Impact of cognition-related single nucleotide polymorphisms on brain imaging phenotype in Parkinson’s disease
Ting Shen , Jia-Li Pu , Ya-Si Jiang Yu-Mei Yue, Ting-Ting He , Bo-Yi Qu , Shuai Zhao Ya-Ping Yan Hsin-Yi Lai , , ,Bao-Rong Zhang
Abstract Multiple single nucleotide polymorphisms may contribute to cognitive decline in Parkinson’s disease.However, the mechanism by which these single nucleotide polymorphisms modify brain imaging phenotype remains unclear.The aim of this study was to investigate the potential effects of multiple single nucleotide polymorphisms on brain imaging phenotype in Parkinson’s disease.Forty-eight Parkinson’s disease patients and 39 matched healthy controls underwent genotyping and 7T magnetic resonance imaging.A cognitive-weighted polygenic risk score model was designed, in which the effect sizes were determined individually for 36 single nucleotide polymorphisms.The correlations between polygenic risk score, neuroimaging features, and clinical data were analyzed.Furthermore, individual single nucleotide polymorphism analysis was performed to explore the main effects of genotypes and their interactive effects with Parkinson’s disease diagnosis.We found that, in Parkinson’s disease, the polygenic risk score was correlated with the neural activity of the hippocampus,parahippocampus, and fusiform gyrus, and with hippocampal-prefrontal and fusiform-temporal connectivity, as well as with gray matter alterations in the orbitofrontal cortex.In addition, we found that single nucleotide polymorphisms in α-synuclein (SNCA) were associated with white matter microstructural changes in the superior corona radiata, corpus callosum, and external capsule.A single nucleotide polymorphism in catechol-O-methyltransferase was associated with the neural activities of the lingual, fusiform, and occipital gyri, which are involved in visual cognitive dysfunction.Furthermore, DRD3 was associated with frontal and temporal lobe function and structure.In conclusion, imaging genetics is useful for providing a better understanding of the genetic pathways involved in the pathophysiologic processes underlying Parkinson’s disease.This study provides evidence of an association between genetic factors,cognitive functions, and multi-modality neuroimaging biomarkers in Parkinson’s disease.
Key Words: cognition; imaging genetics; magnetic resonance imaging; multi-modality; Parkinson’s disease; polygenic risk score; single nucleotide polymorphism; ultra-high field,
Introduction
Parkinson’s disease (PD) is primarily a movement disorder accompanied by a battery of non-motor symptoms.Cognitive decline is one of the most disabling non-motor symptoms of PD.PD-related cognitive decline is commonly characterized by executive dysfunction, with a lower incidence of impairment in attention, memory, language, and visuospatial domains (Emre, 2003).Executive function is primarily regulated by the prefrontal cortex (Alvarez and Emory, 2006).Impaired attention in PD is considered to be controlled by the prefrontal cortex and some regions of the parietal visual cortex(Baldauf and Desimone, 2014).Language function depends on the auditory,visual, and motor systems (Friederici, 2011).Moreover, the hippocampus,cingulate, and neocortex are involved in maintaining memory (Fang et al.,2020).Furthermore, higher visual processing regions, including the occipital and parietal regions, are affected in PD (Weil et al., 2016).Studying cognition deficits may provide insight into the pathological mechanisms of PD.
Both genetic and environmental factors contribute to the risk of PD.Risk variants identified in genome-wide association studies (GWASs) could explain 16–36% of the observed heritability in PD (Nalls et al., 2019).Similarly, PD-related cognitive decline also has a certain genetic predisposition, and a series of possible genetic risk loci have been identified that convey different degrees of risk.Single nucleotide polymorphisms (SNPs) in genes such as α-synuclein(SNCA
), catechol-O-methyltransferase (COMT
), brain-derived neurotrophic factor (BDNF
), and the dopamine receptor gene are related to an increased risk of PD-related cognitive decline.These genes are involved in a variety ofneural physiological processes and participate in regulating cognitive function(Barnett et al., 2009; Canivet et al., 2015; Campelo et al., 2017; Papenberg et al., 2017; Yin et al., 2019).TheSNCA
gene encodes α-synuclein, which is a soluble protein expressed at presynaptic terminals in the central nervous system (Chung et al., 2019).Mutations inSNCA
lead to the generation of misfolded and aggregated proteins, which are an important component of Lewy bodies, the main pathological hallmark of PD.COMT
is crucial for regulating dopamine flux in the prefrontal cortex, as well as for prefrontal cortex-dependent cognition and activation (Meyer-Lindenberg et al., 2006).TheCOMT
-rs4680 polymorphism modulates executive function interactively with levodopa therapy upon activation of the frontoparietal network (Nombela et al., 2014).It is also associated with an increase in hippocampal formation and ventrolateral prefrontal cortex activation (Drabant et al., 2006).Dopamine plays an important role in memory.When the amount or activity of dopamine receptors in the prefrontal cortex increases, dopamine transmitter levels and dopaminergic pathway activation also increase, leading to an increase in prefrontal cortex activity and memory.The dopamine receptor genes are also vital for memory function (Papenberg et al., 2017); for example, the rs6280 polymorphism results in the production of variant protein with higher affinity for dopamine that affects cognitive function (Ji-Ze et al., 2013).These genetic variations may have relatively small effect sizes in the development of cognitive decline.Cognitive function in patients with PD may be modified by the cumulative influence of multiple genetic variations that individually have only weak effects (Lill, 2016).The polygenic risk score(PRS) is a powerful tool that can be used to evaluate the additive effect of multiple SNPs on clinical phenotypes by calculating the sum of the number of risk alleles (Porter et al., 2018).Indeed, a PRS generated based on 46 PD-related SNPs had high predictive value for risk of developing PD and age at onset (Li et al., 2019).Moreover, a mitochondrial-specific PRS showed that the cumulative effect of 14 variants involved in mitophagy was also associated with PD risk (Billingsley et al., 2019).In addition, recent studies have linked genetic factors to brain imaging phenotypes.Previous studies have investigated the correlations between disease-related PRSs and imaging features, such as white matter microstructure (Simoes et al., 2019) and gray matter volume (Wang et al., 2019), to identify brain imaging phenotypegenotype associations.
