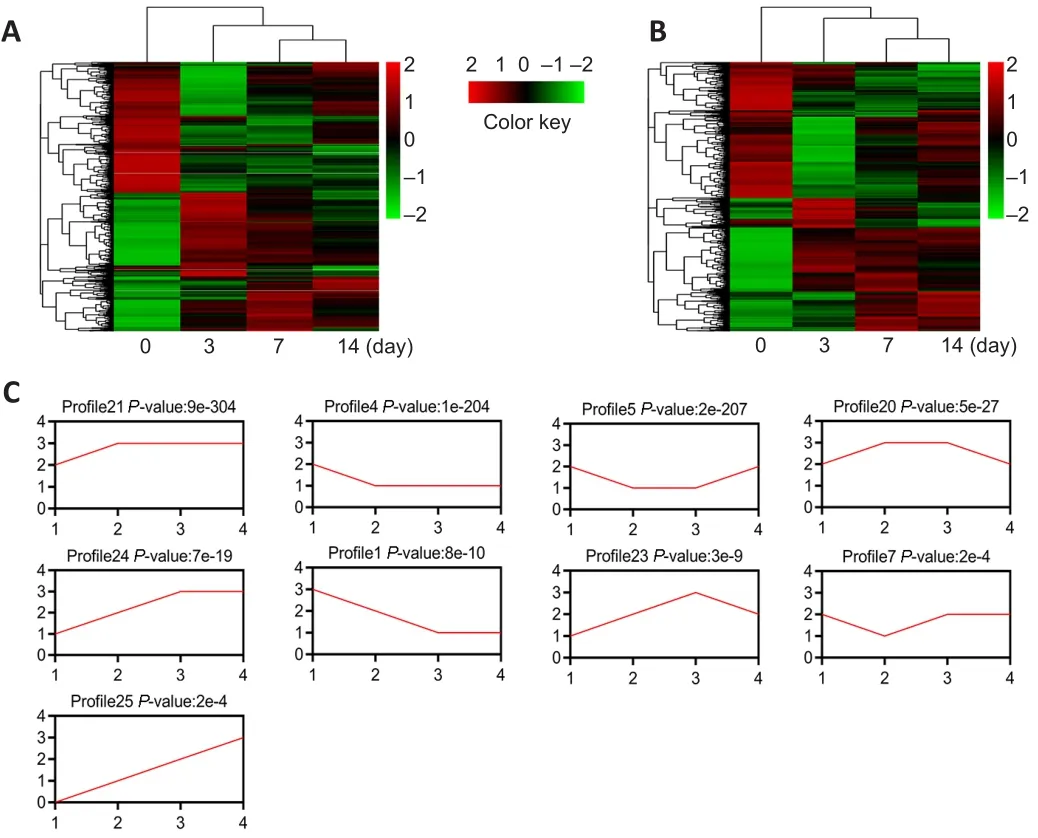
Figure 2|Expression profile analysis of lncRNAs and mRNAs in mice after peripheral nerve injury.