Magnetic resonance imaging (MRI) is an important tool for understanding the neural basis of cognitive decline.PD patients with cognitive decline exhibit widespread atrophy and cortical thinning in the frontal, temporal, parietal,and limbic areas (Sasikumar and Strafella, 2020), disruptions in the frontostriatal and parieto-temporal pathways (Sasikumar and Strafella, 2020), and impaired white matter microstructure in the insular cortices, anterior and inferior fronto-occipital, uncinate, superior longitudinal fasciculi, corona radiata, and corpus callosum (CC) (Gorges et al., 2019).Thus, we selected these neuroimaging features as intermediate brain imaging phenotypes to investigate the effect of genetic risk variants.We analyzed these features using ultra-high field 7T MRI with the expectation that its high spatial resolution, signal-to-noise ratio, and detection power would help us to detect previously unknown brain alterations that occur due to weak SNP effects.
To gain a better understanding of the pathophysiological mechanism of cognitive decline in PD, we introduced a weighted PRS model representing cognitive decline in PD.This model enabled us to assess the cumulative effect of multiple SNPs on brain structure and function and verify the influence of individual SNPs.
Methods
Participants and clinical assessments
This was an observational cross-sectional study (Figure 1) that was approved by the Ethics Committee of the Second Affiliated Hospital of Zhejiang University School of Medicine (approval No.2015-081) on January 14, 2016 and was conducted in accordance with the Declaration of Helsinki.This study is reported according to the STrengthening the Reporting of OBservational studies in Epidemiology (STROBE) statement (von Elm et al., 2007).We recruited 48 patients with PD from the outpatient clinic of the Department of Neurology of Second Affiliated Hospital of Zhejiang University School of Medicine, as well as 39 age- and sex-matched healthy controls (HCs) from neighboring communities from July 2017 to July 2019.This sample size was estimated using G*Power and was calculated to exceed 80% power to detect an effect size ofD
= 0.4 atα
= 0.05 (Szucs and Ioannidis, 2020).Written informed consent was obtained from all of the participants.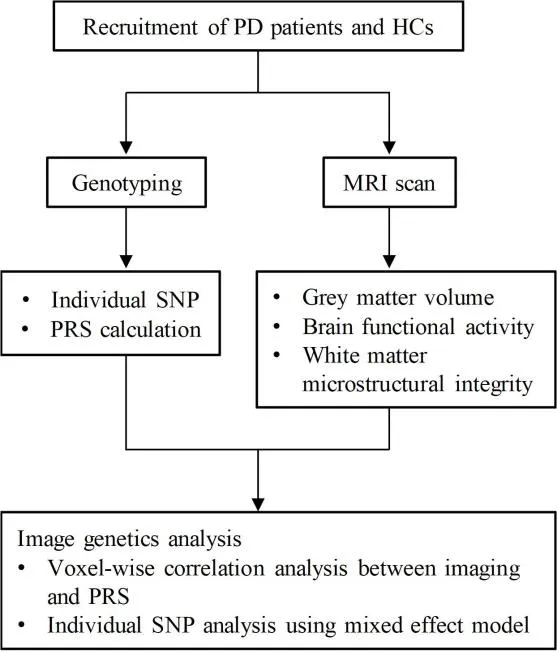
Figure 1|Study design.
Patients with PD were diagnosed according to the Movement Disorder Society(MDS) Clinical Diagnostic Criteria for PD (Postuma et al., 2015).Patients with other neurological disorders or injuries that affect cognitive functions were excluded.The HCs had no history of neurological or psychiatric diseases and no family history of PD or related neurodegenerative disorders.PD disease severity was assessed using the Unified Parkinson’s Disease Rating Scale (UPDRS) (Ramaker et al., 2002) and Hoehn & Yahr stage (H&Y stage)(Hoehn and Yahr, 1967) during defined off-stages (PD medications on hold for a minimum of 12 hours) to reach an appropriate washout of levodopa.Cognitive function was evaluated using the Mini-Mental State Examination(MMSE) (Pangman et al., 2000), Montreal Cognitive Assessment (MoCA)(Nasreddine et al., 2005), and Parkinson’s Neuropsychometric Dementia Assessment (PANDA) (Kalbe et al., 2008), covering all five cognition domains:attention, executive function, language, memory, and visuospatial function.Cognitive testing was performed in HCs and patients with PD during the onstage.As shown in Additional Table 1, the devised cognitive domain index scores were calculated by adding all of the corresponding items together for these five cognition domains (Julayanont et al., 2014).Individual cognitive domain index scores were transformed into z-scores using the mean and standard deviation of the whole sample according to the following formula:(individual raw score - mean score)/standard deviation (Liu et al., 2017).