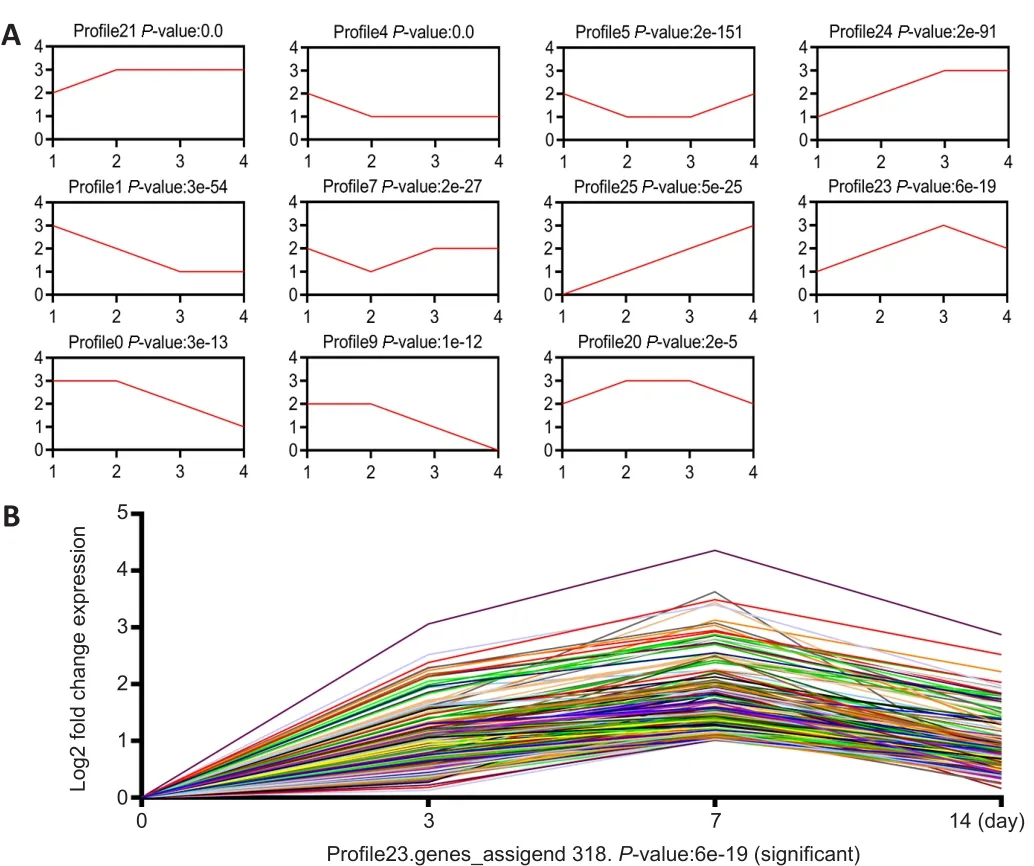
Figure 3| Expression profile analysis of lncRNAs in mice after peripheral nerve injury.
These results indicate that lncRNA Pvt1 may regulate the expression of c-Jun by competitively binding to miR-214 in Schwann cells.
lncRNA Pvt1 can promote the proliferation and migration of Schwann cells in vitro
To examine the role of lncRNA Pvt1 on Schwann cell proliferation, we performed EdU incorporation and CCK8 assays.EdU assays showed that the proliferative ability of Schwann cells in the Pvt1group was clearly higher compared with that of controls (n
= 6; Figure 7A and B).However, cotransfection with miR-214 reduced proliferation to levels similar to those of controls (P
> 0.05).Similarly, CCK8 assays showed increased proliferation of the Pvt1group; there was no significant difference in proliferation between the Pvt1/miR-214and control groups at any time point (n
= 6,P
> 0.05;Figure 7C).These results show that lncRNA Pvt1 can promote the proliferation of Schwann cellsin vitro
, but miR-214 overexpressing eliminates the effect of Pvt1 on Schwann cell proliferation.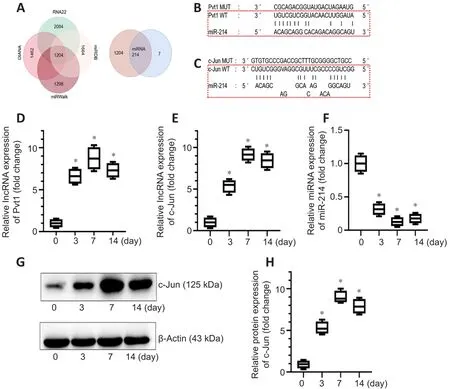
Figure 4|The expression trend of lncRNA Pvt1, c-Jun and miR-214 after peripheral nerve injury.

Figure 5| Identification of Schwann cells and luciferase reporter assay.