Genotyping and polygenic risk score calculation
Peripheral blood samples were collected at the outpatient clinic of the Department of Neurology of the Second Affiliated Hospital of Zhejiang University School of Medicine, and genomic DNA was extracted.On the basis of a literature review and prior evidence, we selected SNPs that were potentially associated with cognitive function in PD (Additional file 1).The selected SNPs were genotyped using time-of-flight mass spectrometry.SNPs with a minor allele frequency greater than 5% and SNP missingness lower than 0.02 were included (Li et al., 2019).To avoid PRS inflation by strong linkage disequilibrium (r
> 0.8), only one marker per locus was selected.Finally, 36 SNPs in 28 candidate genes were included in Hardy-Weinberg equilibrium in both groups (Additional Table 2).For each SNP, the allele showing a higher frequency in PD was considered to be the risk allele, and an individual with one risk allele was assigned a value of 1 (no risk allele =0, one risk allele = 1, and two risk alleles = 2) (Hettige et al., 2016).The PRS was defined as the summation of the risk allele counts of SNPs per individual weighted by their corresponding effect sizes.According to a previous study,regression coefficients estimated from the multivariate linear regression model based on the present study were regarded as effect sizes for all SNPs,indicating the strength of relationships between selected SNPs and cognitive capacity (sum of three scales) (Porter et al., 2018).The mathematical representation of multiple linear regression is: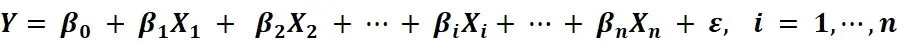
Y
is the cognitive score,β
is the ith coefficient representing the effect size of each SNP, andβ
is a constant.X
is the summation of the risk allele counts of the ith SNP, andn
is the number of included SNPs.As PD and HCs did not differ in all cognitive scales (Table 1), the effect sizes were calculated in all participants (Additional Table 2).MRI image acquisition and processing
MRI images were acquired using a 7T MRI research system (Magnetom,Siemens Healthcare, Erlangen, Germany).The MRI acquisition protocol included: (1) magnetization-prepared 2 rapid gradient echoes (MP2RAGE)sequence (voxel size: 0.7 × 0.7 × 0.7 mm, repetition time (TR) = 5000 ms, TI1/TI2 = 900/2750 ms, echo time (TE) = 2.3 ms, α1/α2 = 5°/3°) for high-resolution anatomical imaging, (2) multiband accelerated echo-planar imaging (EPI)sequence (voxel size: 1.5 × 1.5 × 1.5 mm, TR = 2500 ms, TE = 20.4 ms) for resting-state functional MRI (rs-fMRI), and (3) multiband EPI sequence (voxel size: 1.5 × 1.5 × 1.5 mm, TR = 6000 ms, TE = 87.4 ms, applying diffusionsensitive gradient in 130 directions) for diffusion tensor imaging (DTI).PD patients were scanned after overnight (at least 12-hour) withdrawal from anti-parkinsonism medication.
The rsfMRI data were processed with Data Processing and Analysis for Brain Imaging (DPABI) software (http://www.rfmri.org/dpabi) (Yan et al.,2016) through standard preprocessing steps, including motion correction,normalization to Montreal Neurological Institute (MNI) space, nuisance regression, detrend, smoothing with a 4-mm full width at half maximum(FWHM), and band-pass filtering (0.01–0.08 Hz).For head motion estimate,the framewise displacement value was calculated, and this value did not differ between the two groups.Amplitude of low-frequency fluctuation (ALFF)and regional homogeneity (ReHo) values were generated from the rsfMRI data.ALFF was used to reflect the intensity of regional spontaneous brain activity, and ReHo was used to evaluate local synchronization of spontaneous neural function (Yue et al., 2020).In addition, we extracted brain regions with significant results as regions of interest (ROIs) for seed-based functional connectivity (FC) analysis.
The high-resolution MP2RAGE anatomical images were processed using the voxel-based morphometry (VBM8) toolbox (http://vbm.neuro.uni-jena.de/vbm) (Ashburner, 2009) function of the Statistical Parametric Mapping(SPM8) software program (Wellcome Trust Centre for Neuroimaging, UCL,London, UK; https://www.fil.ion.ucl.ac.uk/spm) (Friston, 2003).All images were warped into MNI space and segmented to obtain a gray matter (GM)map indicating GM volume (GMV).Then, the GM map was smoothed with a 4-mm FWHM for subsequent analysis.The GMV value was extracted from the MP2RAGE images to describe gray matter structure.
The DTI data were processed using FSL 6.0.1 (FMRIB Software Library; www.fmrib.ox.ac.uk/fsl) (Jenkinson et al., 2012) to conduct processing steps including Eddy current correction, DTIFIT, and registration to standard space.The generated fractional anisotropy (FA), axial diffusivity (AD), and radial diffusivity (RD) maps were smoothed with a 4-mm FWHM for subsequent analysis.Fractional anisotropy (FA), axial diffusivity (AD), and radial diffusivity(RD) were generated from the DTI.The FA value was used to assess microstructural characteristics, including fiber density, axonal diameter,myelin sheath thickness, and fiber directionality (Fan et al., 2015).AD and RD values were used to describe axonal injury and myelin damage, respectively(Fan et al., 2015).Thus, we obtained the ALFF, ReHo, FC, GMV, FA, AD and RD maps for subsequent analysis.
Statistical analysis
Statistical analyses were performed using SPSS Statistics software (IBM Corporation, Armonk, NY, USA), and statistical plots were generated using GraphPad Prism software (version 9, GraphPad Inc., San Diego, CA, USA).The Shapiro-Wilk normality test was used to test the normality of the variable distribution.A two-samplet
-test or Mann-WhitneyU
test was applied for comparison of continuous variables with normal or non-normal distribution,respectively.The chi-squared test was used to compare categorical variables.Statistical significance was set atP
< 0.05.Statistical analysis of the MRI data was conducted using DPABI.Voxel-wise correlation analysis was conducted to investigate correlations between cognitive domain index z-scores and MRI features, as well as between MRI features and PRS scores.Moreover, to verify the genotype effect of these SNPs, individual SNP analysis was conducted using the mixed effect model (genotypes × groups; genotypes: risk allele carriers and non-carriers;groups: PD and HC) to analyze the genotype effect and interactive effect of diagnosis and genotype on brain imaging phenotype.Age, sex, and education were entered as covariates because of their potential influence.We applied a multiple comparison correction method, Gaussian random field(GRF) correction, to identify significant clusters (voxelP
< 0.001, clusterP
<0.05).The clusters showing significant results were reported based on the Automated Anatomical Labeling (AAL3) atlas (Rolls et al., 2020).The MRI features of brain areas with statistical significance were extracted.To evaluate the correlations between MRI features and clinical characteristics,Pearson’s or Spearman’s correlation analysis was performed for normally distributed or non-normally distributed data, respectively.Statistical significance was set atP
< 0.05.Results
Participant characteristics
The demographic and clinical characteristics of the participants are summarized in Table 1.No statistically significant differences were noted in age, sex, education, and cognitive scores between the PD and HC groups (P
>0.05); thus, the two groups were well matched and controlled for non-genetic influencing factors.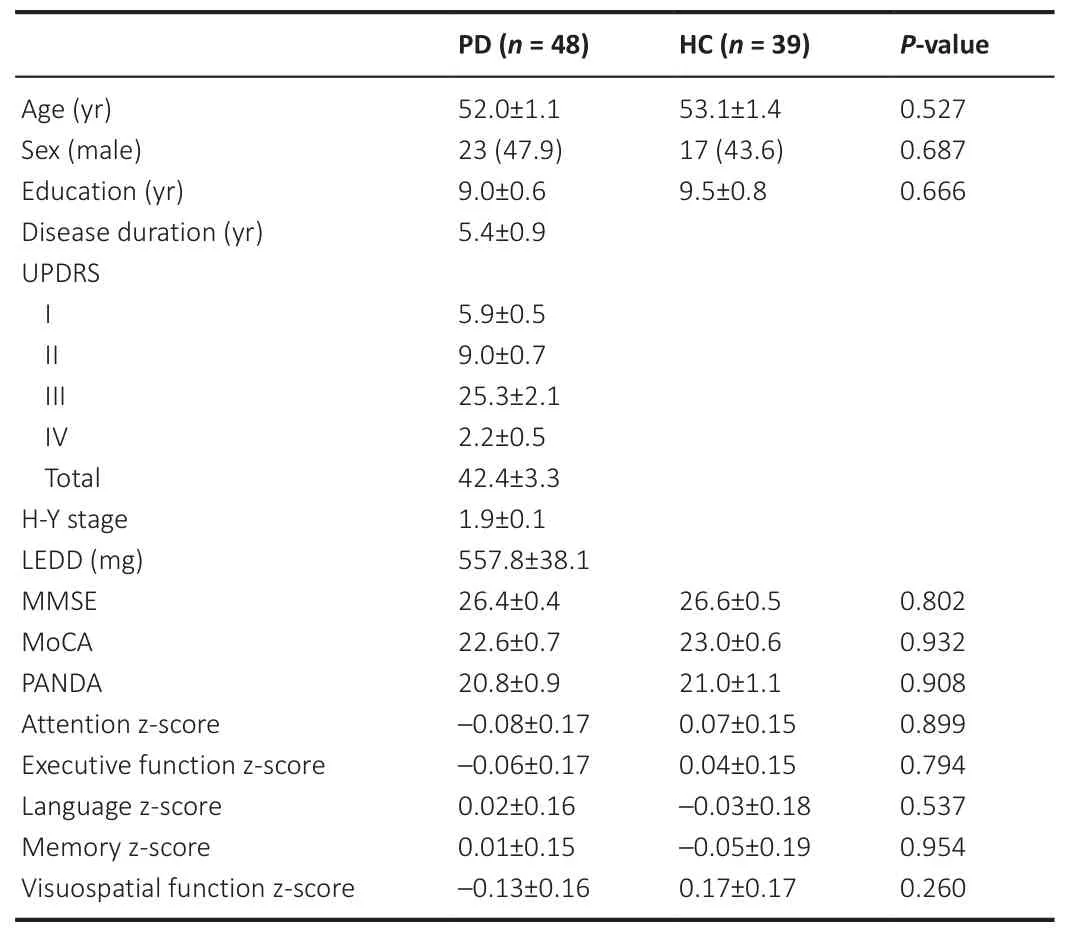
Table 1 |Demographic and clinical characteristics of study participants
Polygenic risk score for cognitive function of PD
No significant difference in PRSs was found between the PD and HC groups(P
> 0.05).Moreover, the PRS was positively correlated with attention(Spearman’sr
= 0.40,P
< 0.01), executive (Pearson’sr
= 0.47,P
< 0.01),language (Spearman’sr
= 0.40,P
< 0.01), memory (Spearman’sr
= 0.43,P
< 0.01), and visuospatial (Spearman’sr
= 0.32,P
< 0.01) cognitive domain z-scores in all participants.Cognitive domain index z-scores are associated with brain imaging features
After correcting for age, sex, and education, the attention z-score of patients with PD was negatively correlated with the ALFF values of the right middle temporal gyrus (MTG) and the right middle occipital gyrus (MOG), which was also reported to be involved in selective spatial attention, indicating that theabnormally increased neural activities in the MTG and MOG were correlated with decreased attention function (Figure 2A).The executive z-score was negatively correlated with the GMV of the right pole of the superior temporal gyrus (STG) and the ALFF values of the right lingual gyrus and right precuneus in PD, and positively correlated with the RD value of the right superior longitudinal fasciculus (Figure 2B).The lingual gyrus is a visual processing area that consistently participates in visual imagery and visual memory encoding,as well as being involved in executive function.The precuneus is a core hub of the default mode network that connects to the core hub region of the executive control network (Beaty et al., 2015).