We further explored the impact of lncRNA Pvt1 on Schwann cell migration using Transwell experiments.The migratory ability of Schwann cells in the Pvt1group was significantly higher than that in the control group; cotransfection with miR-214 reduced migration to levels similar to those of controls (n
= 6,P
> 0.05; Figure 7D and E).These results show that lncRNA Pvt1 can promote the migration of Schwann cellsin vitro
, but miR-214 overexpressing eliminates the effect of Pvt1 on Schwann cell migration.lncRNA Pvt1 promotes Schwann cell migration and peripheral nerve repair in the in vivo environment
To elucidate the effect of Pvt1 on Schwann cell activation and nerve regenerationin vivo
, we generated a PNI model and Schwann cells expressing pcDNA3.1-Pvt1 or vector controls were injected into injured sites of mice.Two weeks later, the nerve tissues in the damaged area were collected and analyzed.The proliferative ability of Schwann cells in the injured nerve segment in the Pvt1group was significantly higher than that in the control group (n
= 5,P
< 0.05; Figure 8A and B).In addition, the number of axons across the injured sites was greater in the Pvt1group than that in the control group (n
= 5,P
< 0.05; Figure 8A and C).These results demonstrated that Pvt1 overexpressing Schwann cells enhanced the regenerative ability of injured nerves following PNI.The function of the mouse sciatic nerve was assessed by SFI.Our results showed that SFI following injury dropped rapidly from –10 (normal function)to –100 (complete loss of function) and gradually recovered on day 2 (Figure 8D).Notably, on day 4, the SFI of mice from the Pvt1group was significantly higher than that in the control group (n
= 5,P
< 0.05).These experiments demonstrated that Pvt1 can activate Schwann cells, therefore promoting nerve regeneration after injury.
Figure 6| lncRNA Pvt1 may regulate c-Jun expression as ceRNA by sponging miR-214 in Schwann cells.
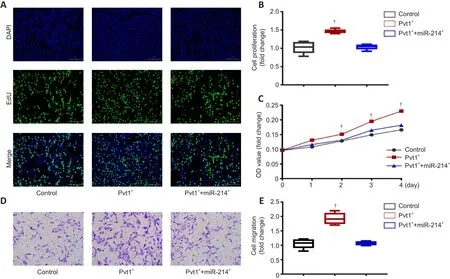
Figure 7| lncRNA Pvt1 can promote the proliferation and migration of Schwann cells in vitro.
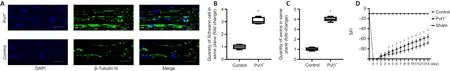
Figure 8|Pvt1 promotes Schwann cell migration and peripheral nerve repair in the in vivo environment.
Discussion
PNI remains a challenging clinical problem.Traditional direct nerve suture repair technique offers poor surgical effects, while autologous nerve grafts are limited by the availability of human tissues (Li, 2014).The peripheral nervous system has a certain degree of elasticity, which can enable effective regeneration depending on the injury site and severity (Xia et al., 2020; Qu et al., 2021).Increasing evidence suggests that Schwann cells are involved in nerve regeneration after PNI (Jessen et al., 2015).However, the regulatory mechanism of Schwann cells is not yet clear.
After PNI, Schwann cells undergo dedifferentiation before redifferentiation,which generates special repair cells and contributes to proliferation.These special cells participate in the regrowth of injured axons and help with functional recovery of the damaged nerve.c-Jun is a key protein for Schwann cell repair and creating a repair cell to guide regeneration.However, the regulatory mechanism of c-Jun has not been fully explained.
We analyzed the expression of lncRNAs after PNI using microarray analysis and identified differentially expressed lncRNAs in the distal segments of the injured site.Through our analyses, we identified a potential Pvt1/miR-214/c-Jun axis and revealed the regulatory interactions among lncRNA Pvt1, miR-214, and c-Jun using molecular biology and biological cytology.Our findings indicated that lncRNA Pvt1 may increase c-Jun expression by sponging miR-214 in Schwann cells following PNI.
Research has demonstrated that lncRNAs play roles in nervous system diseases (Tan et al., 2020).lncRNA Vof-16 knockout was shown to improve the recovery of hypoxic-ischemic brain injury in newborn rats.In ischemic stroke,lncRNA GAS5 silencing inhibits DNMT3B-dependent MAP4K4 methylation and inhibits neuronal apoptosis and nerve injury (Deng et al., 2020).In recent years, more research has established a connection between lncRNAs and PNI repair.Several studies have found that lncRNAs are differentially expressed after PNI (Pan et al., 2017b; Ma et al., 2020).lncRNA MEG3 can promotes nerve regeneration in rats by regulating both the migratory and proliferative activities of Schwann cells.