Moreover, the language z-score was found to be inversely related to the GMV value of the right pole of the STG and the ReHo values of the right parahippocampus, right lingual, and fusiform gyri, and positively related to the AD value of the right posterior corona radiata in patients with PD (Figure 2C).In addition, the memory z-score was negatively correlated with the GMV values of the right STG, while it was positively correlated with the left calcarine in patients with PD.Meanwhile, the memory z-score was positively associated with the FA values of the right posterior thalamic radiation and right sagittal stratum, the AD values of the bilateral posterior corona radiata and posterior thalamic radiation, and the RD values of the bilateral posterior corona radiata,right posterior thalamic radiation, and right superior longitudinal fasciculus(Figure 2D).Thus, abnormal white matter microstructure in the sagittal stratum and posterior thalamic radiation correlated with memory dysfunction.Furthermore, the visuospatial z-score was also positively associated with the AD and RD values of the bilateral posterior corona radiata and the RD value of the right superior longitudinal fasciculus in patients with PD (Figure 2E).The posterior corona radiata is associated with the language, memory,and visuospatial domains, is connected to the precuneus, and is involved in verbal comprehension and working memory (Long et al., 2018).The posterior thalamic radiation connects to visual memory-associated cortical regions(Menegaux et al., 2017).The superior longitudinal fasciculus has also been reported to be associated with memory function, especially spatial working memory (Vestergaard et al., 2011).
Cognitive-weighted PRS are associated with brain imaging phenotype
With regard to structural brain features, the PRS was negatively correlated with the GMV of the right orbitofrontal cortex (OFC), including the right orbital part of the middle frontal gyrus (ORBsup) and the orbital part of the superior frontal gyrus (ORBmid) in patients with PD (Figure 3B).These orbitofrontal subdivisions of the frontal lobe play an important role in executive behavior and cognition, especially in decision-making and processing visual, spatial, and emotional information (Burks et al., 2018).As for functional MRI features, negative correlations between the PRS and ALFF values of the right fusiform gyrus, hippocampus, and parahippocampus were found in the PD group (Figure 3A).The fusiform gyrus is a key region for high-level visual processing.The hippocampus supports functions such as verbal memory, spatio-temporal organization, autobiographical memory,and narrative memory (Burgess et al., 2002), while the parahippocampus is involved in associative, source, and spatial memory, scene perception, and navigation (Aminoff et al., 2013).
Brain regions with a significant correlation between the ALFF and the PRS were selected as the three seed ROIs.The results of the FC analysis are shown in Figure 4.With an increase in PRS, patients with PD showed increased FCs between the right hippocampus and the left superior frontal gyrus (SFG) and left middle frontal gyrus (MFG), as well as decreased FCs between the right fusiform gyrus and the right STG and right MTG.
The imaging features of the significant brain regions identified above were extracted.As shown in Figure 3C and 3D, the ALFF value of the right fusiform gyrus was negatively related to attention z-score in both patients with PD and HCs, and negatively related to executive, language, and visuospatial z-scores in HCs, which only showed negative trends in patients with PD.Moreover,the ALFF values of the hippocampus and parahippocampus were inversely associated with the attention z-score, while they only showed negative trends with other cognitive domain scores in patients with PD.In HCs, the ALFF values of the hippocampus and parahippocampus were also inversely associated with attention, language, and visuospatial z-scores, and showed negative trends with executive and memory z-scores.Furthermore, the FC between the right hippocampus and the left SFG and left MFG was positively associated with the visuospatial z-score in patients with PD (Figure 4A), while the associations with attention, executive, language, and memory cognitive domain z-scores only showed a positive trend.The FC between the right fusiform gyrus and the right STG and right MTG was inversely correlated with all five cognitive domain scores in PD (Figure 4B).However, no significant correlations were detected in HCs.