lncRNAs play critical roles in various cellular processes by functioning as ceRNAs.In previous reports, mRNA and lncRNA co-expression networks were identified and mRNA-miRNA-lncRNA-ceRNA networks after PNI were described (Qian et al., 2018).A recent study showed that lncRNA Arrl1 acts as a ceRNA in PNI and can sponge miR-761, upregulate the expression of the cyclin-dependent kinase inhibitor 2B, and regulate nerve regeneration(Wang et al., 2020b).Yao et al.(2021) reported that lncRNA Loc680254 acts as a ceRNA in PNI and enhances the migratory and proliferative activities of Schwann cells.MSTRG.24008.1 prevents the nerve damage repaired through the miR-331-3p/NLRP3/MAL axis (Yin et al., 2021), and lncRNA BC083743 regulates the proliferative ability of Schwann cells via the miR-103-3p/BDNF axis (Gao et al., 2020).In this study, we identified lncRNA Pvt1 as a differentially expressed lncRNA after PNI and describe its function as a ceRNA.Previous studies showed that the lncRNA Pvt1 plays a role in several diseases.Pvt1 regulates the expression of HK2 in gallbladder cancer by competitively binding miR-143 and promoting cell proliferation and migration (Chen et al., 2019).Fu et al.(2018) reported that lncRNA Pvt1 regulates GREM1 and BMP expression by sponging miR-128-3p and promotes tumorigenesis and progression of glioma.However, lncRNA Pvt1 had not been investigated in PNI.Our results suggest that lncRNA Pvt1 targets c-Jun by sponging miR-214 as a ceRNA following PNI.We further showed that miR-214 can regulate c-Jun expression.As an important miRNA, miR-214 has been linked to different diseases (Li et al., 2020, 2021).For example, miR-214 is involved in the proliferation and migration of gallbladder cancer and also inhibits the proliferation and migration of esophageal squamous cell carcinoma.
Our molecular biology and cell biology experiments verified the regulatory relationships of lncRNA Pvt1, miR-214, and c-Jun in Schwann cells after PNI.We also injected Pvt1-overexpressing Schwann cells into mice with PNI and the recovery of injured axon and nerve function was remarkably better in mice with PNI than in mice from the control group.
In conclusion, our findings indicate that lncRNA Pvt1 can help injured nerve to regenerate by promoting the proliferation and migration of Schwann cells following PNI, and its functional mechanism may be realized through increasing c-Jun expression by sponging microRNA-214 in Schwann cells.Our results have improved our understanding of the ceRNA regulatory mechanism after PNI and highlighted Pvt1 as a new potential target for treating PNI.
Peripheral nerve regeneration is a complex process that requires multiple factors such as Schwann cells, macrophages, and neurotrophic factors.In this study, we mainly focused on the activation and mechanism of lncRNA Pvt1 on Schwann cells after PNI.One limitation of the study is that we did not verify the effects of Schwann cells overexpressing Pvt1 and miR-214 on nerve regenerationin vivo
.Ourin vivo
results showed that Schwann cells overexpressing Pvt1 promoted axon regeneration and restored motor function of experimental mice within 14 days.However, further experimental research is needed to confirm our findings.Author contributions:
BP, MHC and HF designed the experiments.BP and DG performed the cellular biology assays and analyzed the data.BP, DG, LJ and KL conducted the animal assay.ZWL, XG and WZ provided many suggestions during manuscript preparation, BP, DG, XL and GL wrote the manuscript.All authors read and approved the final version of the manuscript.
Conflicts of interest:
The authors declare that the research have no conflict of interest.
Open access statement:
This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
- 中国神经再生研究(英文版)的其它文章
- Patient-specific monocyte-derived microglia as a screening tool for neurodegenerative diseases
- Molecular hallmarks of long non-coding RNAs in aging and its significant effect on aging-associated diseases
- Inflammation in diabetic retinopathy: possible roles in pathogenesis and potential implications for therapy
- Targeting the nitric oxide/cGMP signaling pathway to treat chronic pain
- Neurosteroids as stress modulators and neurotherapeutics: lessons from the retina
- Myelinosome organelles in pathological retinas:ubiquitous presence and dual role in ocular proteostasis maintenance