Impact of individual SNPs on brain imaging phenotype
TheSNCA
-rs3910105 variant interacted with PD status to affect the FA values of the body of the corpus callosum, the left superior corona radiata, and the right external capsule.The C and T alleles differentially influenced FA values in patients with PD and HCs: PD–C-carriers had higher FA values in the corpus callosum, superior corona radiata, and external capsule than PD–non-carriers,while HC–C-carriers had lower FA values than HC–non-carriers (Figure 5A).Regarding theSNCA
-rs356181 variant, the A carriers showed lower RD values in the left anterior corona radiata, and this variant interacted with PD status to affect the AD value of the left anterior corona radiata (Figure 5B).PD–A-carriers had higher AD values in the left anterior corona radiata than PD–non-carriers, while HC–A-carriers showed lower AD values than HC–noncarriers.Irrespective of PD diagnosis, theCOMT
-rs4680 risk allele A carriers showed lower ALFF values in the right lingual and fusiform gyri than did non-carriers,while no interactive effect betweenCOMT
-rs4680 and PD diagnosis was found(Figure 5C).Subjects withDRD3
-rs6280 G alleles had lower ALFF values in the right STG,as well as higher GMV values for the right rectus and OFC, including the right ORBsup, left ORBmid, and left orbital part of the inferior frontal gyrus (ORBinf)(Figure 5D).Moreover, theDRD3
-rs6280 variant and PD status had interactive effects on both the AD and RD values of the right external capsule.The G and A alleles differentially affected AD and RD values in patients with PD and HCs:PD–G-carriers had higher AD and RD values in the right external capsule than PD–non-carriers, while HC–G-carriers had lower AD and RD values than HC–non-carriers (Figure 5D).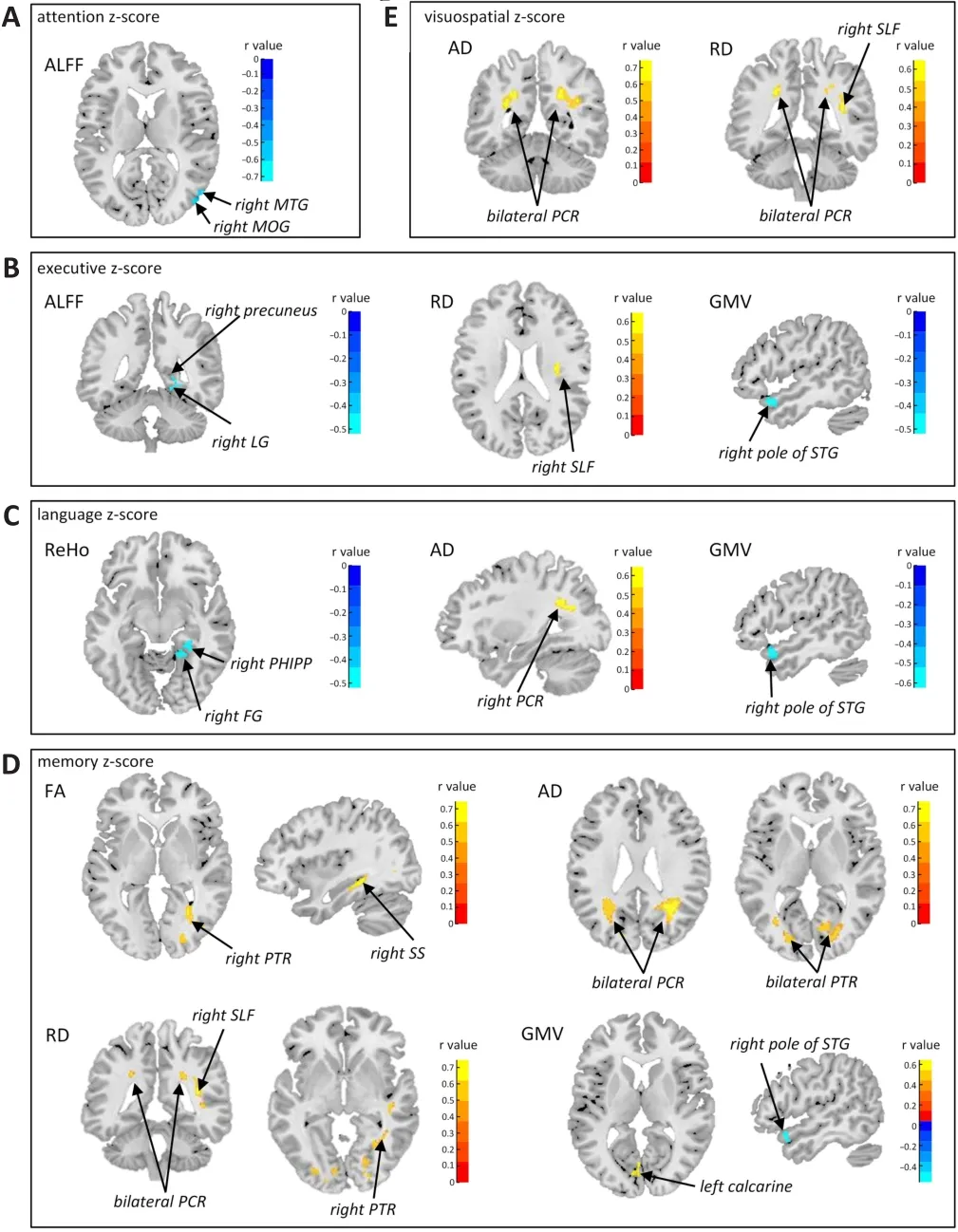
Figure 2 | Voxel-wise correlation analysis between cognitive domain index z-scores and brain imaging features in PD.
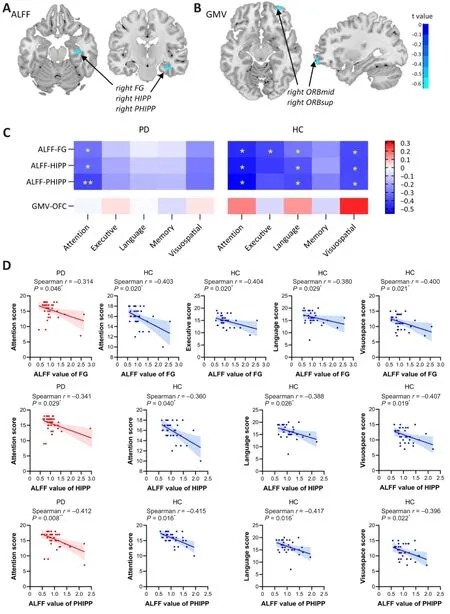
Figure 3|Voxel-wise correlation analysis between PRS, imaging features, and cognitive scores.

Figure 4 | Voxel-wise correlation analysis between FCs and cognitive domain scores.
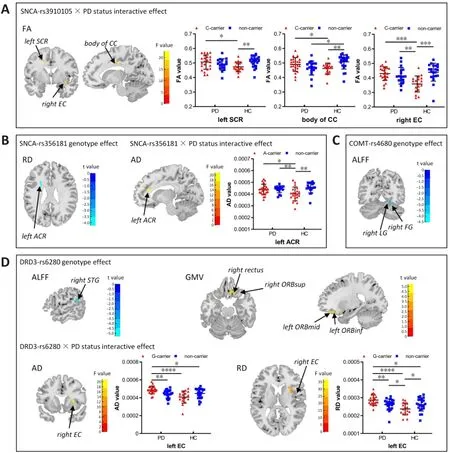
Figure 5 | Effect of individual SNPs and interactive genotype × PD status effect on imaging features.
Discussion
Owing to the complex genetic background, numerous genetic variations are associated with the decline in cognition seen in PD.Patients with PD also exhibit disease-specific brain imaging phenotypes.However, details of the association between brain imaging phenotypes and genetic variations remain unclear.This study investigated the potentially cumulative effects of multiple SNPs, as well as the effects of individual SNPs, on the brain in PD.Multi-modality imaging biomarkers were applied to assess brain imaging phenotypes, including gray matter volume/density, white matter microstructural integrity, water molecular diffusivity, and local neural activity.The associations between PRS, brain imaging phenotype, and clinical features were further analyzed to identify pathophysiological pathways associated with PD-related cognitive decline.
Cognitive domain-related imaging features
To enhance our understanding of impairments in different cognitive domains in PD, the correlations between five cognitive domains and multimodal imaging features were explored.The attention function was found to decrease along with increased neural activity of the middle temporal and middle occipital gyri, which also participated in selective spatial attention.Thus, attention deficits in PD might be the result of dysfunction of these two brain regions.Executive dysfunction is the predominant cognitive impairment reported in PD (Parker et al., 2013).In this study, executive function was found to decrease along with increased neural activity in the precuneus and lingual gyrus.Moreover, the FC of the lingual gyrus might be an early alteration related to the future development of cognitive decline in PD (Yoneyama et al., 2018).It follows that the precuneus and lingual gyrus might contribute to executive dysfunction in PD.
In PD, language function was found to be associated with local synchronization of the lingual and fusiform gyri, which are involved in processing visual information, and especially in identifying and recognizing words (Mechelli et al., 2000).Language function is also related to gray matter structural alteration of the STG, which is involved in language comprehension, as well as white matter structural alteration of the posterior corona radiate, which is involved in verbal comprehension (Long et al., 2018).Moreover, memory capacity was found to be associated with changes in memory-related white matter fibers, the posterior thalamic radiation, posterior corona radiate, and superior longitudinal fasciculus.Visuospatial impairment is also commonly observed in PD, manifesting as the development of visual hallucinations and gait dysfunction (Specketer et al., 2019).Visuospatial function was found to be associated with increased diffusivity of the posterior corona radiata and superior longitudinal fasciculus fiber tracts in PD.In general, these findings help map the relationships between different cognitive domains and multi-modality imaging biomarkers.Attention and executive function were mainly related to brain function; memory and visuospatial functions were mainly associated with white matter microstructural changes; and language function was related to gray matter and white matter structure, as well as to brain function.
Genetic effects contribute to brain imaging phenotype
To reveal the cumulative effects of genetic risk factors on the brain, we introduced a cognitive-related PRS model and analyzed its correlation with imaging features.A higher PRS was found to be related to lower neural activity in the hippocampus and parahippocampus in patients with PD.Meanwhile, hippocampal-prefrontal functional connectivity increased along with the increased cognitive-related PRS.The hippocampal-prefrontal circuit plays an important role in cognitive and emotional processes and memory consolidation.A previous study showed that hippocampal and parahippocampal atrophy is linked to memory impairment in PD (Burton et al., 2004).Altered neural activities in the hippocampus and parahippocampus have been demonstrated, indicating increased local field potential activities in PD.Accordingly, increased parahippocampal neural activities were found to be related to a decline in cognitive capacity in the current study, reflecting a functional compensatory mechanism for the decreased hippocampalprefrontal connectivity.Generally, it was speculated that the alterations within these circuits might be ascribed to the cumulative impact of these genetic variations to some extent.
Altered FC of the fusiform gyrus is commonly seen in subjects demonstrating cognitive decline (Cai et al., 2015).Patients with PD also exhibit visuospatial impairment, which could be regarded as a predictor of cognitive decline in PD (Weil et al., 2016).Previous studies have also reported increased neural activity in the fusiform gyrus in PD (Pan et al., 2017).As expected,the neural activity of the fusiform gyrus, as well as its FC with the temporal lobe, changed with the change in PRS.Meanwhile, the neural activity of the fusiform gyrus and fusiform-temporal connectivity increased along with the decline in cognitive capacity, suggesting a compensatory mechanism for cognitive decline, especially in the visuospatial domain.Therefore, these genetic variations may also contribute to cognitive decline through visual cognitive dysfunction.
Regarding the structural imaging features, relationships between PRS and GMV values in ORBsup and ORBmid were found in patients with PD,suggesting that the genetic effects described above also influence the gray matter structure of the orbitofrontal cortex.As mentioned above,executive dysfunction is a major domain of cognitive dysfunction in PD.The orbitofrontal cortex exhibits a relationship between compensatory dopamine levels and movement preparation in PD (Marinelli et al., 2015).Thus, the reduced gray matter volume of the orbitofrontal cortex is related to increased genetic factors, providing more evidence for the compensatory mechanism in the dopaminergic pathways of cognition.
Individual SNPs affect brain imaging phenotype
Individual SNP analysis was performed to investigate these phenomena and verify the specific genetic effects of certain SNPs.Revealing the detailed effects of these genetic variations might provide valuable insight into the mechanism of PD.
Patients with PD who haveSNCA
mutations often exhibit cognitive dysfunction(Compta et al., 2011).This study showed that the SNCA-rs356181 variant mainly contributes to axonal injury and myelin damage in the anterior corona radiata, which contains associative fibers connecting to the frontal lobe and correlates with both processing speed and working memory (Kochunov et al., 2017).TheSNCA
-rs3910105 variant interacted with PD disease status to influence the white matter microstructure of the superior corona radiata,corpus callosum, and external capsule.The superior corona radiata connects the cortical region, basal ganglia, and thalamus, contributing to cognitive speed and flexibility (Birdsill et al., 2014).The corpus callosum connects the bilateral hemispheres, especially the genu of the corpus callosum, which is the main channel of cognitive function between the two hemispheres.Moreover, white matter changes in external capsule cholinergic fiber tracts have been found to be associated with executive dysfunction even in nondemented elderly individuals (Nolze-Charron et al., 2020).Interactive effects on the brain revealed thatSNCA
gene activity had opposite effects in patients with PD and in HCs.Therefore,SNCA
might promote the development of PD cognitive impairment to a certain extent by causing white matter microstructural changes.Another SNP of interest wasCOMT
-rs4680.Our current findings further support the effects of theCOMT
A allele on the intensity of spontaneous brain activity in the visual processing regions, including the lingual, fusiform gyri, and occipital lobe, functional alteration of which may be an early sign of cognitive decline in PD (Yoneyama et al., 2018).Interestingly, previous studies also showed varied functional connectivity of the lingual and fusiform gyri among differentCOMT
-rs4680 genotypes (Jaspar et al., 2016).Therefore, theCOMT
variant may also contribute to cognitive decline through impairment of visual cognitive function.On the basis of our findings, as a gene that directly affects the dopaminergic level of the brain,COMT
polymorphisms could contribute to the development of cognitive impairment in PD by affecting brain structure and function.DRD3
-rs6280 correlates with brain activity intensity in the temporal lobe, as well as the gray matter structure of the orbitofrontal cortex.A previous study also reported similar results, in that a significant interaction betweenDRD3
-rs6280 and PD status was observed in spontaneous brain activity intensity in the right medial frontal gyrus (Zhi et al., 2019).The results indicate thatDRD3
could play an important role in modifying cognitive function by affecting the frontal and temporal lobes.This study had several limitations.First, the sample size was fairly small.Although the study was adequately powered to detect statistical significance,and ultra-high-field MRI increased the sensitivity of detecting imaging abnormalities, the results should be confirmed in a larger study in the future.Second, SNP selection was based on prior knowledge, and data-driven approaches might provide more information.In addition, the weighting of each SNP used to calculate the PRS was generated based on the present study, and might be more rigorous if the sample size were expanded.
In conclusion, this cognitive-weighted PRS is useful for evaluating cognitive function in PD and identified structural and functional features of brain regions related to executive, memory, and visuospatial domains.Individual SNP analysis further provided evidence that SNCA might promote the development of cognitive impairment in patients with PD by causing change in white matter microstructure.COMT may contribute to cognitive decline in PD through visual cognitive dysfunction–related regions, including the lingual region, fusiform gyri, and occipital lobe.DRD3 potentially plays a role in modifying cognitive function by affecting the frontal and temporal lobes.Our results demonstrate that neuroimaging features, genetic information,and clinical data may provide complementary information about PD-related neurodegenerative processes and help identify pathways and mechanisms associated with cognitive decline in PD.
Acknowledgments:
We would like to thank all patients and heathy controls that participating in the study.And we also would like to acknowledge Zhejiang University 7T Brain Imaging Research Center for all the help and support.
Author contributions:
TS, HYL and BRZ designed this study.JLP, YMY, YPY and BRZ were responsible for diagnosis and clinical evaluation.YSJ, YMY, TTH and SZ collected MRI data.TS, YSJ and BYQ analyzed MRI data.TS wrote the manuscript.HYL and JLP contributed to the revision of the manuscript.
Conflicts of interest:
There are no conflicts of interest.Editor note: HYL and TS are Editorial Board members of Neural Regeneration Research.They are blinded from reviewing or making decisions on the manuscript.The article was subject to the journal’s standard procedures, with peer review handled independently of this Editorial Board member and their research groups.
Availability of data and materials:
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open access statement:
This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional files:
Subitems for each cognition domain extracted from three cognitive scales.
Information of SNPs included in the calculation of the polygenic risk score.
Supplementary methods.
- 中国神经再生研究(英文版)的其它文章
- Patient-specific monocyte-derived microglia as a screening tool for neurodegenerative diseases
- Molecular hallmarks of long non-coding RNAs in aging and its significant effect on aging-associated diseases
- Inflammation in diabetic retinopathy: possible roles in pathogenesis and potential implications for therapy
- Targeting the nitric oxide/cGMP signaling pathway to treat chronic pain
- Neurosteroids as stress modulators and neurotherapeutics: lessons from the retina
- Myelinosome organelles in pathological retinas:ubiquitous presence and dual role in ocular proteostasis maintenance